Enhanced Segmentation of Glioma Subregions via Modality-Aware Encoding and Channel-Wise Attention in Multimodal MRI
Abstract
1. Introduction
- We introduce a multi-encoder architecture for fine-grained segmentation of glioma subregions, namely, ET, NCR, and ED, which are clinically relevant yet often underrepresented in standard segmentation protocols.
- We leverage both MRI modality-specific encoding and modular attention-based refinement to effectively capture complementary information across modalities. This allows us to improve segmentation performance while keeping a moderate number of parameters, making the model suitable for clinical scenarios with constrained computational resources.
- We benchmark the performance of our model against a state-of-the-art transformer-based approach, obtaining statistically significant improvements on all tumor subregions.
- We test our model on two public external validation datasets, achieving competitive results in the segmentation of all tumor subregions and demonstrating the strong generalization capacity of our model.
2. Related Work
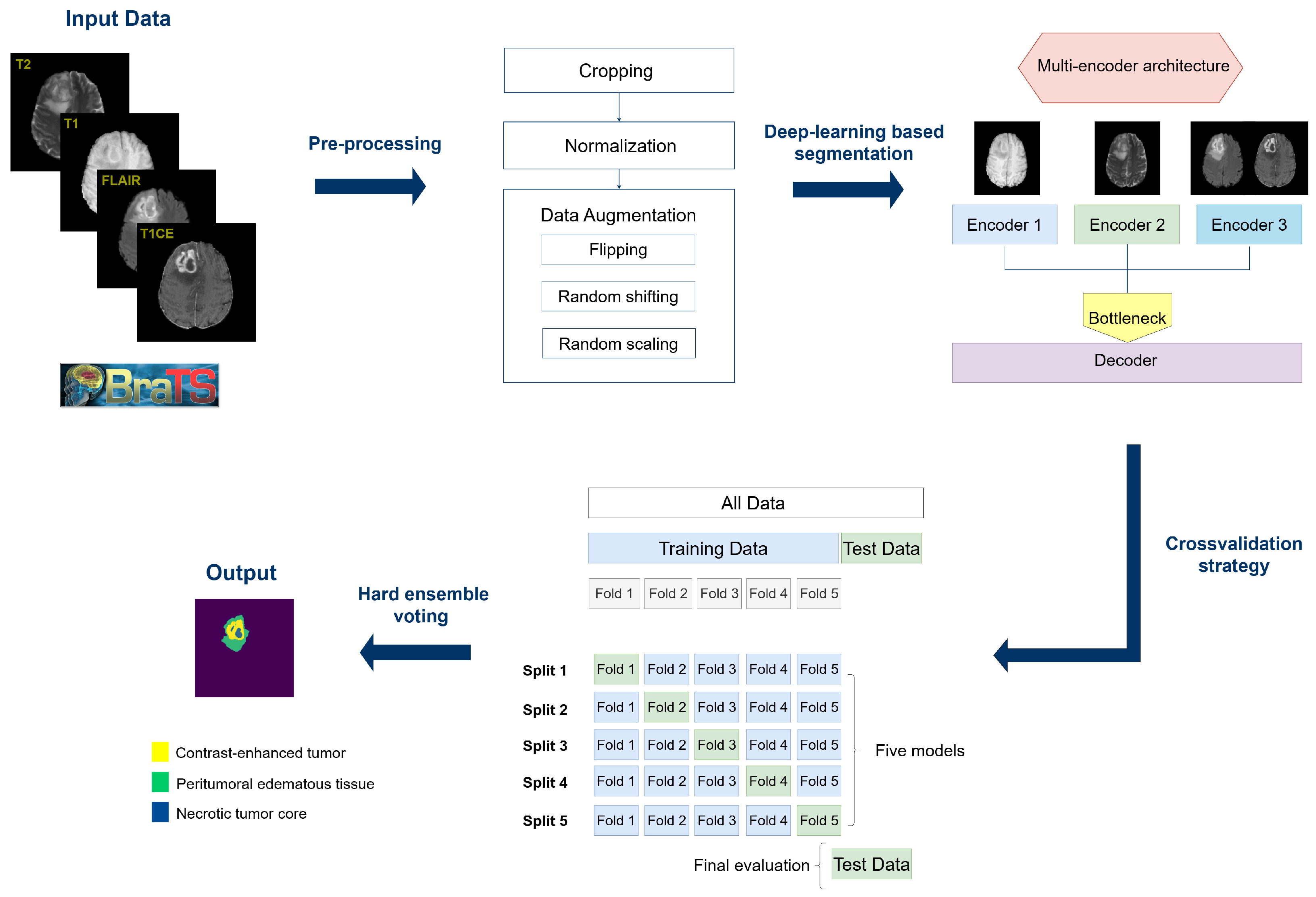
3. Materials and Methods
3.1. BraTS-2023 Dataset
- Contrast-enhanced tumor (ET): Regions that appear strongly highlighted in MRI after contrast medium administration.
- Necrotic tumor core (NCR): Tumor regions which appear hypointense in T1CE sequences.
- Peritumoral edematous/invaded tissue (ED): Areas of diffuse hyperintensity in FLAIR sequences, which includes the infiltrative non-enhancing tumor as well as vasogenic edema in the peritumoral region.
3.2. External Validation Datasets
3.3. Preprocessing
3.4. Multi-Encoder Architecture
3.5. Experimental Details
3.6. Model Evaluation
4. Results
4.1. Results on BraTS Dataset
4.2. Results on External Validation Datasets
5. Discussion
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van den Bent, M.J.; Geurts, M.; French, P.J.; Smits, M.; Capper, D.; Bromberg, J.E.; Chang, S.M. Primary brain tumours in adults. Lancet 2023, 402, 1564–1579. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.; Louis, D.; Poussaint, T.; Linscott, L.; Salzman, K. The 2021 World Health Organization Classification of Tumors of the Central Nervous System: What Neuroradiologists Need to Know. Am. J. Neuroradiol. 2022, 43, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Trinh, D.L.; Kim, S.H.; Yang, H.J.; Lee, G.S. The efficacy of shape radiomics and deep features for glioblastoma survival prediction by deep learning. Electronics 2022, 11, 1038. [Google Scholar] [CrossRef]
- Wadhwa, A.; Bhardwaj, A.; Verma, V.S. A review on brain tumor segmentation of MRI images. Magn. Reson. Imaging 2019, 61, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Daimary, D.; Bora, M.B.; Amitab, K.; Kandar, D. Brain tumor segmentation from MRI images using hybrid convolutional neural networks. Procedia Comput. Sci. 2020, 167, 2419–2428. [Google Scholar] [CrossRef]
- Rayed, M.E.; Islam, S.S.; Niha, S.I.; Jim, J.R.; Kabir, M.M.; Mridha, M. Deep learning for medical image segmentation: State-of-the-art advancements and challenges. Inform. Med. Unlocked 2024, 47, 101504. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Wang, P.; Yang, Q.; He, Z.; Yuan, Y. Vision transformers in multi-modal brain tumor MRI segmentation: A review. Meta Radiol. 2023, 1, 100004. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zeng, S.; Xie, D.; Zeng, W.; Huang, Y.; Mazu, L.; Zhu, N.; Yang, Z.; Chu, J.; Zhao, J. Looking through the imaging perspective: The importance of imaging necrosis in glioma diagnosis and prognostic prediction–single centre experience. Radiol. Oncol. 2024, 58, 23. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Shu, T.; Luo, P.; Shao, Y.; Lin, L.; Tu, Z.; Zhu, X.; Wu, L. The peritumoral edema index and related mechanisms influence the prognosis of GBM patients. Front. Oncol. 2024, 14, 1417208. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.K.T.; Mizumoto, M.; Ishikawa, E.; Matsuda, M.; Tanaka, K.; Kohzuki, H.; Numajiri, H.; Oshiro, Y.; Okumura, T.; Matsumura, A.; et al. Peritumoral edema status of glioblastoma identifies patients reaching long-term disease control with specific progression patterns after tumor resection and high-dose proton boost. J. Cancer Res. Clin. Oncol. 2021, 147, 3503–3516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ye, F.; Su, M.; Cui, M.; Chen, H.; Ma, X. The prognostic role of peritumoral edema in patients with newly diagnosed glioblastoma: A retrospective analysis. J. Clin. Neurosci. 2021, 89, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Schoenegger, K.; Oberndorfer, S.; Wuschitz, B.; Struhal, W.; Hainfellner, J.; Prayer, D.; Heinzl, H.; Lahrmann, H.; Marosi, C.; Grisold, W. Peritumoral edema on MRI at initial diagnosis: An independent prognostic factor for glioblastoma? Eur. J. Neurol. 2009, 16, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.; Navard, P.; Yilmaz, A. Segformer3d: An efficient transformer for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 4981–4988. [Google Scholar]
- Kamnitsas, K.; Ferrante, E.; Parisot, S.; Ledig, C.; Nori, A.V.; Criminisi, A.; Rueckert, D.; Glocker, B. DeepMedic for brain tumor segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Second InternationalWorkshop, BrainLes 2016, with the Challenges on BRATS, ISLES and mTOP 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, 17 October 2016; pp. 138–149. [Google Scholar]
- Ghaffari, M.; Sowmya, A.; Oliver, R. Automated brain tumor segmentation using multimodal brain scans: A survey based on models submitted to the BraTS 2012–2018 challenges. IEEE Rev. Biomed. Eng. 2019, 13, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, S.; Chen, H.; Luo, L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front. Neurosci. 2019, 13, 810. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. arXiv 2018, arXiv:1802.10508. [Google Scholar] [CrossRef]
- Henry, T.; Carré, A.; Lerousseau, M.; Estienne, T.; Robert, C.; Paragios, N.; Deutsch, E. Brain tumor segmentation with self-ensembled, deeply-supervised 3D U-net neural networks: A BraTS 2020 challenge solution. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; pp. 327–339. [Google Scholar]
- Sachdeva, J.; Sharma, D.; Ahuja, C.K. Multiscale segmentation net for segregating heterogeneous brain tumors: Gliomas on multimodal MR images. Image Vis. Comput. 2024, 149, 105191. [Google Scholar] [CrossRef]
- Ahuja, S.; Panigrahi, B.; Gandhi, T.K. Fully automatic brain tumor segmentation using DeepLabv3+ with variable loss functions. In Proceedings of the 2021 8th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 26–27 August 2021; pp. 522–526. [Google Scholar]
- Isensee, F.; Jäger, P.F.; Full, P.M.; Vollmuth, P.; Maier-Hein, K.H. nnU-Net for brain tumor segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; pp. 118–132. [Google Scholar]
- Luu, H.M.; Park, S.H. Extending nn-UNet for brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Virtual Event, 27 September 2021; pp. 173–186. [Google Scholar]
- Zeineldin, R.A.; Karar, M.E.; Coburger, J.; Wirtz, C.R.; Burgert, O. DeepSeg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Chen, Y.; He, X.; Zhuang, Z.; Wang, T.; Huang, H.; Wang, X.; Fu, X. DeepScan: Exploiting deep learning for malicious account detection in location-based social networks. IEEE Commun. Mag. 2018, 56, 21–27. [Google Scholar] [CrossRef]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, R.A.; Karar, M.E.; Burgert, O.; Mathis-Ullrich, F. Multimodal CNN networks for brain tumor segmentation in MRI: A BraTS 2022 challenge solution. In Proceedings of the International MICCAI Brainlesion Workshop, Singapore, 18 September 2022; pp. 127–137. [Google Scholar]
- Ferreira, A.; Solak, N.; Li, J.; Dammann, P.; Kleesiek, J.; Alves, V.; Egger, J. How we won brats 2023 adult glioma challenge? Just faking it! Enhanced synthetic data augmentation and model ensemble for brain tumour segmentation. arXiv 2024, arXiv:2402.17317. [Google Scholar] [CrossRef]
- Liang, J.; Yang, C.; Zhong, J.; Ye, X. BTSwin-Unet: 3D U-shaped symmetrical Swin transformer-based network for brain tumor segmentation with self-supervised pre-training. Neural Process. Lett. 2023, 55, 3695–3713. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J. TSEUnet: A 3D neural network with fused Transformer and SE-Attention for brain tumor segmentation. In Proceedings of the 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS), Shenzen, China, 21–23 July 2022; pp. 131–136. [Google Scholar]
- Xing, Z.; Yu, L.; Wan, L.; Han, T.; Zhu, L. NestedFormer: Nested modality-aware transformer for brain tumor segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 140–150. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 568–578. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Mejía, G.; Moreno, D.; Ruiz, D.; Aparicio, N. Hirni: Segmentation of Brain Tumors in Multi-parametric Magnetic Resonance Imaging Scans. In Proceedings of the 2021 IEEE 2nd International Congress of Biomedical Engineering and Bioengineering (CI-IB&BI), Bogotá, Colombia, 13–15 October 2021; pp. 1–4. [Google Scholar]
- Beser-Robles, M.; Castellá-Malonda, J.; Martínez-Gironés, P.M.; Galiana-Bordera, A.; Ferrer-Lozano, J.; Ribas-Despuig, G.; Teruel-Coll, R.; Cerdá-Alberich, L.; Martí-Bonmatí, L. Deep learning automatic semantic segmentation of glioblastoma multiforme regions on multimodal magnetic resonance images. Int. J. Comput. Assist. Radiol. Surg. 2024, 19, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation Labels for the Pre-Operative Scans of the TCGA-GBM Collection. 2017. [Data Set]. Available online: https://www.cancerimagingarchive.net/analysis-result/brats-tcga-gbm/ (accessed on 9 July 2025).
- Bakas, S.; Sako, C.; Akbari, H.; Bilello, M.; Sotiras, A.; Shukla, G.; Rudie, J.; Flores Santamaria, N.; Fathi Kazerooni, A.; Pati, S.; et al. Multi-parametric magnetic resonance imaging (mpMRI) scans for de novo Glioblastoma (GBM) patients from the University of Pennsylvania Health System (UPENN-GBM). Cancer Imaging Arch. 2021, 9, 453. [Google Scholar]
- Cardoso, M.J.; Li, W.; Brown, R.; Ma, N.; Kerfoot, E.; Wang, Y.; Murrey, B.; Myronenko, A.; Zhao, C.; Yang, D.; et al. MONAI: An open-source framework for deep learning in healthcare. arXiv 2022, arXiv:2211.02701. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dowdell, B.; Engelder, T.; Pulmano, Z.; Osa, N.; Barman, A. Glioblastoma tumor segmentation using an ensemble of vision transformers. In Proceedings of the Medical Imaging 2025: Computer-Aided Diagnosis, San Diego, CA, USA, 17–20 February 2025; Volume 13407, pp. 487–496. [Google Scholar]
- Myronenko, A. 3D MRI brain tumor segmentation using autoencoder regularization. arXiv 2018, arXiv:1810.11654. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, B.; Peng, Y.; Chen, F.; Li, W. Segmentation of glioblastomas via 3D FusionNet. Front. Oncol. 2024, 14, 1488616. [Google Scholar] [CrossRef] [PubMed]



| Subregion | SegFormer3D | Multi-Encoder | ||||
|---|---|---|---|---|---|---|
| Dice | Precision | Recall | Dice | Precision | Recall | |
| NCR * | 0.66 ± 0.29 | 0.68 ± 0.28 | 0.74 ± 0.26 | 0.78 ± 0.28 | 0.84 ± 0.24 | 0.79 ± 0.27 |
| ED * | 0.78 ± 0.18 | 0.77 ± 0.18 | 0.82 ± 0.18 | 0.86 ± 0.14 | 0.87 ± 0.15 | 0.87 ± 0.14 |
| ET * | 0.79 ± 0.18 | 0.80 ± 0.17 | 0.80 ± 0.19 | 0.88 ± 0.15 | 0.89 ± 0.12 | 0.89 ± 0.15 |
| TC * | 0.81 ± 0.26 | 0.82 ± 0.29 | 0.87 ± 0.15 | 0.91 ± 0.13 | 0.93 ± 0.13 | 0.90 ± 0.16 |
| WT * | 0.84 ± 0.19 | 0.84 ± 0.22 | 0.90 ± 0.11 | 0.92 ± 0.08 | 0.94 ± 0.07 | 0.92 ± 0.10 |
| Subregion | UPENN-GBM | TCGA-GBM | ||||
|---|---|---|---|---|---|---|
| Dice | Precision | Recall | Dice | Precision | Recall | |
| NCR | 0.76 ± 0.19 | 0.86 ± 0.16 | 0.72 ± 0.21 | 0.74 ± 0.22 | 0.82 ± 0.22 | 0.73 ± 0.22 |
| ED | 0.83 ± 0.13 | 0.83 ± 0.15 | 0.85 ± 0.12 | 0.86 ± 0.13 | 0.88 ± 0.12 | 0.86 ± 0.15 |
| ET | 0.85 ± 0.11 | 0.83 ± 0.13 | 0.88 ± 0.12 | 0.88 ± 0.12 | 0.88 ± 0.15 | 0.91 ± 0.12 |
| TC | 0.91 ± 0.11 | 0.93 ± 0.11 | 0.89 ± 0.12 | 0.92 ± 0.12 | 0.94 ± 0.13 | 0.91 ± 0.11 |
| WT | 0.90 ± 0.09 | 0.91 ± 0.10 | 0.90 ± 0.08 | 0.94 ± 0.10 | 0.95 ± 0.10 | 0.92 ± 0.11 |
| Model | Mean Dice ET | Mean Dice NCR | Mean Dice ED | Inference Time (s) | Peak Memory (MB) | Parameters (M) |
|---|---|---|---|---|---|---|
| SegFormer3D | 0.79 | 0.66 | 0.78 | 0.65 | 1939.5 | ∼4.5 |
| Multi-encoder | 0.88 | 0.78 | 0.86 | 1.41 | 5487.6 | ∼11.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cariola, A.; Sibilano, E.; Brunetti, A.; Buongiorno, D.; Guerriero, A.; Bevilacqua, V. Enhanced Segmentation of Glioma Subregions via Modality-Aware Encoding and Channel-Wise Attention in Multimodal MRI. Appl. Sci. 2025, 15, 8061. https://doi.org/10.3390/app15148061
Cariola A, Sibilano E, Brunetti A, Buongiorno D, Guerriero A, Bevilacqua V. Enhanced Segmentation of Glioma Subregions via Modality-Aware Encoding and Channel-Wise Attention in Multimodal MRI. Applied Sciences. 2025; 15(14):8061. https://doi.org/10.3390/app15148061
Chicago/Turabian StyleCariola, Annachiara, Elena Sibilano, Antonio Brunetti, Domenico Buongiorno, Andrea Guerriero, and Vitoantonio Bevilacqua. 2025. "Enhanced Segmentation of Glioma Subregions via Modality-Aware Encoding and Channel-Wise Attention in Multimodal MRI" Applied Sciences 15, no. 14: 8061. https://doi.org/10.3390/app15148061
APA StyleCariola, A., Sibilano, E., Brunetti, A., Buongiorno, D., Guerriero, A., & Bevilacqua, V. (2025). Enhanced Segmentation of Glioma Subregions via Modality-Aware Encoding and Channel-Wise Attention in Multimodal MRI. Applied Sciences, 15(14), 8061. https://doi.org/10.3390/app15148061









