A Nested Named Entity Recognition Model Robust in Few-Shot Learning Environments Using Label Description Information
Abstract
1. Introduction
2. Related Works
- By utilizing comprehensive label descriptions rather than single words, our model captures deeper semantic relationships between entities, making it more robust in few-shot scenarios where limited examples are available.
- Our method enables effective transfer learning across different domains and datasets, including between nested and flat NER tasks, demonstrating greater versatility than traditional classification-based approaches.
- Extensive experiments across multiple datasets (GENIA, ACE 2004, ACE 2005, and CoNLL 2003) demonstrate that our approach maintains high performance across various entity types, including the challenging nested relationships, where previous methods often struggle in few-shot environments.
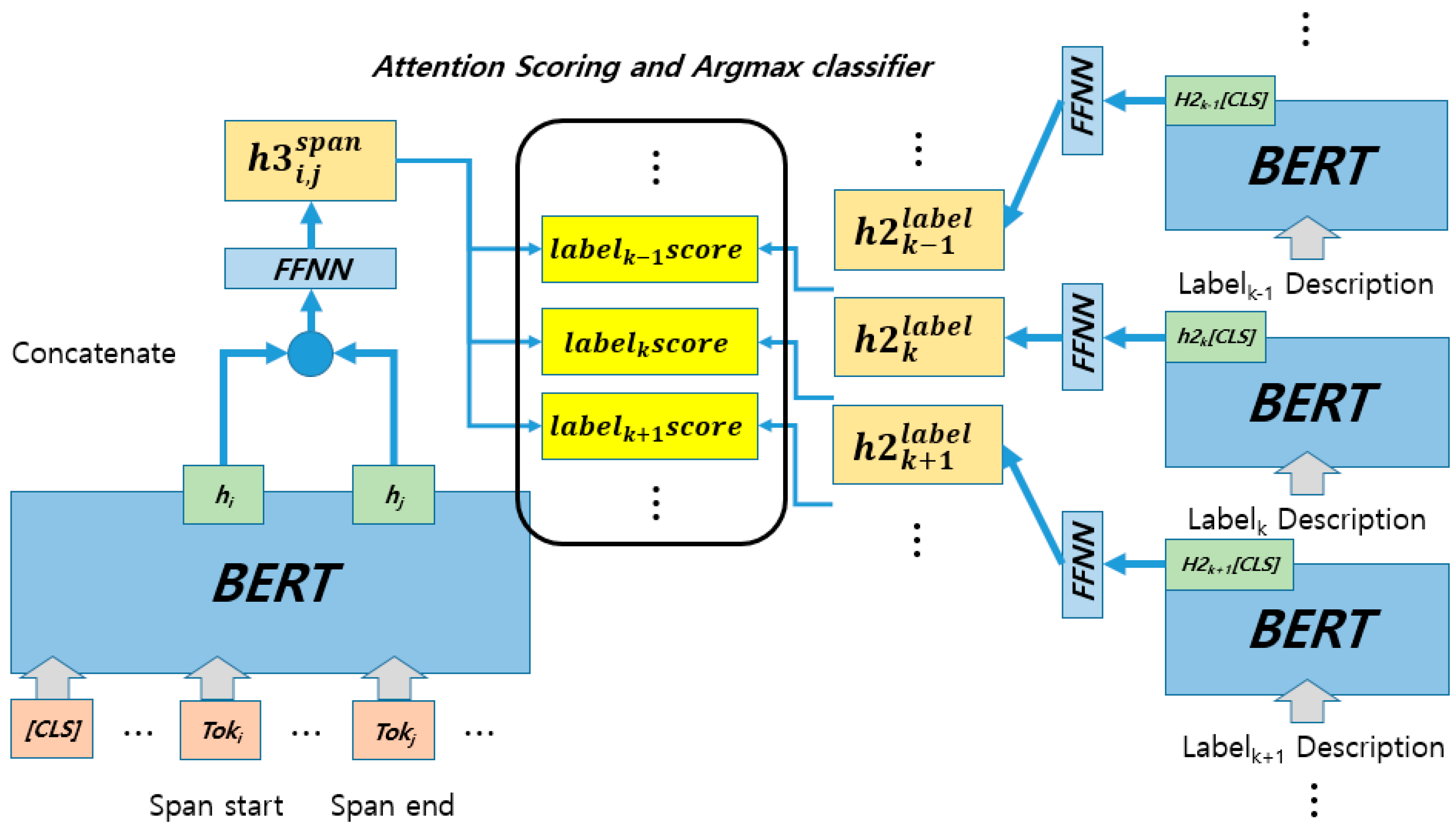
3. Span-Based Nested Named Entity Recognition Model
3.1. Biaffine-Based Nested Named Entity Recognition Model
3.2. Label Description, Embedding Model Using Label Information
3.3. Methodological Overview and Key Differences
- Nested NER Focus: Unlike SpanNER [14] and similar approaches designed primarily for flat NER, our span-based architecture inherently handles hierarchical entity relationships while maintaining the benefits of label semantic information.
4. Experiments and Results
4.1. Detailed Experimental Settings
4.2. Results
4.3. Ablation Study on Label Description Components
- Biaffine: Our baseline model without any label embedding components.
- LDE with Trainable Label Features (LDEw Label Feature): Replaces BERT-encoded label information with simple trainable embedding vectors.
- LDE with Label Words Only (LDEw Label Word): Uses only the entity type words (e.g., “person”, “organization”) without descriptive context.
- LDE with Full Label Descriptions (LDEw Label Description): Our complete proposed model using detailed semantic descriptions of entity types.
| ACE 2005 (Nested) | |||||
|---|---|---|---|---|---|
| Avg. Tokens Per Label Description | 1-Shot | 5-Shot | 10-Shot | 20-Shot | |
| Biaffine | 3.17 ± 1.84 | 32.38 ± 2.96 | 49.50 ± 2.34 | 58.21 ± 0.97 | |
| LDEw Label Feature | 0.00 | 15.71 ± 10.01 | 49.01 ± 1.76 | 58.10 ± 0.63 | |
| LDEw Label Word | 4.0 | 1.96 ± 1.62 | 37.75 ± 3.28 | 54.27 ± 2.27 | 63.42 ± 1.09 |
| LDEw Label Description | 43.13 | 5.54 ± 3.27 | 44.24 ± 2.01 | 57.19 ± 1.27 | 64.50 ± 1.02 |
4.4. Transfer Learning
4.5. Additional Analysis
- Recognize subtle entity mentions that might be overlooked by other approaches;
- Correctly identify hierarchical relationships between nested entities;
- Distinguish between semantically related entity types (e.g., facility vs. location);
- Handle rare entity types with minimal training data;
- Maintain precision in complex nested structures.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Prompt |
|---|
| {Document} Refer to the above document and create descriptions for each object recognition tag {TAG1, TAG2, TAG3, …}. Write a relatively long sentence for each description. |
| NER Tag | Description |
|---|---|
| O | O: Outside of named entities. |
| G#DNA | G#DNA: DNA is a fundamental molecule in the biomedical domain, serving as the genetic blueprint of all living organisms, carrying hereditary information, enabling genomics research, facilitating personalized medicine, aiding in diagnostics and forensics, and offering insights into evolutionary biology and gene editing for disease treatments. |
| G#protein | G#protein: Proteins are fundamental biomolecules in the biomedical domain, serving as essential building blocks of cells and tissues, catalysts for biochemical reactions, and key regulators of biological processes, playing crucial roles in health and disease. |
| G#cell_type | G#cell_type: In the biomedical domain, cell type refers to a specific class or category of cells sharing similar morphological, functional, and genetic characteristics within a particular organism or tissue. |
| G#cell_line | G#cell_line: A cell line in the biomedical domain refers to a population of cells derived from a single source and cultured in a laboratory setting, providing a valuable tool for studying various biological processes and testing experimental treatments. |
| G#RNA | G#RNA: RNA (Ribonucleic acid) in the biomedical domain is a versatile molecule responsible for translating genetic information from DNA to proteins, regulating gene expression, and serving as a potential therapeutic target in various diseases. |
| NER Tag | Description |
|---|---|
| O | O: Outside of named entities. |
| ORG | ORG: Organizations indicate companies, subdivisions of companies, brands, political movements, government bodies, publications, musical companies, public organizations, and other collections of people within the text data. |
| MISC | MISC: Miscellaneous includes adjectives and derivations from terms associated with locations, organizations, individuals, or general concepts, as well as encompassing entities indicating religions, political ideologies, nationalities, languages, events, wars, sports-related names, titles, slogans, eras, or objects types within the text data. |
| PER | PER: Persons indicate the first, middle, and last names of people, animals and fictional characters, and aliases within the text data. |
| LOC | LOC: Locations are entities that indicate specific places, such as roads, trajectories, regions, structures, natural locations, public places, commercial places, assorted buildings, countries, or landmarks, within the text data. |
| NER Tag | Description |
|---|---|
| O | O: Outside of named entities. |
| ORG | ORG: Organizations are entities that indicate government agencies, commercial companies, educational institutions, non-profit organizations, and other structured groups of people, encompassing various subtypes like government, commercial, educational, non-profit entities within the text data. |
| GPE | GPE: Geo-Political Entities are complex entities that represent geographical regions, political entities, or their combinations, including nations, states, cities, and other politically defined locations that have both a physical and administrative aspect within the text data. |
| PER | PER: Persons are entities that indicate human beings through named mentions (proper names), nominal mentions (descriptions), or pronominal mentions (pronouns), including individual names, titles, roles, and references to people as individuals or groups within the text data. |
| LOC | LOC: Locations are entities that indicate purely geographical or physical places without political significance, such as mountains, rivers, oceans, regions, continents, and other natural or artificial geographical features within the text data. |
| FAC | FAC: Facilities are entities that indicate human-made structures, buildings, architectural features, and infrastructure elements like bridges, airports, highways, and other constructed spaces within the text data. |
| VEH | VEH: Vehicles are entities that indicate any means of transportation, including cars, planes, ships, spacecraft, and other mobile machines designed for carrying and transporting within the text data. |
| WEA | WEA: Weapons are entities that indicate instruments designed for combat or defense, including conventional weapons, military equipment, and other tools specifically designed for warfare or combat within the text data. |
- GENIA: 16,691 sentences in the training set and 1855 sentences in the test set.
- ACE2004: 6198 sentences in the training set and 809 sentences in the test set.
- ACE2005: 7294 sentences in the training set and 1057 sentences in the test set.
- CoNLL 2003 English: 14,041 sentences in the training set and 3453 sentences in the test set.
| Groups | GENIA | |||||
|---|---|---|---|---|---|---|
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 0.00% | 22.83% | 18.54% | 22.92% | 17.97% | 21.73% |
| #2 | 12.50% | 21.62% | 21.50% | 18.50% | - | - |
| #3 | 53.33% | 27.72% | 12.26% | 26.51% | - | - |
| #4 | 21.05% | 24.44% | 28.57% | 18.53% | - | - |
| #5 | 42.86% | 24.74% | 26.84% | 24.12% | - | - |
| Groups | ACE 2004 | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 62.50% | 58.25% | 54.50% | 53.53% | 45.81% | 46.75% |
| #2 | 48.65% | 46.07% | 57.39% | 58.49% | - | - |
| #3 | 57.89% | 53.85% | 52.78% | 57.16% | - | - |
| #4 | 60.61% | 65.48% | 52.27% | 58.22% | - | - |
| #5 | 56.25% | 58.99% | 60.10% | 53.00% | - | - |
| Groups | ACE 2005 | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 70.21% | 50.78% | 46.90% | 49.67% | 40.66% | 39.56% |
| #2 | 53.49% | 50.28% | 55.65% | 46.66% | - | - |
| #3 | 48.08% | 46.06% | 45.66% | 47.98% | - | - |
| #4 | 40.00% | 47.67% | 50.15% | 49.30% | - | - |
| #5 | 66.67% | 53.99% | 50.40% | 45.30% | - | - |
| Groups | GENIA | |||||
|---|---|---|---|---|---|---|
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 3.00 | 3.68 | 3.56 | 3.36 | 3.08 | 3.02 |
| #2 | 3.20 | 4.44 | 4.00 | 4.00 | - | - |
| #3 | 3.00 | 4.04 | 4.24 | 4.15 | - | - |
| #4 | 3.80 | 3.60 | 3.64 | 4.21 | - | - |
| #5 | 4.20 | 3.88 | 3.80 | 3.98 | - | - |
| Groups | ACE 2004 | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 5.71 | 5.89 | 6.03 | 5.26 | 3.58 | 3.75 |
| #2 | 5.29 | 5.46 | 5.70 | 5.30 | - | - |
| #3 | 5.43 | 5.57 | 5.14 | 5.09 | - | - |
| #4 | 4.71 | 4.80 | 5.36 | 5.78 | - | - |
| #5 | 4.57 | 5.09 | 5.51 | 5.35 | - | - |
| Groups | ACE 2005 | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 6.71 | 5.51 | 4.84 | 5.48 | 3.40 | 2.88 |
| #2 | 6.14 | 5.17 | 5.31 | 5.02 | - | - |
| #3 | 7.43 | 4.71 | 4.44 | 4.96 | - | - |
| #4 | 4.29 | 4.91 | 4.61 | 5.11 | - | - |
| #5 | 6.86 | 4.66 | 5.41 | 4.94 | - | - |
| Groups | CoNLL 2003 English | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | Train (100%) | Test | |
| #1 | 3.00 | 2.45 | 2.28 | 2.59 | 1.67 | 1.64 |
| #2 | 3.25 | 2.65 | 2.90 | 2.48 | - | - |
| #3 | 1.50 | 2.40 | 2.43 | 2.18 | - | - |
| #4 | 1.50 | 3.15 | 2.25 | 2.61 | - | - |
| #5 | 3.25 | 1.85 | 2.45 | 2.59 | - | - |
| Parameter | Value |
|---|---|
| BiLSTM size (Only BERT-LSTM-CRF) | 256 |
| FFNN size | {200, 400, 600, 800, 1200, 1600} |
| Drop out | 0.1 |
| Optimizer | AdamW |
| Learning rate | 5 × 10−5 |
| Weight decay | 0.1 |
References
- Yang, J.; Zhang, T.; Tsai, C.-Y.; Lu, Y.; Yao, L. Evolution and emerging trends of named entity recognition: Bibliometric analysis from 2000 to 2023. Heliyon 2024, 10, e30053. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, J.; Meng, Y.; Han, Q.; Wu, F.; Li, J. A Unified MRC Framework for Named Entity Recognition. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020. [Google Scholar]
- Yu, J.; Bohnet, B.; Poesio, M. Named Entity Recognition as Dependency Parsing. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020. [Google Scholar]
- Dozat, T.; Manning, C.D. Deep Biaffine Attention for Neural Dependency Parsing. In Proceedings of the International Conference on Learning Representations, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Yuan, Z.; Tan, C.; Huang, S.; Huang, F. Fusing Heterogeneous Factors with Triaffine Mechanism for Nested Named Entity Recognition. In Findings of the Association for Computational Linguistics: ACL 2022, Dublin, Ireland, 22–27 May 2022; Association for Computational Linguistics: Dublin, Ireland, 2022. [Google Scholar]
- Straková, J.; Straka, M.; Hajič, J. Neural architectures for nested NER through linearization. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; pp. 5326–5331. [Google Scholar]
- Xu, Y.; Yang, Z.; Zhang, L.; Zhou, D.; Wu, T.; Zhou, R. Focusing, bridging and prompting for few-shot nested named entity recognition. In Findings of the Association for Computational Linguistics: ACL 2023, Toronto, ON, Canada, 9–14 July 2023; Association for Computational Linguistics: Toronto, ON, Canada, 2023; pp. 2621–2637. [Google Scholar]
- Yang, J.; Zhu, Z.; Ming, H.; Jiang, L.; An, N. LPNER: Label Prompt for Few-shot Nested Named Entity Recognition. In Proceedings of the 16th Asian Conference on Machine Learning, Hanoi, Vietnam, 11–14 November 2024. [Google Scholar]
- Kim, H.; Kim, J.-E.; Kim, H. Exploring Nested Named Entity Recognition with Large Language Models: Methods, Challenges, and Insights. In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, Miami, FL, USA, 15–20 November 2024. [Google Scholar]
- Akata, Z.; Perronnin, F.; Harchaoui, Z.; Schmid, C. Label-embedding for image classification. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhang, Y. Hierarchically-Refined Label Attention Network for Sequence Labeling. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, Hong Kong, China, 3–7 November 2019. [Google Scholar]
- Kim, H.; Kim, H. Integrated Model for Morphological Analysis and Named Entity Recognition Based on Label Attention Networks in Korean. Appl. Sci. 2020, 10, 3740. [Google Scholar] [CrossRef]
- Li, J.; Chiu, B.; Feng, S.; Wang, H. Few-Shot Named Entity Recognition via Meta-Learning. IEEE Trans. Knowl. Data Eng. 2022, 34, 4245–4256. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, H.; Zhang, C.; Gao, J. Learning from Language Description: Low-shot Named Entity Recognition via Decomposed Framework. In Findings of the Association for Computational Linguistics: EMNLP 2021, Punta Cana, Dominican Republic, 7–11 November 2021; Association for Computational Linguistics: Punta Cana, Dominican Republic, 2021; pp. 1618–1630. [Google Scholar]
- Ma, J.; Ballesteros, M.; Doss, S.; Anubhai, R.; Mallya, S.; Al-Onaizan, Y.; Roth, D. Label Semantics for Few Shot Named Entity Recognition. In Findings of the Association for Computational Linguistics: ACL 2022, Dublin, Ireland, 22–27 May 2022; Association for Computational Linguistics: Dublin, Ireland, 2022. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019. [Google Scholar]
- Luong, T.; Pham, H.; Manning, C.D. Effective Approaches to Attention-based Neural Machine Translation. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015. [Google Scholar]
- Kim, J.D.; Ohta, T.; Tateisi, Y.; Tsujii, J. GENIA corpus—A semantically annotated corpus for bio-textmining. Bioinformatics 2003, 19, i180–i182. [Google Scholar] [CrossRef] [PubMed]
- Doddington, G.; Mitchell, A.; Przybocki, M.; Ramshaw, L.; Strassel, S.; Weischedel, R. The Automatic Content Extraction (ACE) program-tasks, data, and evaluation. In Proceedings of the 4th International Conference on Language Resources and Evaluation, Lisbon, Portugal, 26–28 May 2004; pp. 837–840. [Google Scholar]
- Walker, C.; Strassel, S.; Medero, J.; Maeda, K. ACE 2005 Multilingual Training Corpus; Linguistic Data Consortium: Philadelphia, PA, USA, 2006; Available online: https://catalog.ldc.upenn.edu/LDC2006T06 (accessed on 16 January 2025).
- Tjong Kim Sang, E.F.; De Meulder, F. Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. In Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003, Edmonton, AB, Canada, 31 May–1 June 2003; pp. 142–147. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Bravo-Marquez, F.; Dunstan, J. Simple Yet Powerful: An Overlooked Architecture for Nested Named Entity Recognition. In Proceedings of the 29th International Conference on Computational Linguistics, Gyeongju, Republic of Korea, 12–17 October 2022; pp. 2108–2117. [Google Scholar]

| GENIA (Nested) | |||||
|---|---|---|---|---|---|
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | 100% | |
| FewNER [13] | 23.24 ± 0.73 | 29.19 ± 0.64 | |||
| FIT [7] | 34.43 ± 9.06 | 44.98 ± 3.38 | 51.26 ± 3.96 | ||
| LPNER [8] | 26.32 ± 3.88 | 44.99 ± 2.20 | |||
| BERT-LSTM-CRF (our) | 17.71 ± 8.33 | 17.91 ± 8.39 | 29.16 ± 1.79 | 42.12 ± 8.46 | 76.82 |
| Biaffine (our) | 5.75 ± 2.58 | 30.74 ± 2.95 | 31.93 ± 1.93 | 50.65 ± 2.39 | 78.20 |
| LDE (our) | 11.60 ± 2.51 | 45.07 ± 3.57 | 47.90 ± 2.27 | 61.46 ± 1.62 | 79.01 |
| ACE 2004 (Nested) | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | 100% | |
| FIT [7] | 35.87 ± 4.92 | 44.88 ± 4.82 | 53.92 ± 2.99 | ||
| LPNER [8] | 25.67 ± 7.05 | 42.67 ± 7.55 | |||
| BERT-LSTM-CRF (our) | 10.84 ± 7.17 | 18.94 ± 9.28 | 34.19 ± 4.30 | 49.70 ± 2.96 | 82.38 |
| Biaffine (our) | 3.52 ± 1.95 | 28.10 ± 1.91 | 47.25 ± 1.10 | 55.98 ± 0.88 | 85.77 |
| LDE (our) | 4.06 ± 3.04 | 42.23 ± 2.30 | 57.04 ± 1.19 | 62.86 ± 0.56 | 85.82 |
| ACE 2005 (Nested) | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | 100% | |
| FIT [7] | 37.74 ± 5.33 | 42.25 ± 10.65 | 52.71 ± 2.55 | ||
| LPNER [8] | 25.01 ± 10.83 | 46.62 ± 5.82 | |||
| BERT-LSTM-CRF (our) | 5.74 ± 6.49 | 30.80 ± 5.89 | 41.46 ± 1.49 | 51.88 ± 1.24 | 80.94 |
| Biaffine (our) | 3.17 ± 1.84 | 32.38 ± 2.96 | 49.50 ± 2.34 | 58.21 ± 0.97 | 83.95 |
| LDE (our) | 5.54 ± 3.27 | 44.24 ± 2.01 | 57.19 ± 1.27 | 64.50 ± 1.02 | 84.23 |
| CoNLL 2003 English (Flat) | |||||
| 1-Shot | 5-Shot | 10-Shot | 20-Shot | 100% | |
| SpanNER [14] | 71.1 ± 0.4 | ||||
| BERT-LSTM-CRF (our) | 26.55 ± 8.55 | 46.64 ± 1.67 | 49.74 ± 3.82 | 63.61 ± 1.85 | 89.86 |
| Biaffine (our) | 9.22 ± 9.30 | 36.95 ± 4.00 | 23.15 ± 6.85 | 50.82 ± 2.56 | 91.81 |
| LDE (our) | 6.61 ± 7.50 | 42.53 ± 7.04 | 45.05 ± 2.38 | 63.85 ± 1.85 | 92.06 |
| ACE 2005 (Nested) | |||||
|---|---|---|---|---|---|
| Source Domain | 1-Shot | 5-Shot | 10-Shot | 20-Shot | |
| LDE | - | 5.54 ± 3.27 | 44.24 ± 2.01 | 57.19 ± 1.27 | 64.50 ± 1.02 |
| LDE | GENIA | 14.28 ± 7.26 | 50.94 ± 1.59 | 59.92 ± 1.96 | 66.99 ± 0.75 |
| LDE | CoNLL 2003 English | 33.22 ± 7.33 | 58.06 ± 0.57 | 62.44 ± 1.21 | 68.13 ± 0.80 |
| GENIA (Nested) | |||||
| Source Domain | 1-Shot | 5-Shot | 10-Shot | 20-Shot | |
| LDE | - | 11.60 ± 2.51 | 45.07 ± 3.57 | 47.90 ± 2.27 | 61.46 ± 1.62 |
| LDE | CoNLL 2003 English | 35.14 ± 0.72 | 49.98 ± 1.42 | 57.51 ± 0.63 | 64.75 ± 0.56 |
| LDE | ACE 2005 | 31.67 ± 1.95 | 48.52 ± 1.69 | 58.88 ± 1.56 | 65.35 ± 0.35 |
| ACE 2005 (Nested) 5-Shot | ||||
|---|---|---|---|---|
| Standard | Flat | Nested | Nesting | |
| In-context Learning (GPT-4) [9] | 34.75 | 38.29 | 6.63 | |
| BERT-LSTM-CRF | 33.85 | 36.56 | 25.74 | 0.00 |
| Biaffine | 31.65 | 40.48 | 17.11 | 0.33 |
| LDEw Label Feature | 27.31 | 37.02 | 15.11 | 0.54 |
| LDEw Label Word | 39.92 | 47.59 | 27.69 | 3.51 |
| LDEw Label Description | 43.98 | 54.48 | 28.31 | 2.62 |
| Case 1 | |
|---|---|
| Model | Result |
| Sentence | [CLS] We have been so damned busy with the holidays (that’s what we call December at our house) that I just haven’t had time. [SEP] |
| Gold | (1, 1, ‘We’, ‘PER’) (15, 15, ‘we’, ‘PER’) (19, 19, ‘our’, ‘PER’) (19, 20, ‘our house’, ‘FAC’) (23, 23, ‘I’, ‘PER’) |
| Biaffine | (1, 1, ‘We’, ‘ORG’) (15, 15, ‘we’, ‘PER’) (15, 23, ‘we call December at our house) that I’, ‘PER’) (23, 23, ‘I’, ‘PER’) |
| LDEw Label Feature | (1, 1, ‘We’, ‘PER’) (1, 15, “We have been so damned busy with the holidays (that ‘ s what we”, ‘PER’) (1, 23, “We have been so damned busy with the holidays (that ‘ s what we call December at our house) that I”, ‘PER’) (15, 15, ‘we’, ‘PER’) (15, 23, ‘we call December at our house) that I’, ‘PER’) (23, 23, ‘I’, ‘PER’) |
| LDEw Label Word | (1, 1, ‘We’, ‘ORG’) (15, 15, ‘we’, ‘PER’) |
| LDEw Label Description | (1, 1, ‘We’, ‘ORG’) (15, 15, ‘we’, ‘PER’) (19, 19, ‘our’, ‘GPE’) (23, 23, ‘I’, ‘PER’) |
| Case 2 | |
| Model | Result |
| Sentence | [CLS] He found the lane to the farm and drove up into the farm ##yard, where he was met by the farmer [SEP] |
| Gold | (1, 1, ‘He’, ‘PER’) (3, 7, ‘the lane to the farm’, ‘FAC’) (6, 7, ‘the farm’, ‘FAC’) (12, 22, ‘the farm ##yard, where he was met by the farmer’, ‘FAC’) (16, 16, ‘where’, ‘FAC’) (17, 17, ‘he’, ‘PER’) (21, 22, ‘the farmer’, ‘PER’) |
| Biaffine | (1, 1, ‘He’, ‘PER’) (16, 16, ‘where’, ‘LOC’) (17, 17, ‘he’, ‘PER’) |
| LDEw Label Feature | (1, 1, ‘He’, ‘PER’) (1, 17, ‘He found the lane to the farm and drove up into the farm ##yard, where he’, ‘PER’) |
| LDEw Label Word | (1, 1, ‘He’, ‘PER’) (16, 16, ‘where’, ‘LOC’) (17, 17, ‘he’, ‘PER’) (21, 22, ‘the farmer’, ‘PER’) |
| LDEw Label Description | (1, 1, ‘He’, ‘PER’) (3, 7, ‘the lane to the farm’, ‘FAC’) (6, 7, ‘the farm’, ‘LOC’) (16, 16, ‘where’, ‘LOC’) (17, 17, ‘he’, ‘PER’) (21, 22, ‘the farmer’, ‘PER’) |
| Case 3 | |
| Model | Result |
| Sentence | [CLS] You see that barge down there on the river ? [SEP] |
| Gold | (1, 1, ‘You’, ‘PER’) (3, 9, ‘that barge down there on the river’, ‘VEH’) (8, 9, ‘the river’, ‘LOC’) |
| Biaffine | (6, 6, ‘there’, ‘LOC’) |
| LDEw Label Feature | |
| LDEw Label Word | |
| LDEw Label Description | (3, 9, ‘that barge down there on the river’, ‘FAC’) (6, 6, ‘there’, ‘LOC’) (8, 9, ‘the river’, ‘LOC’) |
| Case 4 | |
| Model | Result |
| Sentence | [CLS] All ##egation ##s have come to light that several OS ##U players received illegal benefits including cash, access to cars, etc. [SEP] |
| Gold | (9, 12, ‘several OS ##U players’, ‘PER’) (10, 11, ‘OS ##U’, ‘ORG’) (21, 21, ‘cars’, ‘VEH’) |
| Biaffine | |
| LDEw Label Feature | (9, 11, ‘several OS ##U’, ‘PER’) (9, 12, ‘several OS ##U players’, ‘PER’) |
| LDEw Label Word | (9, 12, ‘several OS ##U players’, ‘PER’) |
| LDEw Label Description | (9, 12, ‘several OS ##U players’, ‘PER’) (10, 11, ‘OS ##U’, ‘ORG’) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, H.; Jung, Y.; Lee, C.; Go, W. A Nested Named Entity Recognition Model Robust in Few-Shot Learning Environments Using Label Description Information. Appl. Sci. 2025, 15, 8255. https://doi.org/10.3390/app15158255
Hwang H, Jung Y, Lee C, Go W. A Nested Named Entity Recognition Model Robust in Few-Shot Learning Environments Using Label Description Information. Applied Sciences. 2025; 15(15):8255. https://doi.org/10.3390/app15158255
Chicago/Turabian StyleHwang, Hyunsun, Youngjun Jung, Changki Lee, and Wooyoung Go. 2025. "A Nested Named Entity Recognition Model Robust in Few-Shot Learning Environments Using Label Description Information" Applied Sciences 15, no. 15: 8255. https://doi.org/10.3390/app15158255
APA StyleHwang, H., Jung, Y., Lee, C., & Go, W. (2025). A Nested Named Entity Recognition Model Robust in Few-Shot Learning Environments Using Label Description Information. Applied Sciences, 15(15), 8255. https://doi.org/10.3390/app15158255







