Graphs are extremely general and consequential data structures. Programmers can use them to model, analyze, and process various phenomena and concepts by providing natural, machine-readable representations [
1]. They model many elements of human knowledge through a mathematical abstraction. In particular, graphs can be excellent representations of relationships between objects and are incredibly versatile, showing up in various fields. For instance, they are crucial in chemistry [
2] for understanding molecular structures, in analyzing social networks [
3] to reveal connections, and in powering web searches [
4] like Google’s PageRank algorithm. You will also find them in security threat detection [
5], in modeling dependencies between software components [
6], and even in hardware testing and functional test programs [
7].
1.1. Related Works
The MCS problem remains an active area of research due to its computational intractability and broad applicability. Researchers have presented algorithms to find the MCS in the literature since the seventies [
8,
9]. Among the less recent notable algorithms, we may recall conversions to the Maximum Common Clique problem [
10], integer linear programming [
11], and constraint programming [
12,
13]. Conversely, more recent efforts have focused on improving exact solvers or estimation heuristics, exploring novel approaches like machine learning [
14,
15] and quantum computing, or facing the problem of trying different perspectives (like graph simplification) [
16,
17]. Regarding the work related to our approach, we describe the following in more detail.
Barrow et al. [
9] propose solving the subgraph isomorphism problem by transforming it into a maximal clique problem in a specially constructed “association graph.” The core idea is that if graph
G is isomorphic to a subgraph of graph
H, then a clique of a specific size exists in the product graph. One main contribution is the transformation method to convert subgraph isomorphism into a maximal clique problem. It is unlikely that the constructed product graph can become very large, and the transformation does not fundamentally change the NP-complete inherent complexity of the problem.
Guidobene et al. [
18] introduce new heuristics for the MCS problem by reformulating it as the Maximum Clique Problem and its complement, the Maximum Independent Set. The authors apply heuristics for the Maximum Independent Set problem to efficiently reduce graph sizes, enabling faster computation. Unfortunately, the proposed heuristics aim for near-optimal solutions, implying they might not always find the exact maximum common subgraph. Moreover, the authors tested primarily on randomly generated Erdös–R‘enyi graph pairs, which may not fully represent the diversity and complexity of real-world graphs. Finally, they rely on local optima convergence, which might still become stuck in suboptimal solutions depending on the initialization and annealing schedule.
Yu et al. [
19] introduce RRSplit, a new backtracking algorithm aimed at improving practical efficiency and providing theoretical guarantees on worst-case running times. The algorithm leverages new reduction techniques and refined upper bounds to minimize redundant computations during the search. Unfortunately, the theoretical guarantees may still indicate high complexity for extremely large or dense graphs, given the NP-hard nature of MCS. Moreover, the complexity of implementing the new reductions and upper bounds might be higher than that of simpler backtracking methods.
Roy et al. [
20] tackle the graph retrieval problem where the goal is to find graphs similar to a query graph in a large corpus, with similarity being scored by the Maximum Common Edge Subgraph (MCES) or Maximum Common Connected Subgraph (MCCS). They design fast and trainable neural functions that approximate MCES and MCCS using late and early interaction network architectures. Unfortunately, as approximation methods, their techniques do not guarantee exact MCS results, which may be crucial for some applications. Moreover, the effectiveness of the neural networks heavily relies on the quality and diversity of the training data.
Xu et al. [
21] propose Qwalk, a novel quantum algorithm for approximately solving the MCS problem. They leverage discrete-time quantum walks to detect isomorphic neighborhood group matches efficiently through destructive interference. They demonstrate better accuracy, universality, and robustness to noise compared to state-of-the-art approximate classical MCS methods. Like in previous approaches, they provide approximate solutions rather than exact ones, and they rely on quantum computing technology, which is still in its early stages of development and faces significant challenges in terms of qubit count, error rates, and hardware availability.
Dilkas [
22] uses machine learning to perform algorithm selection in MCS problems. The approach does not develop a new MCS algorithm itself but instead optimizes the use of existing ones, arguing that different MCS algorithms perform best on various types of graphs. The algorithm requires a comprehensive dataset of graphs and the running times of multiple MCS algorithms to train the selection model effectively. The best algorithm can change drastically with the number of distinct labels or other graph properties, making generalizability challenging.
McCreesh et al. [
23] describe a partitioning algorithm tailored for the MCS problem, aiming to reduce the computational complexity by introducing a compact representation and a smart upper bound to prune the search space. Researchers have developed many notable variants of McSplit to improve on the original algorithm in terms of the heuristic used to select the graph vertices or prune the state space. Some scholars also extended McSplit with elements coming from the deep learning domain.
Trimble [
24], in his Ph.D. thesis, explores partitioning algorithms as a strategy to solve problems related to induced subgraphs. The thesis likely develops methods to decompose a large graph into smaller, more manageable parts, solve the induced subgraph problem on these partitions, and finally combine the results. However, combining solutions from partitions into a global solution can be complex and might introduce computational overhead. Moreover, finding an optimal partitioning strategy that minimizes computational cost is a complex problem, and partitioning might inadvertently break crucial global graph structures.
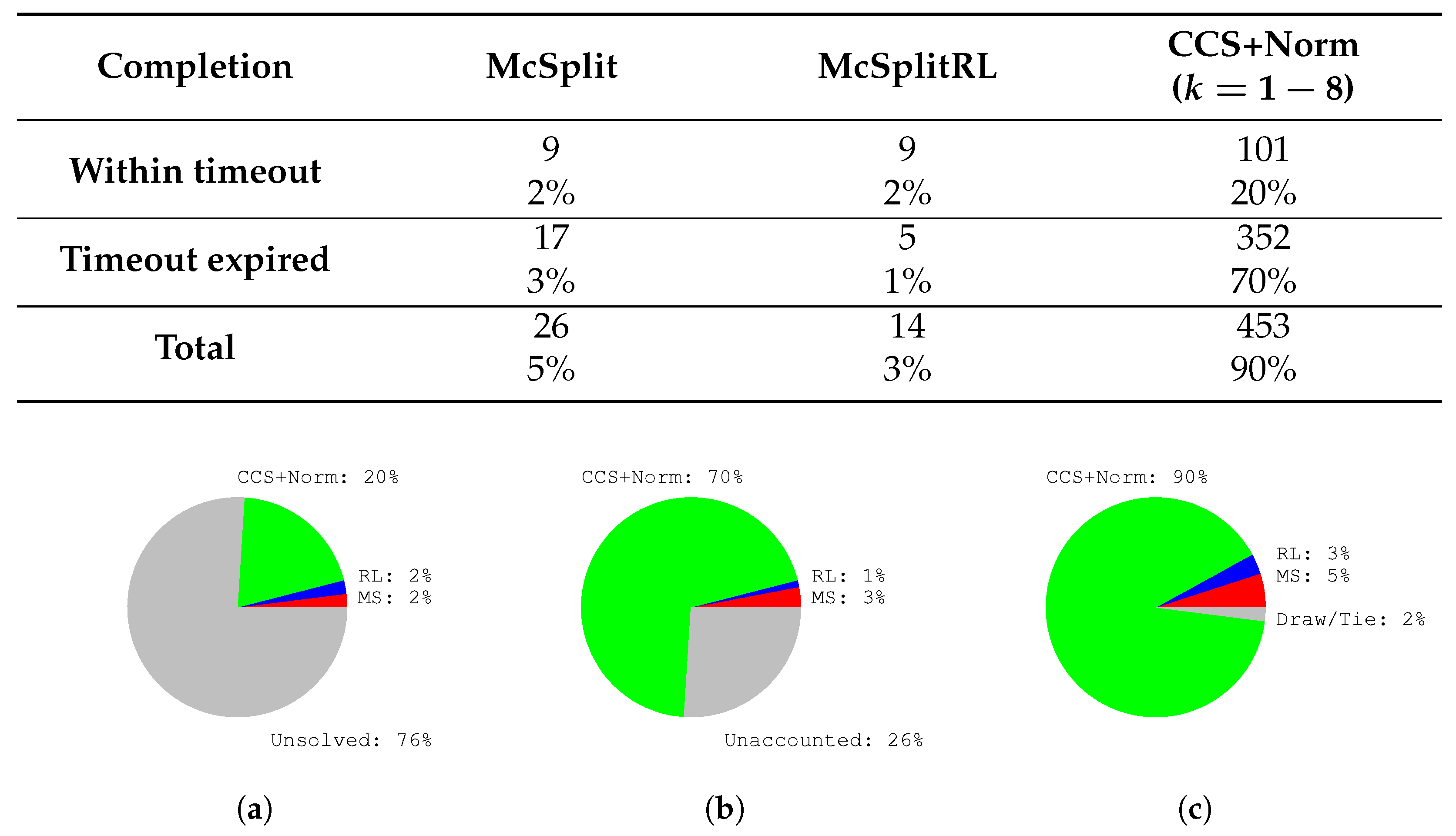
Liu et al. [
25] propose McSplitRL, extending McSplit with Reinforcement Learning. In McSplitRL, the current solution represents the system’s state, selecting a new vertex pair corresponds to an action, and reaching a leaf of the search tree as fast as possible is the goal. The authors define the score as the ability to decrease the bound quickly, since the quicker the bound reduction, the faster the algorithm convergence. Although this strategy successfully reduces the average time required to find an extensive solution, the approach presents two main limitations. The first one is that, since there is no training stage, the scores must be re-initialized (starting from zero) for each problem instance. The second one is that McSplitRL breaks ties between vertices using the original heuristic based on the nodes’ degree. Consequently, the initial recursive steps of McSplitRL closely follow the ones of the original version of McSplit.
Zhou et al. [
26], starting from McSplitRL, build McSplitLL. This procedure presents an enhanced branch and bound algorithm designed for the Maximum Common (Connected) Subgraph. Their solution introduces a new heuristic called Long Short Memory (LSM) and a method called Leaf Vertex Union Match (LUM). The strengthened aspects often mean more intricate data structures and logic despite improvements, potentially increasing implementation complexity.
Liu et al. [
27] introduce McSplitDAL, built upon McSplitRL and McSplitLL. The authors use a new function called Domain Action Learning (DAL) and a hybrid learning policy for choosing the next vertex that will be matched. The hybrid branching policy of this approach has the primary goal of overcoming a possible “Matthew effect,” which causes the algorithm to continue branching on a subset of nodes with very high rewards, getting trapped in a local optimum.
Bai et al. [
28] propose a neural network-based MCS detection approach incorporating a guided subgraph extraction mechanism. Instead of relying solely on traditional search algorithms, this method uses a neural model to learn how to identify common substructures and then guides the extraction of these subgraphs. Unfortunately, like many neural network models, the exact reasoning behind its decisions for MCS detection might be challenging to interpret. Moreover, the algorithm requires substantial labeled graph data to train the neural network effectively. Finally, neural approaches typically provide approximate solutions and may not guarantee optimality.
Bai et al. [
29], extending [
28], build a GNN-based Deep Q-Network, i.e., a deep reinforcement network model, called GLSearch. GLSearch acts as an encoder and computes an embedding for each graph node. It then aggregates embeddings to design a practical function representing the expected rewards for an action in a given state. GLsearch also adds a training phase, which is independent of the order of the input graphs. The main goal of GLSearch is not to reduce the size of the search tree, but rather to reach a good solution in a short time. Unfortunately, GLSearch works at its best with supervision, is susceptible to local optima, and is specifically trained for solving the connected version of the MCS problem. Moreover, the learned search policy might be optimized for specific graph characteristics and might not transfer well to diverse graph datasets without retraining. Finally, achieving the right balance between the learned heuristics and the underlying search algorithm can be tricky. Too much reliance on one or the other could lead to suboptimal performance.
Quer et al. [
30] address the MCS by proposing a parallel and multi-engine approach: The authors leverage parallel computing techniques and potentially integrate multiple different MCS algorithms or heuristics (engines) to work concurrently. The goal is to significantly speed up the computation, especially for large graphs that are computationally demanding for single-threaded or single-algorithm approaches. Unfortunately, coordinating multiple engines or parallel processes can introduce significant overhead, potentially diminishing the benefits of parallelization. Moreover, effectively distributing the workload among various engines or threads can be challenging, especially for irregular graph structures.
Dalke et al. [
2] introduce a Fast Multiple Common Subgraph (FCSM) algorithm to solve the multiple MCS. Unlike typical MCS algorithms, FMCS aims to find a common subgraph in multiple input graphs. The problem is significantly more complex than the pairwise MCS, and even with optimized algorithms, it can face substantial computational challenges for many graphs or huge graphs. Moreover, the definition of common subgraph in multiple contexts can be ambiguous, as it should be present in all graphs, or a certain percentage, and the algorithm’s specific interpretation might limit its flexibility.
Ying et al. [
31] propose a neural network-based approach for subgraph matching. The paper likely designs a neural architecture that can learn to identify occurrences of a query subgraph within a larger target graph, potentially offering a more scalable or approximate solution compared to traditional exact algorithms. However, as in other neural network-based approaches, they typically provide approximate solutions, require substantial labeled data for practical training, and the decision-making process can be less transparent than that of a rule-based algorithm.
Our method significantly advances the previous works by integrating novel deep learning techniques with a state-of-the-art branch-and-bound algorithm. Our approach introduces dynamic, intelligence-driven node selection, unlike previous methods that often rely on static heuristics, approximate solutions, or face scalability issues with large and complex graphs. We transform an uninformed depth-first search into a highly effective best-first search by leveraging graph neural networks and node embeddings. This choice enables the algorithm to find superior solutions within a limited budget, drastically improving time efficiency and applicability on larger, more challenging graphs. Our experimental analysis demonstrates that our approaches consistently outperform the original McSplit algorithm and its reinforcement learning-based evolution, McSplitRL, yielding improved solution sizes and significantly reducing the number of recursions required for convergence. Our approach marks a substantial step forward in the effectiveness and scalability of MCS discovery.
1.2. Our Proposal
We present an improvement to McSplit [
23], a prominent state-of-the-art branch-and-bound algorithm for the Maximum Common Subgraph (MCS) problem. McSplit has seen considerable interest and various extensions in recent years. Notable advancements include McSplitRL [
25], which refines vertex selection order using a Reinforcement Learning approach based on exploration knowledge. McSplitLL [
26], an extension of McSplitRL, surpasses its predecessor by incorporating Long Short-Term Memory, a technique well-suited for specific node characteristics. Moreover, McSplitDAL [
27] builds on McSplitLL by implementing Dynamic Action Learning to improve the reward function initially used in McSplitRL. Conversely, we adopt dynamic heuristics based on embedding norms and cosine similarity. The original version of McSplit considers all possible vertex pairs, one vertex from the first graph and one from the second, sorting vertices based on their degree. Thus, the order is computed statically at the beginning of the process and not modified during the entire procedure. This strategy is quite efficient regarding computational resources, but it is the most impairing element of McSplit. In large graphs, many vertices have identical degrees, making it impossible to discriminate between pairs, and there is no way to prioritize a promising pair discovered during the algorithm’s execution. In our proposed approach, we exploit the core of McSplit, but we replace such a static heuristic with sharper strategies. Specifically, we first use a neural network to compute a node embedding for all subgraphs centered around every possible node of the graph and with a diameter equal to a pre-defined value
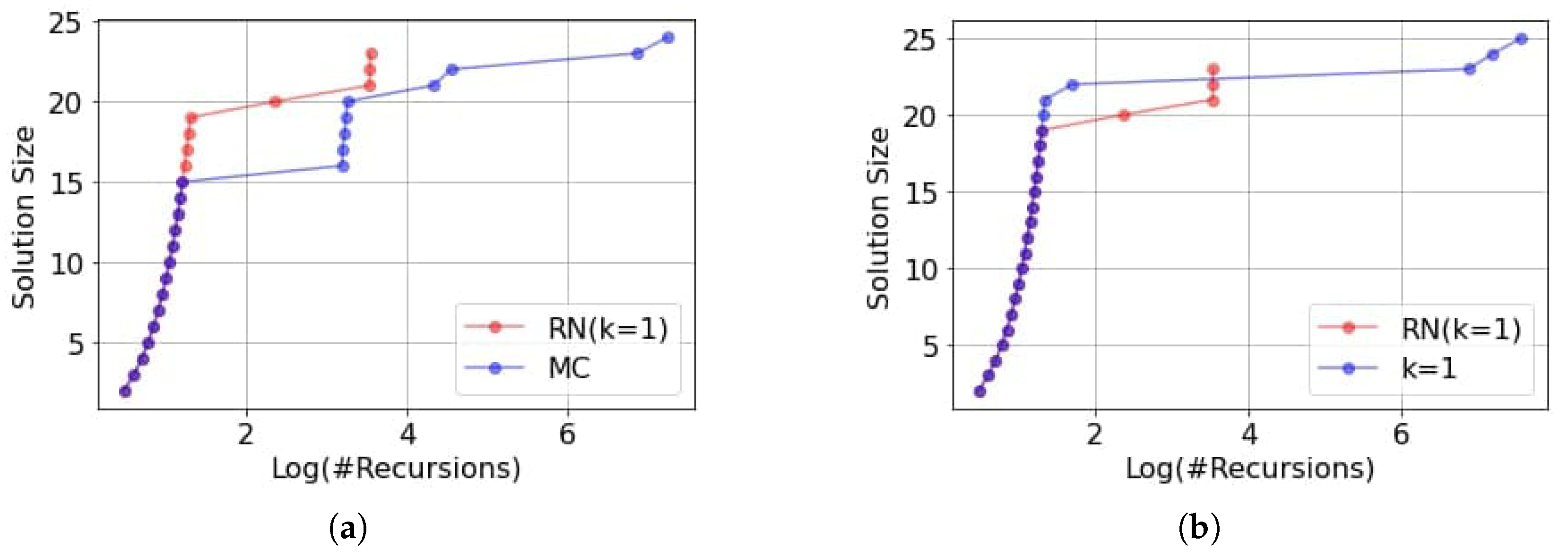
k. After that, once node embeddings have been calculated, we use either the L2-norm or the cumulative cosine similarity as a heuristic to select the vertices to pair within the McSplit procedure. We use McSplit’s efficient brute-force search, adding intelligence to the node selection heuristic with a neural network. From a high-level point of view, we transform an uninformed depth-first search with a relatively efficient bound computation to prune useless paths into a best-first search that exploits a heuristic to be as fast as possible to reach large solutions and improve scalability.
We compare the proposed methodology against the original McSplit algorithm and its direct evolution, McSplitRL, based on reinforcement learning [
25]. We run our experiments using publicly available benchmarks and analyze our approach’s behavior for different values of
k, trading off the prediction accuracy and the evaluation time. We illustrate how to trim our algorithms at their best and discuss the behavior of a parallel version of the tool executing different sorting heuristics in parallel. Our results show that the cosine similarity heuristic outperforms McSplit, the embedding norm heuristic, and McSplitRL in finding the MCS faster or a more extensive solution within a slotted time. Often, we drastically reduce the number of recursions required to reach the final results, suggesting new possibilities to push forward the scalability of MCS approaches on larger graphs.
1.5. Background
1.5.1. Graphs and Maximum Common Subgraphs
We consider unweighted graphs, either directed or undirected, , where V is a finite set of vertices and E is a binary relation on V representing a finite set of edges. We indicate the number of vertices with , and with , the number of edges.
Given a graph
G, if we select a subset of the vertex set
, we define an associated induced subgraph
of
G such that
that is, we keep in
the edges of
G between vertices included in
.
We define the k-hop neighborhood of a given node as the subgraph induced by the set of nodes that includes v and all nodes that v can reach with a path shorter than k.
Given two graphs, G and H, the former is a subgraph of the latter, i.e., , if all its vertices and edges are also vertices and edges of the second. If we remove nodes, we obtain an induced subgraph but include all the edges among the remaining vertices. On the contrary, we obtain a non-induced or partial subgraph by removing nodes and arcs, i.e., some edges may be missing given the selected vertices.
Given two graphs, G and H, the graph S is a common subgraph of G and H, if S is simultaneously isomorphic to a subgraph of G and a subgraph of H. A Maximum Common induced subgraph between G and H (we will indicate it as ) is the largest possible common induced subgraph, i.e., the common subgraph with as many vertices as possible.
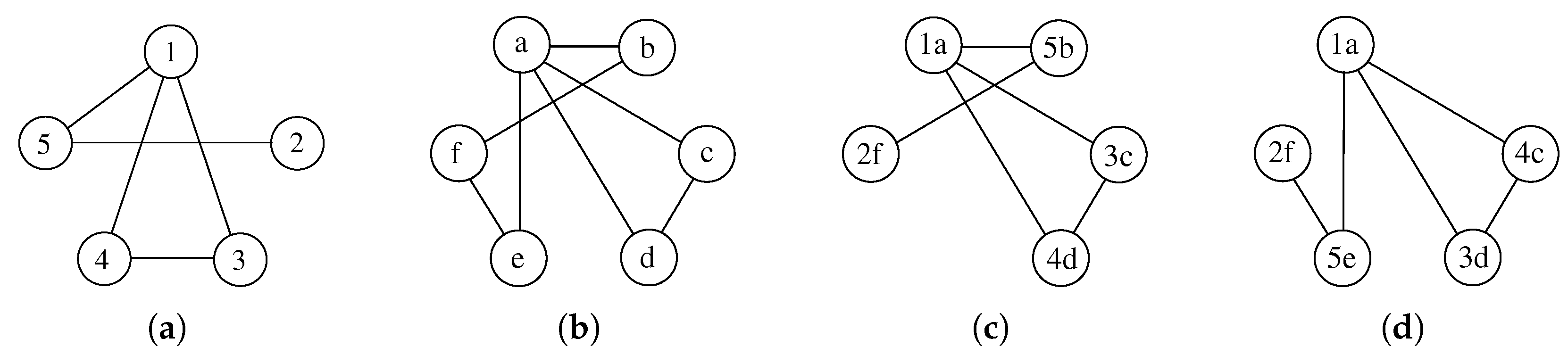
Figure 1 shows a graphical example of two graphs and some possible MCSs. Notice again that node labels, i.e.,
and
, are reported to identify all vertices and their pairing uniquely, but are of no use to match vertices.
1.5.2. The McSplit Algorithm
McSplit is a remarkable recursive branch-and-bound algorithm used to find the MCS of two graphs. Given two graphs G and H, McSplit exhaustively checks all possible matches between one vertex of G and one vertex of H, and it returns the best possible MCS. The algorithm considers a new vertex pair only if it satisfies two conditions:
The vertices within the pair are connected similarly to all the vertices belonging to the same graph previously selected.
The new pair can potentially lead to an MCS larger (in terms of number of nodes) than the current solution.
The resulting procedure is quite efficient in pruning the search space, even if its final cost enormously depends on the vertex selection order and the bound computation.
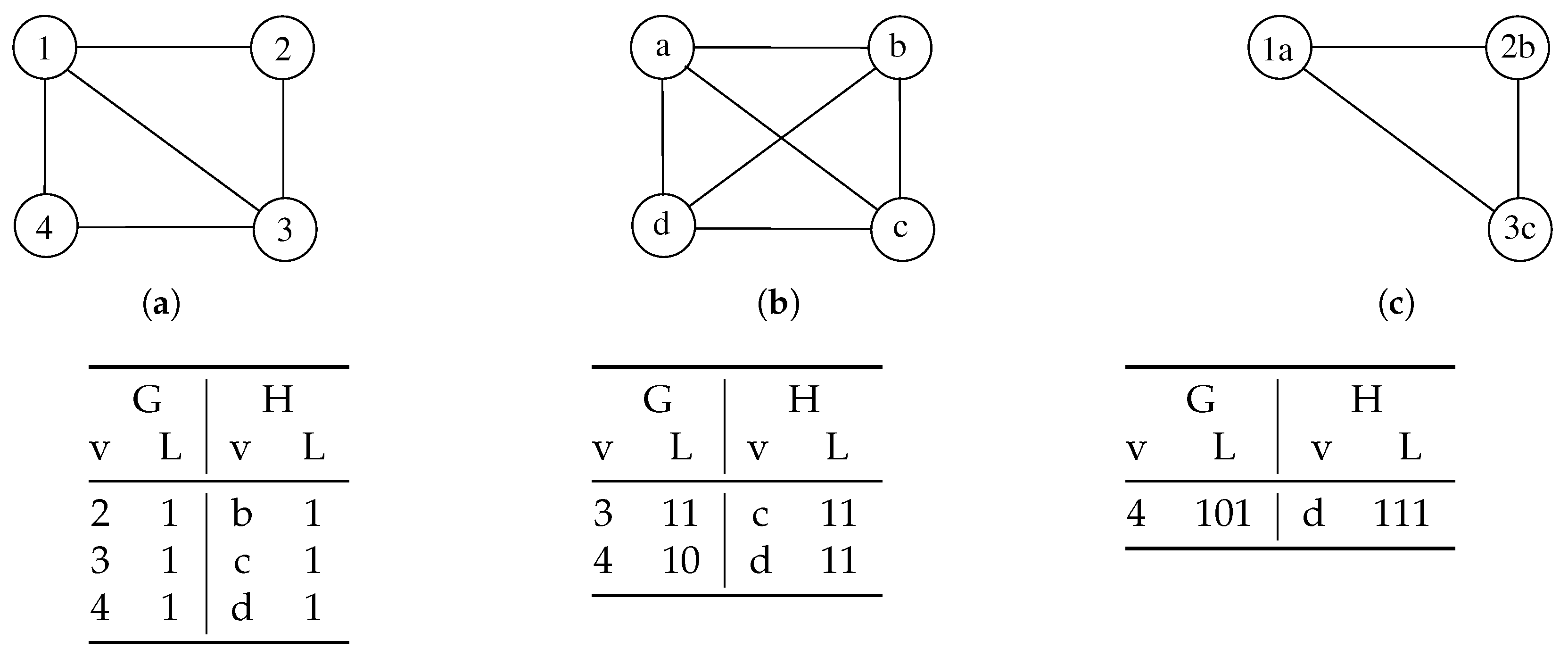
The original work adopts two heuristics to choose a new pair of nodes at each recursive call. The authors based these two heuristics on the concept of bidomain. Given the set of all labels
L and a single label
l (with
), a bidomain
is a set of pairs of vertices sharing the same label:
where
(
) are the sets of nodes from
G (
H) labeled with
l. The first heuristic selects the bidomain with the smallest value of
. The second heuristic chooses the nodes with the highest node degree in each previously selected bidomain.
The algorithm also computes an upper bound before each new pair selection to prune the space search effectively. While parsing a branch using a recursive call, it also evaluates the following bound:
where
is the cardinality of the current mapping and
L is the actual set of labels. If the bound is smaller than the size of the current mapping, there is no reason to follow that path of the decision tree, as it is impossible to find a more extensive matching set than the current one within the path itself. This way, the algorithm prunes consistent branches of the decision tree, drastically reducing the computation effort.
1.5.3. Graph Neural Networks and NeuroMatch
NeuroMatch is a Graph Neural Network (GNN) that can compute graph embeddings [
31]. More in detail, given a graph
and one of its nodes
, NeuroMatch (
https://github.com/snap-stanford/neural-subgraph-learning-GNN, accessed on 1 July 2024). learns the embedding of
v by extracting its
k-hop neighborhood. The
k-hop neighborhood of a given node
is the subgraph induced by the set of nodes that includes
v and all nodes that can be reached from
v with a path shorter than
k. We can find neighborhoods using a Breadth-First Search (BFS) of
G starting from
v, the “anchor node” in NeuroMatch’s terminology. Graph embeddings are learned, enforcing an order constraint, as the geometry of the embeddings represents the relationship between subgraphs, and this also allows computing matchings by just comparing the components of two embeddings. Each embedding represents the
k-hop neighborhood subgraph of
v in
G, enabling fast subgraph matchings.
NeuroMatch satisfies four properties necessary for subgraph relationships; given three graphs G, H, and L:
Transitivity: If G is a subgraph of H and H is a subgraph of L, then G is a subgraph of L.
Anti-symmetry: G is a subgraph H and H is a subgraph of G iff they are isomorphic.
Intersection set: The intersection of G and H contains all common subgraphs of G and H.
Non-trivial intersection: The trivial graph, i.e., a graph with one node and no edge, is contained in the intersection between any two graphs.
In practice, a graph
G which has an embedding vector
with
D dimensions is classified as being a subgraph of a graph
H with embedding vector
if each component of
is less than the corresponding component in
:
This equation shows that the result is an ordered embedding space, i.e., a graph
G is contained in a bigger graph
H, if
G is found in the lower-left side of
H in the order embedding space. To ensure the already mentioned order constraint, NeuroMatch is trained using the Negative Log likelihood loss with the mean reduction (
https://github.com/snap-stanford/neural-subgraph-learning-GNN/blob/master/subgraph_matching/train.py, accessed on 1 July 2024, line 124):
Positive samples are graph pairs in which the first graph is a subgraph of the second. Negative samples are graph pairs in which the previous condition does not hold. We minimize the loss when the subgraph relationship, defined by
E, holds in the case of pairs in
P and is violated by at least
pairs in
N. We further enhance the curriculum learning scheme with the training procedure, i.e., we first train the model on easier instances, which progressively get harder when the model loss stabilizes.
1.5.4. Embedding Computation
We need to map every node into a d-dimensional feature vector. Such a vector is called an embedding. Similar nodes in the graph should also be close in the embedding space. Nodes that are not identical in the graph should not be comparable in the embedding space. We need to define the following:
A notion of similarity between nodes in the graph.
An encoder, i.e., a function that maps nodes to the embedding space.
A decoder, i.e., a function that maps embeddings to a similarity score (usually the dot product).
The encoder parameters must be optimized so that the similarity function on the embedding space is as close as possible to the similarity function in the graph. Usually, an embedding vector dimension goes from 64 to 1000, but we also have a “shallow” embedding method called node2vec.
The basic idea of node2vec [
32] is to compute a matrix. A column of the matrix represents a specific node embedding. We aim to find the optimal matrix, i.e., a set of embeddings such that the dot product between them represents our notion of similarity. The procedure node2vec computes embeddings by resorting to Random Walks. A random walk is generated with some random strategy incorporating local and high-order neighborhood information. We are interested in node pairs that co-occur in a random walk and want those nodes to be close in the embedding space. In summary, the main steps to optimize the embedding space using random walks are the following:
Running a fixed number of random walks starting from each node.
Collecting the neighborhood for each node. In this case, the neighborhood is the multiset of the nodes visited using random walks.
Optimizing the embedding space according to its ability to predict a node’s neighborhood.