Artificial Intelligence in Dermatology: A Review of Methods, Clinical Applications, and Perspectives
Abstract
1. Introduction
2. AI Methods
2.1. Machine Learning
2.2. Deep Learning
2.3. Applications of Neural Networks
2.4. New Trends in 2023–2025
2.5. Limitations of AI Methods in Dermatology
3. Decision-Making Modeling in Clinical Issues
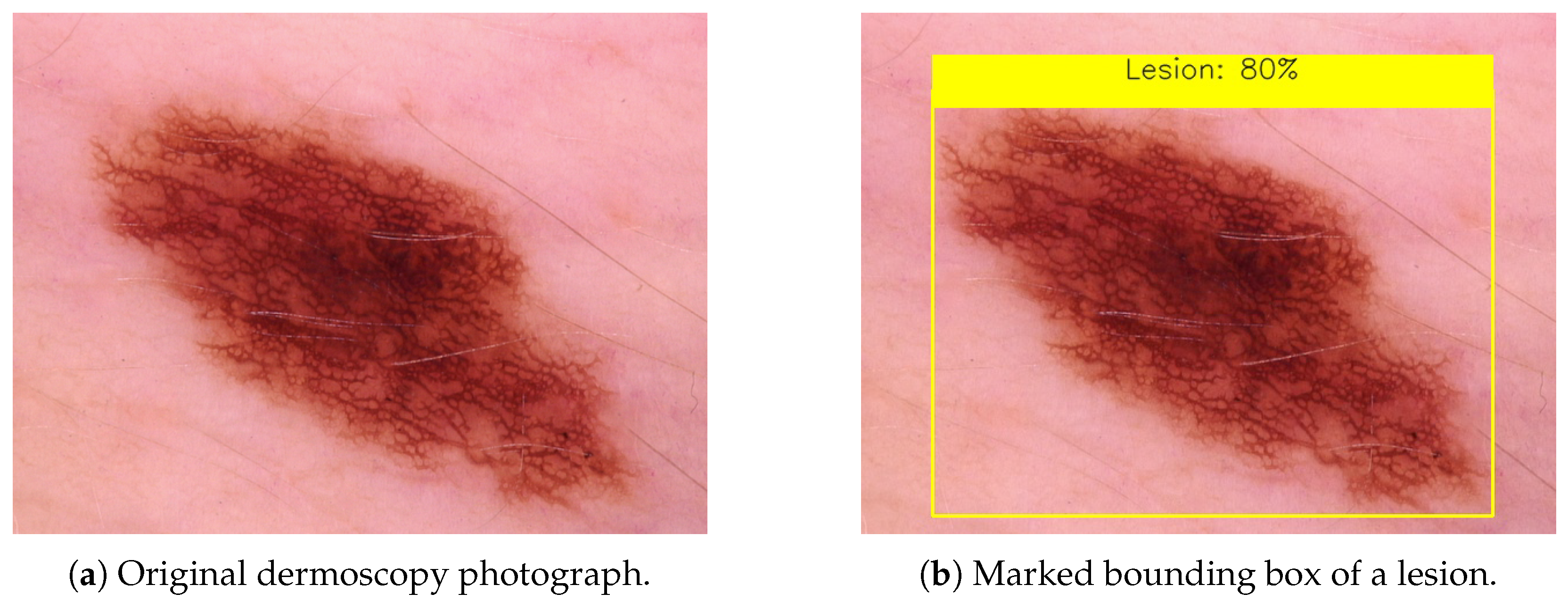
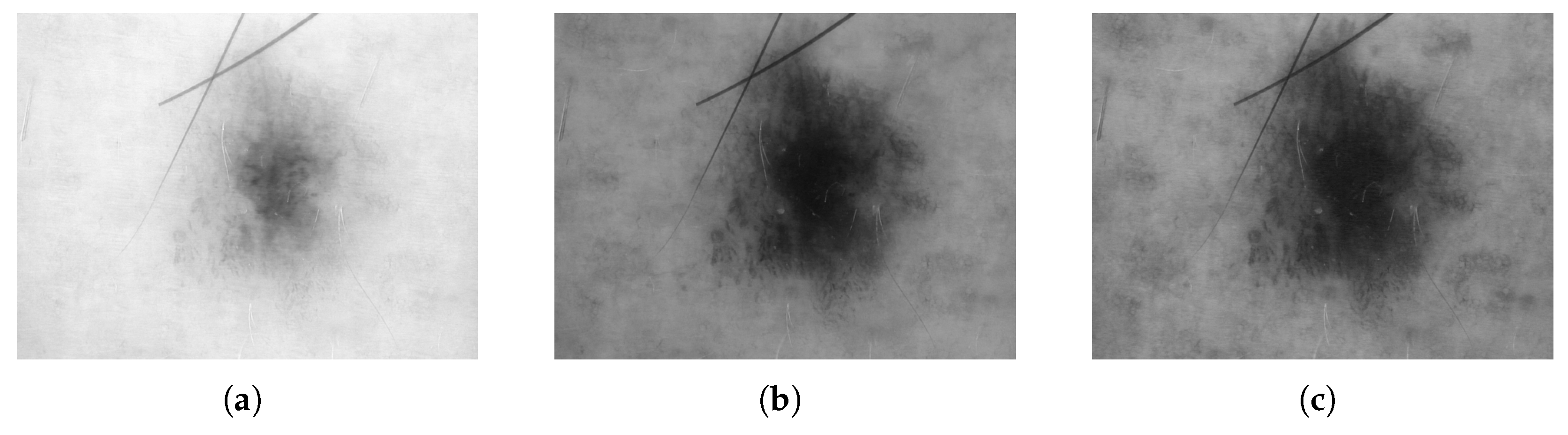
3.1. Digital Images
3.2. Dermoscopy
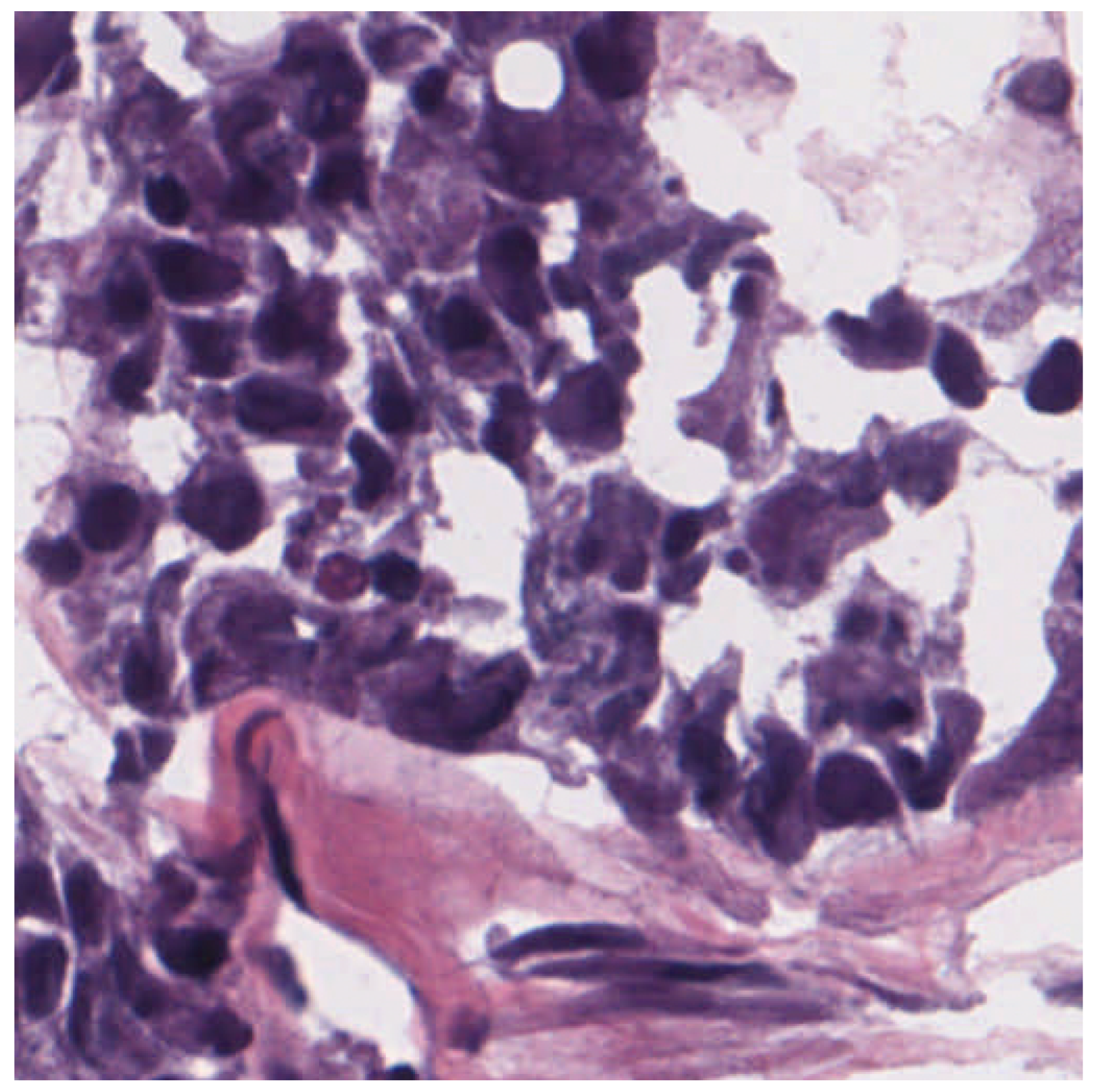
3.3. Dermatopathology
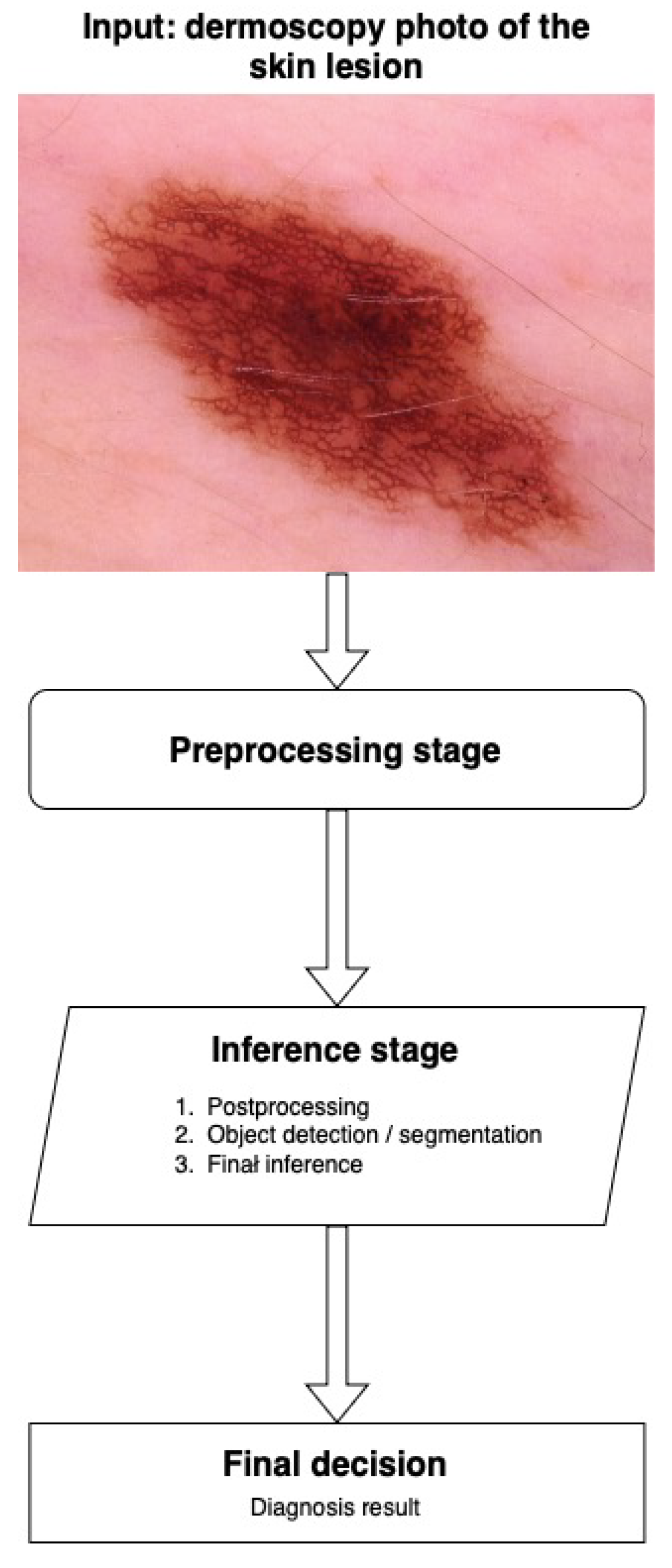
3.4. Inference and Decision
3.5. Classification Quality Measures
- true positives (TP)—classified positively by both the model and specialists;
- false positives (FP)—classified positively by the model but negatively by specialists;
- false negatives (FN)—classified negatively by the model but positively by specialists;
- true negatives (TN)—classified negatively by both the model and specialists.
4. Methods
4.1. Protocol and Registration
4.2. Inclusion and Exclusion Criteria
4.3. Search Strategy
5. AI Regulations, Verification, and Ethical Problems
5.1. Regulations on AI in Medicine
5.2. Verification of the Model
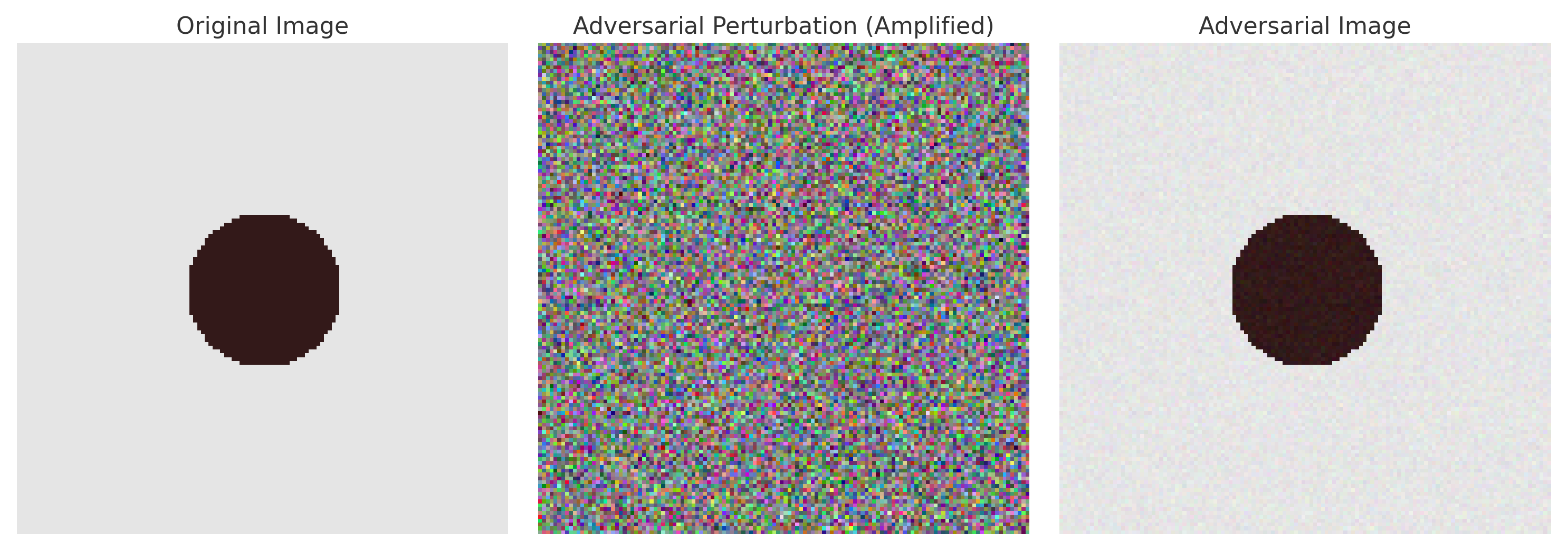
5.2.1. Adversarial Attacks
5.2.2. Explainability
5.3. Ethical Implications
5.3.1. Informed Consent and Data Privacy
5.3.2. Bias and Fairness
5.3.3. Legal Liability and Accountability
5.4. Human–AI Interaction
6. Bibliometric Analysis of Articles on AI Applications in Dermatology
7. Overview of the Major Applications of AI in Dermatology
7.1. Core Applications
7.2. Applications in Clinical Practice
7.3. Future Research Directions
8. Comparative Analysis of Neural Network Architectures and Datasets in Skin Diagnostics
8.1. Comparison of Neural Network Architectures
8.2. Comparison of Datasets
9. Key Observations and Recommendations
9.1. Key Observations
- Dominance of CNNs in diagnostic imaging. Most works (∼65%) use convolutional neural networks, with transfer learning architectures (e.g., ResNet, DenseNet) being the most common.
- Emergence of Transformer-based models. Recent studies have introduced Vision Transformer architectures (e.g., Swin Transformer, ViT variants) that achieve state-of-the-art accuracy in skin lesion classification, albeit with substantially higher computational and data requirements. This marks a new trend that challenges the CNN’s dominance.
- Adoption of ensemble learning for performance gains. A few works combine multiple models to boost the accuracy—for example, ensembling lightweight CNNs (as in SkinNet) significantly improved the classification metrics (achieving AUC ≈ 0.96). Such ensembles leverage complementary strengths at the cost of greater complexity.
- Growing role of data augmentation. The use of GAN-generated images and augmented imaging (rotation, scaling, colour shifts) has become common, improving the classification accuracy, on average, by 3–7%. It helps to address data scarcity and imbalances in training sets.
- Persistent class imbalance challenges. Skewed datasets (e.g., few melanomas among many nevi) remain problematic. Specialised strategies to rebalance training data (for instance, the approach used in DSCC_Net to equalise class representation) have been shown to markedly enhance the model AUC and sensitivity, underlining the importance of addressing rare lesion classes.
- Insufficient standardisation of evaluations. A lack of uniform protocols for training/test splits and performance metrics (such as cross-validation vs. random splits and varying datasets) makes it difficult to compare results across studies. This variability hampers the objective benchmarking of different AI models in dermatology.
- Limited interpretability of models. Few publications consider model interpretability (e.g., saliency maps like Grad-CAM or LIME), which raises concerns about clinical trust. The black-box nature of many AI models remains a significant obstacle to their acceptance in dermatological practice.
9.2. Key Recommendations
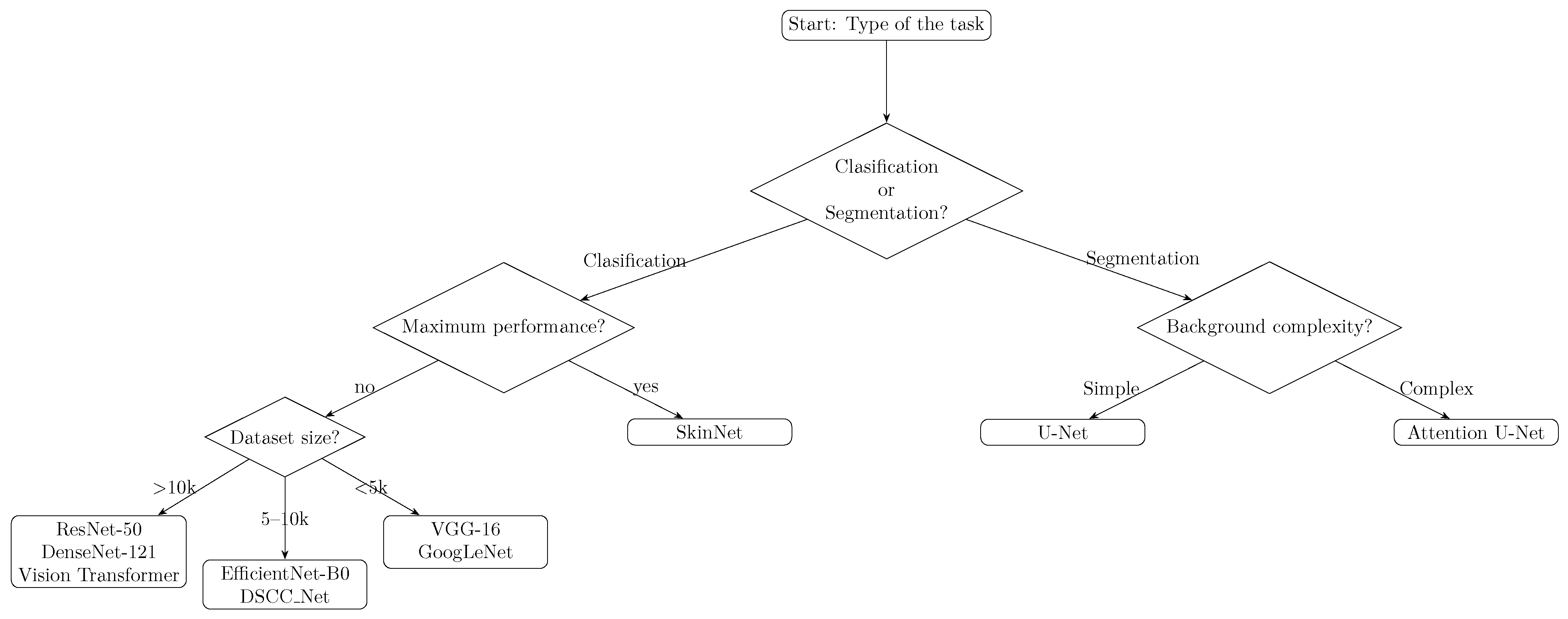
- Selection of the appropriate architecture
- –
- Melanoma classification: We recommend DenseNet-121 as a base in the transfer learning approach, which provides a good compromise between network depth and limiting overfitting.
- –
- Skin lesion segmentation: We recommend using the UNet model enriched with an attention module (Attention UNet), which enables the more precise extraction of lesion boundaries, especially with limited data.
- –
- Large-scale lesion classification: If a large quantity of data (>10k images) and computing resources are available, consider advanced architectures like Vision Transformers (e.g., Swin Transformer) for potentially higher accuracy. Please note that these Transformer models are resource-intensive and require longer training times.
- –
- Maximising diagnostic performance: For critical applications requiring the highest accuracy, an ensemble of models can be used. Combining multiple complementary CNN architectures (as in SkinNet, which fused MobileNet, ResNet18, and VGG11) has demonstrated improved performance (AUC ∼ 0.96), albeit with increased training and deployment complexity.
- Standardisation of experimental protocols
- –
- Use K-fold CV instead of a single random split, which provides a more robust assessment of model generalisation.
- –
- Use publicly available, standardised reference sets such as ISIC or HAM10000 to increase the comparability between studies.
- Augmentation and generative models for data enrichment
- –
- Use traditional techniques (rotation, scaling, brightness changes) in parallel with StyleGAN2 or CycleGAN to generate synthetic images of rare lesion types.
- –
- Regularly verify the impact of newly generated samples on model metrics, with the goal of increasing the sensitivity with a minimal increase in false alarms.
- Ensure interpretability and clinical confidence
- –
- Publish saliency maps (e.g., Grad-CAM, LIME) for each new model, which allows verification that the network focuses on symptomatic parts of the lesion.
- –
- Consider integrating tools such as SHAP to analyse the impact of individual image features on the classifier’s decision.
- Regular bibliometric and statistical analysis
- –
- Update publication trend graphs every six months, taking into account new techniques (e.g., Transformers in computer vision).
- –
- When comparing performance across architectures, use significance tests (e.g., ANOVA) to determine whether differences in accuracy are statistically significant.
9.3. Method Comparison Table
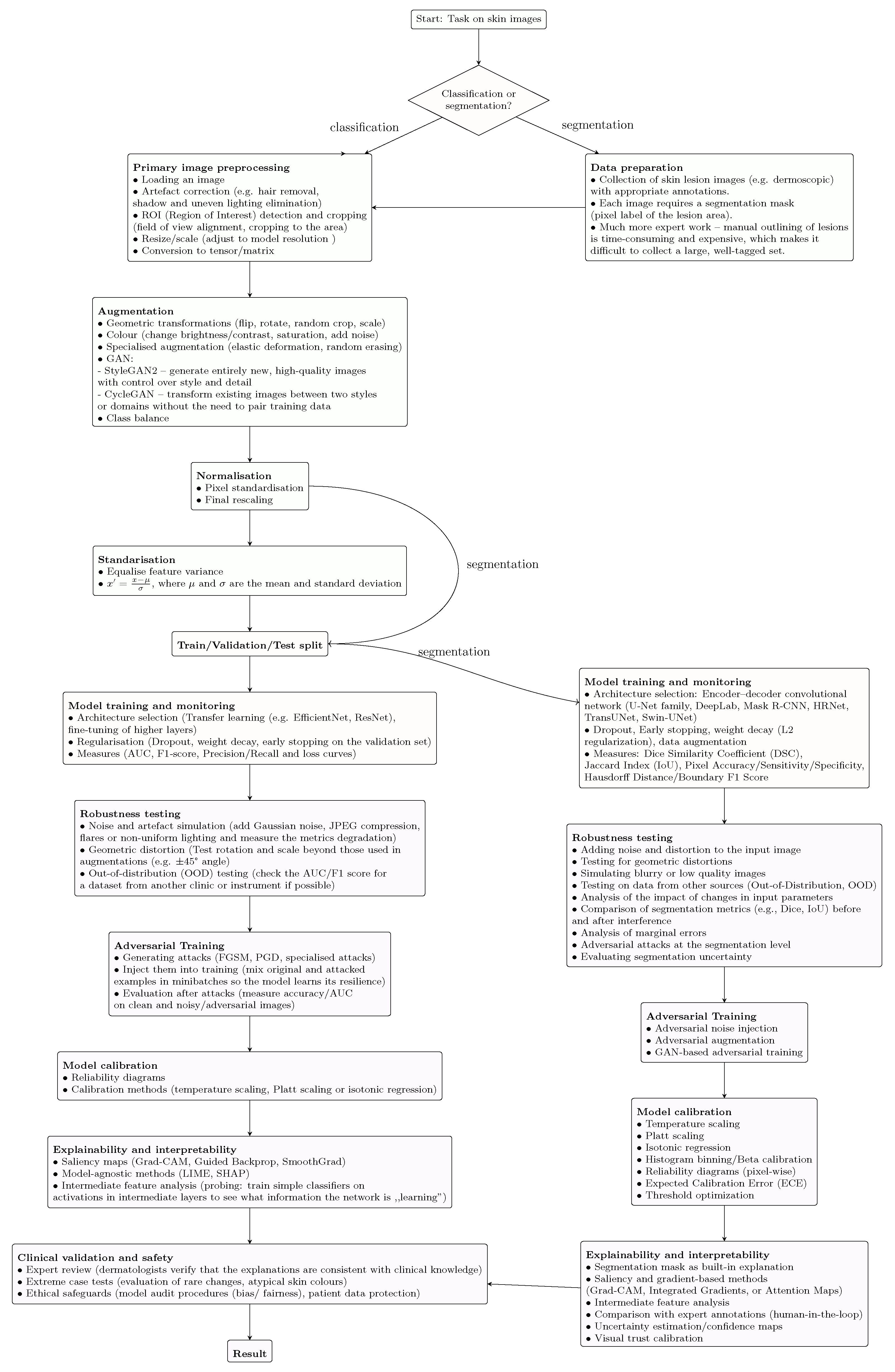
9.4. Machine Learning Flowchart
10. Other Review Perspectives
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, G. History of Artificial Intelligence. In Introduction to the History of Computing; Springer International Publishing: New York, NY, USA, 2016; pp. 249–273. [Google Scholar] [CrossRef]
- Buchanan, B.G.; Shortliffe, E.H. Rule Based Expert Systems: The Mycin Experiments of the Stanford Heuristic Programming Project; The Addison-Wesley Series in Artificial Intelligence; Addison-Wesley Longman Publishing Co., Inc.: Boston, MA, USA, 1984. [Google Scholar]
- AI Funding in Healthcare Through the Years. Available online: https://www.beckershospitalreview.com/digital-health/ai-funding-in-healthcare-through-the-years.html (accessed on 7 November 2024).
- Stypińska, J.; Franke, A. AI revolution in healthcare and medicine and the (re-)emergence of inequalities and disadvantages for ageing population. Front. Sociol. 2023, 7, 1038854. [Google Scholar] [CrossRef] [PubMed]
- Dermatologist, A. AI Dermatologist Skin Scanner. Available online: https://ai-derm.com/ai (accessed on 30 May 2025).
- Liopyris, K.; Gregoriou, S.; Dias, J.; Stratigos, A.J. Artificial Intelligence in Dermatology: Challenges and Perspectives. Dermatol. Ther. 2022, 12, 2637–2651. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Sarda, A.; Gupta, S.; Das, S. Use of artificial intelligence in dermatology. Indian J. Dermatol. 2020, 65, 352. [Google Scholar] [CrossRef]
- 2025. Available online: https://skin-analytics.com/ (accessed on 7 July 2025).
- Renders, J.M.; Simonart, T. Role of Artificial Neural Networks in Dermatology. Dermatology 2009, 219, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Pawlak, Z. Rough sets. Int. J. Comput. Inf. Sci. 1982, 11, 341–356. [Google Scholar] [CrossRef]
- Polkowski, L.; Skowron, A. Rough mereology: A new paradigm for approximate reasoning. Int. J. Approx. Reason. 1996, 15, 333–365. [Google Scholar] [CrossRef]
- Zadeh, L. Fuzzy sets. Inf. Control 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Solomonoff, R.J. An inductive inference machine. In Proceedings of the IRE Convention Record, Section on Information Theory; The Institute of Radio Engineers: New York, NY, USA, 1957. [Google Scholar]
- Alwakid, G.; Gouda, W.; Humayun, M.; Sama, N.U. Melanoma Detection Using Deep Learning-Based Classifications. Healthcare 2022, 10, 2481. [Google Scholar] [CrossRef]
- Ivakhnenko, A.; Lapa, V. Cybernetics and Forecasting Techniques. In Modern Analytic and Computational Methods in Science and Mathematics; American Elsevier Publishing Company: New York, NY, USA, 1967. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, Y. Supervised Learning. In Encyclopedia of the Sciences of Learning; Springer: New York, NY, USA, 2012; pp. 3243–3245. [Google Scholar] [CrossRef]
- Barlow, H. Unsupervised Learning. Neural Comput. 1989, 1, 295–311. [Google Scholar] [CrossRef]
- Kaelbling, L.P.; Littman, M.L.; Moore, A.W. Reinforcement Learning: A Survey. J. Artif. Intell. Res. 1996, 4, 237–285. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, Z.; Jiang, Z.; Huang, K.; Guo, K.; Zhao, S. Background selection schema on deep learning-based classification of dermatological disease. Comput. Biol. Med. 2022, 149, 105966. [Google Scholar] [CrossRef]
- Inthiyaz, S.; Altahan, B.R.; Ahammad, S.H.; Rajesh, V.; Kalangi, R.R.; Smirani, L.K.; Hossain, M.A.; Rashed, A.N.Z. Skin disease detection using deep learning. Adv. Eng. Softw. 2023, 175, 103361. [Google Scholar] [CrossRef]
- Bettuzzi, T.; Hua, C.; Diaz, E.; Colin, A.; Wolkenstein, P.; de Prost, N.; Ingen-Housz-Oro, S. Epidermal necrolysis: Characterization of different phenotypes using an unsupervised clustering analysis. Br. J. Dermatol. 2022, 186, 1037–1039. [Google Scholar] [CrossRef]
- Garcia, J.B.; Tanadini-Lang, S.; Andratschke, N.; Gassner, M.; Braun, R. Suspicious Skin Lesion Detection in Wide-Field Body Images using Deep Learning Outlier Detection. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2928–2932. [Google Scholar] [CrossRef]
- Usmani, U.A.; Watada, J.; Jaafar, J.; Aziz, I.A.; Roy, A. A Reinforcement Learning Algorithm for Automated Detection of Skin Lesions. Appl. Sci. 2021, 11, 9367. [Google Scholar] [CrossRef]
- Barata, C.; Rotemberg, V.; Codella, N.C.F.; Tschandl, P.; Rinner, C.; Akay, B.N.; Apalla, Z.; Argenziano, G.; Halpern, A.; Lallas, A.; et al. A reinforcement learning model for AI-based decision support in skin cancer. Nat. Med. 2023, 29, 1941–1946. [Google Scholar] [CrossRef]
- Piccialli, F.; Somma, V.D.; Giampaolo, F.; Cuomo, S.; Fortino, G. A survey on deep learning in medicine: Why, how and when? Inf. Fusion 2021, 66, 111–137. [Google Scholar] [CrossRef]
- Wolpert, D.; Macready, W. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Eklund, A.; Dufort, P.; Forsberg, D.; LaConte, S.M. Medical image processing on the GPU—Past, present and future. Med. Image Anal. 2013, 17, 1073–1094. [Google Scholar] [CrossRef] [PubMed]
- Cun, Y.L.; Boser, B.; Denker, J.S.; Howard, R.E.; Habbard, W.; Jackel, L.D.; Henderson, D. Handwritten digit recognition with a back-propagation network. In Advances in Neural Information Processing Systems 2; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1990; pp. 396–404. [Google Scholar]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2021, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Chen, K.; Shi, Z.; Guo, Y.; Ye, J. Object Detection in 20 Years: A Survey. Proc. IEEE 2023, 111, 257–276. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.H.; Wei, Z.; Heidari, A.A.; Zheng, N.; Li, Z.; Chen, H.; Hu, H.; Zhou, Q.; Guan, Q. Generative Adversarial Networks in Medical Image augmentation: A review. Comput. Biol. Med. 2022, 144, 105382. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Computer Vision—ECCV 2016; Springer International Publishing: New York, NY, USA, 2016; pp. 21–37. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Springer International Publishing: New York, NY, USA, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. arXiv 2017, arXiv:1703.10593. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; McClelland, J.L. Learning Internal Representations by Error Propagation. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations; MIT Press: Cambridge, MA, USA, 1987; pp. 318–362. [Google Scholar]
- Goyal, A.; Choudhary, A.; Malik, D.; Baliyan, M.S.; Rani, S. Implementing and Analysis of RNN LSTM Model for Stock Market Prediction. In Advances in Data and Information Sciences; Springer: Singapore, 2022; pp. 241–248. [Google Scholar] [CrossRef]
- Zhao, J.; Zeng, D.; Liang, S.; Kang, H.; Liu, Q. Prediction model for stock price trend based on recurrent neural network. J. Ambient Intell. Humaniz. Comput. 2020, 12, 745–753. [Google Scholar] [CrossRef]
- Hou, J.; Tian, Z. Application of recurrent neural network in predicting athletes’ sports achievement. J. Supercomput. 2021, 78, 5507–5525. [Google Scholar] [CrossRef]
- Khurana, D.; Koli, A.; Khatter, K.; Singh, S. Natural language processing: State of the art, current trends and challenges. Multimed. Tools Appl. 2022, 82, 3713–3744. [Google Scholar] [CrossRef]
- Patwardhan, N.; Marrone, S.; Sansone, C. Transformers in the Real World: A Survey on NLP Applications. Information 2023, 14, 242. [Google Scholar] [CrossRef]
- Wankhade, M.; Rao, A.C.S.; Kulkarni, C. A survey on sentiment analysis methods, applications, and challenges. Artif. Intell. Rev. 2022, 55, 5731–5780. [Google Scholar] [CrossRef]
- Bose, P.; Srinivasan, S.; Sleeman, W.C.; Palta, J.; Kapoor, R.; Ghosh, P. A Survey on Recent Named Entity Recognition and Relationship Extraction Techniques on Clinical Texts. Appl. Sci. 2021, 11, 8319. [Google Scholar] [CrossRef]
- Widyassari, A.P.; Rustad, S.; Shidik, G.F.; Noersasongko, E.; Syukur, A.; Affandy, A.; Setiadi, D.R.I.M. Review of automatic text summarization techniques &; methods. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 1029–1046. [Google Scholar] [CrossRef]
- Popel, M.; Tomkova, M.; Tomek, J.; Kaiser, L.; Uszkoreit, J.; Bojar, O.; Žabokrtský, Z. Transforming machine translation: A deep learning system reaches news translation quality comparable to human professionals. Nat. Commun. 2020, 11, 4381. [Google Scholar] [CrossRef]
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L. GPT-4 Technical Report. arXiv 2023, arXiv:2303.08774. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to Sequence Learning with Neural Networks. arXiv 2014, arXiv:1409.3215. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Proceedings of the 2019 Conference of the North; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar] [CrossRef]
- Zhao, W.X.; Zhou, K.; Li, J.; Tang, T.; Wang, X.; Hou, Y.; Min, Y.; Zhang, B.; Zhang, J.; Dong, Z.; et al. A Survey of Large Language Models. arXiv 2025, arXiv:2303.18223. [Google Scholar] [CrossRef]
- Matin, R.N.; Linos, E.; Rajan, N. Leveraging large language models in dermatology. Br. J. Dermatol. 2023, 189, 253–254. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.; Tanveer, M.A.; Aqeel Khan, H. Skin Lesion Classification Using GAN based Data Augmentation. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 916–919. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Y.; Chang, Y. CycleGAN Based Data Augmentation For Melanoma images Classification. In Proceedings of the 2020 3rd International Conference on Artificial Intelligence and Pattern Recognition, AIPR ’20, New York, NY, USA, 26–28 June 2020; pp. 115–119. [Google Scholar] [CrossRef]
- Asaf, M.Z.; Rasul, H.; Akram, M.U.; Hina, T.; Rashid, T.; Shaukat, A. A Modified Deep Semantic Segmentation Model for Analysis of Whole Slide Skin Images. Sci. Rep. 2024, 14, 23489. [Google Scholar] [CrossRef]
- Sarwar, N.; Irshad, A.; Naith, Q.H.; D.Alsufiani, K.; Almalki, F.A. Skin lesion segmentation using deep learning algorithm with ant colony optimization. BMC Med Inform. Decis. Mak. 2024, 24, 265. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Sommella, P.; Carratù, M.; O’Nils, M.; Lundgren, J. A Deep CNN Transformer Hybrid Model for Skin Lesion Classification of Dermoscopic Images Using Focal Loss. Diagnostics 2023, 13, 72. [Google Scholar] [CrossRef]
- Innani, S.; Dutande, P.; Baid, U.; Pokuri, V.; Bakas, S.; Talbar, S.; Baheti, B.; Guntuku, S.C. Generative adversarial networks based skin lesion segmentation. Sci. Rep. 2023, 13, 13467. [Google Scholar] [CrossRef] [PubMed]
- Oda, J.; Takemoto, K. Mobile applications for skin cancer detection are vulnerable to physical camera-based adversarial attacks. Sci. Rep. 2025, 15, 18119. [Google Scholar] [CrossRef]
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. Npj Digit. Med. 2021, 4, 5. [Google Scholar] [CrossRef]
- Phillips, M.; Marsden, H.; Jaffe, W.; Matin, R.N.; Wali, G.N.; Greenhalgh, J.; McGrath, E.; James, R.; Ladoyanni, E.; Bewley, A.; et al. Assessment of Accuracy of an Artificial Intelligence Algorithm to Detect Melanoma in Images of Skin Lesions. JAMA Netw. Open 2019, 2, e1913436. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, H.; Gichoya, J.W.; Katabi, D.; Ghassemi, M. The limits of fair medical imaging AI in real-world generalization. Nat. Med. 2024, 30, 2838–2848. [Google Scholar] [CrossRef]
- Salinas, M.P.; Sepúlveda, J.; Hidalgo, L.; Peirano, D.; Morel, M.; Uribe, P.; Rotemberg, V.; Briones, J.; Mery, D.; Navarrete-Dechent, C. A systematic review and meta-analysis of artificial intelligence versus clinicians for skin cancer diagnosis. Npj Digit. Med. 2024, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.A.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.M.; Bailey, E.E.; Gevaert, O.; et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci. Adv. 2022, 8, 32. [Google Scholar] [CrossRef]
- Montoya, L.N.; Roberts, J.S.; Hidalgo, B.S. Towards Fairness in AI for Melanoma Detection: Systemic Review and Recommendations. arXiv 2024, arXiv:2411.12846. [Google Scholar] [CrossRef]
- Mahbod, A.; Bancher, B.; Ellinger, I. CryoNuSeg. 2021. Available online: https://www.kaggle.com/dsv/1900145 (accessed on 10 May 2025). [CrossRef]
- Kainth, K.; Singh, B. Analysis of CCD and CMOS Sensor Based Images from Technical and Photographic Aspects (January 19, 2020). International Conference of Advance Research & Innovation (ICARI) 2020. Available online: https://ssrn.com/abstract=3559236 (accessed on 10 July 2025).
- Süsstrunk, S.; Buckley, R.; Swen, S. Standard RGB Color Spaces. Color Imaging Conf. 1999, 7, 127–134. [Google Scholar] [CrossRef]
- Fairchild, M.D. Refinement of the RLAB color space. Color Res. Appl. 1996, 21, 338–346. [Google Scholar] [CrossRef]
- Shaik, K.B.; Ganesan, P.; Kalist, V.; Sathish, B.; Jenitha, J.M.M. Comparative Study of Skin Color Detection and Segmentation in HSV and YCbCr Color Space. Procedia Comput. Sci. 2015, 57, 41–48. [Google Scholar] [CrossRef]
- Campos-do-Carmo, G.; Ramos-e-Silva, M. Dermoscopy: Basic concepts. Int. J. Dermatol. 2008, 47, 712–719. [Google Scholar] [CrossRef]
- Alwakid, G.; Gouda, W.; Humayun, M.; Jhanjhi, N.Z. Diagnosing Melanomas in Dermoscopy Images Using Deep Learning. Diagnostics 2023, 13, 1815. [Google Scholar] [CrossRef]
- Deena, D.G.; Prakash, M.B.; Sreekar, C.S.; Rohith, S. A New Approach for Diagnosis of Melanoma from Dermoscopy Images. J. Surv. Fish. Sci. 2023, 10–15. [Google Scholar] [CrossRef]
- Dugonik, B.; Dugonik, A.; Marovt, M.; Golob, M. Image Quality Assessment of Digital Image Capturing Devices for Melanoma Detection. Appl. Sci. 2020, 10, 2876. [Google Scholar] [CrossRef]
- Alves, J.; Moreira, D.; Alves, P.; Rosado, L.; Vasconcelos, M. Automatic Focus Assessment on Dermoscopic Images Acquired with Smartphones. Sensors 2019, 19, 4957. [Google Scholar] [CrossRef]
- Kubinger, W.; Vincze, M.; Ayromlou, M. The role of gamma correction in colour image processing. In Proceedings of the 9th European Signal Processing Conference (EUSIPCO 1998), Rhodes Island, Greece, 8–11 September 1998; pp. 1–4. [Google Scholar]
- Cavalcanti, P.G.; Scharcanski, J.; Lopes, C.B.O. Shading Attenuation in Human Skin Color Images. In Advances in Visual Computing; Springer: Berlin/Heidelberg, Germany, 2010; pp. 190–198. [Google Scholar] [CrossRef]
- Quintana, J.; Garcia, R.; Neumann, L. A novel method for color correction in epiluminescence microscopy. Comput. Med Imaging Graph. 2011, 35, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Glaister, J.; Amelard, R.; Wong, A.; Clausi, D.A. MSIM: Multistage Illumination Modeling of Dermatological Photographs for Illumination-Corrected Skin Lesion Analysis. IEEE Trans. Biomed. Eng. 2013, 60, 1873–1883. [Google Scholar] [CrossRef] [PubMed]
- Pizer, S.; Johnston, R.; Ericksen, J.; Yankaskas, B.; Muller, K. Contrast-limited adaptive histogram equalization: Speed and effectiveness. In Proceedings of the First Conference on Visualization in Biomedical Computing, Atlanta, GA, USA, 22–25 May 1990; pp. 337–345. [Google Scholar] [CrossRef]
- Celebi, M.E.; Iyatomi, H.; Schaefer, G. Contrast enhancement in dermoscopy images by maximizing a histogram bimodality measure. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 2601–2604. [Google Scholar] [CrossRef]
- Abbas, Q.; Garcia, I.F.; Emre Celebi, M.; Ahmad, W.; Mushtaq, Q. A perceptually oriented method for contrast enhancement and segmentation of dermoscopy images. Skin Res. Technol. 2012, 19, e490–e497. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Lee, T.K.; Atkins, M.S. Segmentation of light and dark hair in dermoscopic images: A hybrid approach using a universal kernel. In Proceedings of the Medical Imaging 2010: Image Processing; Dawant, B.M., Haynor, D.R., Eds.; SPIE: Bellingham, WA, USA, 2010; Volume 7623, p. 76234N. [Google Scholar] [CrossRef]
- Abbas, Q.; Celebi, M.; García, I.F. Hair removal methods: A comparative study for dermoscopy images. Biomed. Signal Process. Control 2011, 6, 395–404. [Google Scholar] [CrossRef]
- Abbas, Q.; Garcia, I.F.; Emre Celebi, M.; Ahmad, W. A Feature-Preserving Hair Removal Algorithm for Dermoscopy Images. Skin Res. Technol. 2011, 19, e27–e36. [Google Scholar] [CrossRef]
- Kempf, W.; Hantschke, M.; Kutzner, H. Dermatopathology; Springer International Publishing: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Olsen, T.G.; Jackson, B.H.; Feeser, T.A.; Kent, M.N.; Moad, J.C.; Krishnamurthy, S.; Lunsford, D.D.; Soans, R.E. Diagnostic Performance of Deep Learning Algorithms Applied to Three Common Diagnoses in Dermatopathology. J. Pathol. Inform. 2018, 9, 32. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Qu, D.; Xue, Y.; Bi, X.; Chen, Z. A Deep Learning Approach for Basal Cell Carcinomas and Bowen’s Disease Recognition in Dermatopathology Image. J. Biomater. Tissue Eng. 2022, 12, 879–887. [Google Scholar] [CrossRef]
- Dalal, A.; Moss, R.H.; Stanley, R.J.; Stoecker, W.V.; Gupta, K.; Calcara, D.A.; Xu, J.; Shrestha, B.; Drugge, R.; Malters, J.M.; et al. Concentric decile segmentation of white and hypopigmented areas in dermoscopy images of skin lesions allows discrimination of malignant melanoma. Comput. Med. Imaging Graph. 2011, 35, 148–154. [Google Scholar] [CrossRef]
- Kaur, R.; Albano, P.P.; Cole, J.G.; Hagerty, J.; LeAnder, R.W.; Moss, R.H.; Stoecker, W.V. Real-time supervised detection of pink areas in dermoscopic images of melanoma: Importance of color shades, texture and location. Skin Res. Technol. 2015, 21, 466–473. [Google Scholar] [CrossRef]
- Emre Celebi, M.; Kingravi, H.A.; Iyatomi, H.; Alp Aslandogan, Y.; Stoecker, W.V.; Moss, R.H.; Malters, J.M.; Grichnik, J.M.; Marghoob, A.A.; Rabinovitz, H.S.; et al. Border detection in dermoscopy images using statistical region merging. Skin Res. Technol. 2008, 14, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Scharcanski, J.; Fieguth, P. Automatic Skin Lesion Segmentation via Iterative Stochastic Region Merging. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Mat Said, K.A.; Jambek, A.; Sulaiman, N. A study of image processing using morphological opening and closing processes. Int. J. Control Theory Appl. 2016, 9, 15–21. [Google Scholar]
- Kasmi, R.; Mokrani, K.; Rader, R.K.; Cole, J.G.; Stoecker, W.V. Biologically inspired skin lesion segmentation using a geodesic active contour technique. Skin Res. Technol. 2015, 22, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Serrano, C.; Acha, B. Pattern analysis of dermoscopic images based on Markov random fields. Pattern Recognit. 2009, 42, 1052–1057. [Google Scholar] [CrossRef]
- Celebi, M.E.; Mendonca, T.; Marques, J.S. (Eds.) Dermoscopy Image Analysis; A Bioinspired Color Representation for Dermoscopy Image Analysis; CRC Press: Boca Raton, FL, USA, 2015; p. 44. [Google Scholar] [CrossRef]
- Celebi, M.E.; Mendonca, T.; Marques, J.S. (Eds.) Dermoscopy Image Analysis; Where’s the Lesion?: Variability in Human and Automated Segmentation of Dermoscopy Images of Melanocytic Skin Lesions; CRC Press: Boca Raton, FL, USA, 2015; p. 30. [Google Scholar] [CrossRef]
- Barata, C.; Ruela, M.; Francisco, M.; Mendonca, T.; Marques, J.S. Two Systems for the Detection of Melanomas in Dermoscopy Images Using Texture and Color Features. IEEE Syst. J. 2014, 8, 965–979. [Google Scholar] [CrossRef]
- Ercal, F.; Chawla, A.; Stoecker, W.; Lee, H.C.; Moss, R. Neural network diagnosis of malignant melanoma from color images. IEEE Trans. Biomed. Eng. 1994, 41, 837–845. [Google Scholar] [CrossRef]
- Celebi, M.E.; Kingravi, H.A.; Uddin, B.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H. A methodological approach to the classification of dermoscopy images. Comput. Med. Imaging Graph. 2007, 31, 362–373. [Google Scholar] [CrossRef]
- Kahofer, P.; Hofmann-Wellenhof, R.; Smolle, J. Tissue counter analysis of dermatoscopic images of melanocytic skin tumours: Preliminary findings. Melanoma Res. 2002, 12, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Kadry, S.; Meqdad, M.N.; Crespo, R.G. CNN supported framework for automatic extraction and evaluation of dermoscopy images. J. Supercomput. 2022, 78, 17114–17131. [Google Scholar] [CrossRef]
- Schaefer, G.; Krawczyk, B.; Celebi, M.E.; Iyatomi, H. An ensemble classification approach for melanoma diagnosis. Memetic Comput. 2014, 6, 233–240. [Google Scholar] [CrossRef]
- Marques, J. Streak Detection in Dermoscopic Color Images Using Localized Radial Flux of Principal Intensity Curvature. In Dermoscopy Image Analysis; CRC Press: Boca Raton, FL, USA, 2015; pp. 227–246. [Google Scholar] [CrossRef]
- Rifkin, R.; Klautau, A. In Defense of One-Vs-All Classification. J. Mach. Learn. Res. 2004, 5, 101–141. [Google Scholar]
- Senan, E.M.; Jadhav, M.E. Analysis of dermoscopy images by using ABCD rule for early detection of skin cancer. Glob. Transit. Proc. 2021, 2, 1–7. [Google Scholar] [CrossRef]
- Almattar, W.; Luqman, H.; Khan, F.A. Diabetic retinopathy grading review: Current techniques and future directions. Image Vis. Comput. 2023, 139, 104821. [Google Scholar] [CrossRef]
- Grzybowski, A.; Peeters, F.; Barão, R.C.; Brona, P.; Rommes, S.; Krzywicki, T.; Stalmans, I.; Jacob, J. Evaluating the efficacy of AI systems in diabetic retinopathy detection: A comparative analysis of Mona DR and IDx-DR. Acta Ophthalmol. 2024, 103, 388–395. [Google Scholar] [CrossRef]
- Grzybowski, A.; Brona, P.; Krzywicki, T.; Ruamviboonsuk, P. Diagnostic Accuracy of Automated Diabetic Retinopathy Image Assessment Software: IDx-DR and RetCAD. Ophthalmol. Ther. 2024, 14, 73–84. [Google Scholar] [CrossRef]
- Commision, E. Regulation of the European Parliament and of the Council Laying Down Harmonised Rules on Artificial Intelligence (Artificial Intelligence ACT) and Amending Certain Union Legislative Acts. 2021. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:52021PC0206&from=EN (accessed on 11 May 2025).
- The, U.S. Food and Drug Administration (FDA). Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) Action Plan. 2021. Available online: https://www.fda.gov/media/145022/download (accessed on 11 May 2025).
- Powles, J.; Hodson, H. Google DeepMind and healthcare in an age of algorithms. Health Technol. 2017, 7, 351–367. [Google Scholar] [CrossRef]
- Gerke, S.; Minssen, T.; Cohen, G. Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Elsevier: Amsterdam, The Netherlands, 2020; pp. 295–336. [Google Scholar] [CrossRef]
- Cohen, I.G.; Amarasingham, R.; Shah, A.; Xie, B.; Lo, B. The Legal And Ethical Concerns That Arise From Using Complex Predictive Analytics In Health Care. Health Aff. 2014, 33, 1139–1147. [Google Scholar] [CrossRef]
- Eapen, B. Artificial intelligence in dermatology: A practical introduction to a paradigm shift. Indian Dermatol. Online J. 2020, 11, 881. [Google Scholar] [CrossRef]
- Venkatesh, K.P.; Kadakia, K.T.; Gilbert, S. Learnings from the first AI-enabled skin cancer device for primary care authorized by FDA. Npj Digit. Med. 2024, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Purohit, J.; Shivhare, I.; Jogani, V.; Attari, S.; Surtkar, S. Adversarial Attacks and Defences for Skin Cancer Classification. In Proceedings of the 2023 International Conference for Advancement in Technology (ICONAT), Goa, India, 24–26 January 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Huq, A.; Pervin, M.T. Analysis of Adversarial Attacks on Skin Cancer Recognition. In Proceedings of the 2020 International Conference on Data Science and Its Applications (ICoDSA), Bandung, Indonesia, 5–6 August 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Ma, X.; Niu, Y.; Gu, L.; Wang, Y.; Zhao, Y.; Bailey, J.; Lu, F. Understanding adversarial attacks on deep learning based medical image analysis systems. Pattern Recognit. 2021, 110, 107332. [Google Scholar] [CrossRef]
- Chanda, T.; Hauser, K.; Hobelsberger, S.; Bucher, T.C.; Garcia, C.; Wies, C.; Kittler, H.; Tschandl, P.; Navarrete-Dechent, C.; Podlipnik, S.; et al. Dermatologist-like explainable AI enhances trust and confidence in diagnosing melanoma. arXiv 2023, arXiv:2303.12806. [Google Scholar] [CrossRef]
- Hauser, K.; Kurz, A.; Haggenmüller, S.; Maron, R.C.; von Kalle, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; Sergon, M.; et al. Explainable artificial intelligence in skin cancer recognition: A systematic review. Eur. J. Cancer 2022, 167, 54–69. [Google Scholar] [CrossRef]
- Lucieri, A.; Bajwa, M.N.; Braun, S.A.; Malik, M.I.; Dengel, A.; Ahmed, S. ExAID: A multimodal explanation framework for computer-aided diagnosis of skin lesions. Comput. Methods Programs Biomed. 2022, 215, 106620. [Google Scholar] [CrossRef]
- Madry, A.; Makelov, A.; Schmidt, L.; Tsipras, D.; Vladu, A. Towards Deep Learning Models Resistant to Adversarial Attacks. In Proceedings of the 6th International Conference on Learning Representations, ICLR 2018, Vancouver, BC, Canada, 30 April–3 May 2018. Conference Track Proceedings; OpenReview.net, 2018. [Google Scholar]
- Carlini, N.; Wagner, D. Towards Evaluating the Robustness of Neural Network. arXiv 2017, arXiv:1608.04644. [Google Scholar] [CrossRef]
- Google Inc.; OpenAI and Pennsylvania State University. CleverHans. 2016. Available online: https://github.com/cleverhans-lab/cleverhans (accessed on 14 August 2024).
- LF AI & Data. Adversarial Robustness Toolbox (ART). 2018. Available online: https://github.com/Trusted-AI/adversarial-robustness-toolbox (accessed on 14 August 2024).
- Bethge Lab. Foolbox. 2017. Available online: https://github.com/bethgelab/foolbox (accessed on 14 August 2024).
- Baidu X-Lab. AdvBox. 2018. Available online: https://github.com/advboxes/AdvBox (accessed on 14 August 2024).
- Zhao, M.; Zhang, L.; Ye, J.; Lu, H.; Yin, B.; Wang, X. Adversarial Training: A Survey. arXiv 2024, arXiv:2410.15042. [Google Scholar] [CrossRef]
- Li, S.; Ma, X.; Jiang, S.; Meng, L.; Zhang, S. Adaptive Perturbation-Driven Adversarial Training. In Proceedings of the 2024 5th International Conference on Computer Vision, Image and Deep Learning (CVIDL), Zhuhai, China, 19–21 April 2024; pp. 993–997. [Google Scholar] [CrossRef]
- Tu, W.; Liu, X.; Hu, W.; Pan, Z. Dense-Residual Network with Adversarial Learning for Skin Lesion Segmentation. IEEE Access 2019, 7, 77037–77051. [Google Scholar] [CrossRef]
- Li, X.; Cui, Z.; Wu, Y.; Gu, L.; Harada, T. Estimating and Improving Fairness with Adversarial Learning. arXiv 2021, arXiv:2103.04243. [Google Scholar] [CrossRef]
- Zunair, H.; Ben Hamza, A. Melanoma detection using adversarial training and deep transfer learning. Phys. Med. Biol. 2020, 65, 135005. [Google Scholar] [CrossRef] [PubMed]
- Kanca, E.; Ayas, S.; Kablan, E.B.; Ekinci, M. Implementation of Fast Gradient Sign Adversarial Attack on Vision Transformer Model and Development of Defense Mechanism in Classification of Dermoscopy Images. In Proceedings of the 2023 31st Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 5–8 July 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Finlayson, S.G.; Kohane, I.S.; Beam, A.L. Adversarial Attacks Against Medical Deep Learning Systems. arXiv 2018, arXiv:1804.05296. [Google Scholar] [CrossRef]
- Ali, S.; Abuhmed, T.; El-Sappagh, S.; Muhammad, K.; Alonso-Moral, J.M.; Confalonieri, R.; Guidotti, R.; Del Ser, J.; Díaz-Rodríguez, N.; Herrera, F. Explainable Artificial Intelligence (XAI): What we know and what is left to attain Trustworthy Artificial Intelligence. Inf. Fusion 2023, 99, 101805. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Ser, J.D.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2019, 58, 82–115. [Google Scholar] [CrossRef]
- Markus, A.F.; Kors, J.A.; Rijnbeek, P.R. The role of explainability in creating trustworthy artificial intelligence for health care: A comprehensive survey of the terminology, design choices, and evaluation strategies. J. Biomed. Inform. 2021, 113, 103655. [Google Scholar] [CrossRef]
- Nigar, N.; Umar, M.; Shahzad, M.K.; Islam, S.; Abalo, D. A Deep Learning Approach Based on Explainable Artificial Intelligence for Skin Lesion Classification. IEEE Access 2022, 10, 113715–113725. [Google Scholar] [CrossRef]
- Giavina-Bianchi, M.; Vitor, W.G.; Fornasiero de Paiva, V.; Okita, A.L.; Sousa, R.M.; Machado, B. Explainability agreement between dermatologists and five visual explanations techniques in deep neural networks for melanoma AI classification. Front. Med. 2023, 10, 1241484. [Google Scholar] [CrossRef] [PubMed]
- Jalaboi, R.; Faye, F.; Orbes-Arteaga, M.; Jørgensen, D.; Winther, O.; Galimzianova, A. DermX: An end-to-end framework for explainable automated dermatological diagnosis. Med. Image Anal. 2023, 83, 102647. [Google Scholar] [CrossRef]
- Jalaboi, R.; Winther, O.; Galimzianova, A. Dermatological Diagnosis Explainability Benchmark for Convolutional Neural Networks. arXiv 2023, arXiv:2302.12084. [Google Scholar] [CrossRef]
- Adebayo, J.; Gilmer, J.; Muelly, M.; Goodfellow, I.; Hardt, M.; Kim, B. Sanity Checks for Saliency Maps. arXiv 2020, arXiv:1810.03292. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar] [CrossRef]
- Chanda, T.; Haggenmueller, S.; Bucher, T.C.; Holland-Letz, T.; Kittler, H.; Tschandl, P.; Heppt, M.V.; Berking, C.; Utikal, J.S.; Schilling, B.; et al. Dermatologist-like explainable AI enhances melanoma diagnosis accuracy: Eye-tracking study. Nat. Commun. 2025, 16, 4739. [Google Scholar] [CrossRef]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing Machine Learning in Health Care—Addressing Ethical Challenges. N. Engl. J. Med. 2018, 378, 981–983. [Google Scholar] [CrossRef]
- Mittelstadt, B. Principles alone cannot guarantee ethical AI. Nat. Mach. Intell. 2019, 1, 501–507. [Google Scholar] [CrossRef]
- Gordon, E.R.; Trager, M.H.; Kontos, D.; Weng, C.; Geskin, L.J.; Dugdale, L.S.; Samie, F.H. Ethical considerations for artificial intelligence in dermatology: A scoping review. Br. J. Dermatol. 2024, 190, 789–797. [Google Scholar] [CrossRef]
- Goktas, P.; Grzybowski, A. Assessing the Impact of ChatGPT in Dermatology: A Comprehensive Rapid Review. J. Clin. Med. 2024, 13, 5909. [Google Scholar] [CrossRef]
- Adamson, A.S.; Smith, A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatol. 2018, 154, 1247. [Google Scholar] [CrossRef]
- Daneshjou, R.; Smith, M.P.; Sun, M.D.; Rotemberg, V.; Zou, J. Lack of Transparency and Potential Bias in Artificial Intelligence Data Sets and Algorithms: A Scoping Review. JAMA Dermatol. 2021, 157, 1362. [Google Scholar] [CrossRef]
- Froomkin, A.M.; Kerr, I.; Pineau, J. When AIs outperform doctors: Confronting the challenges of a tort-induced over-reliance on machine learning. Ariz. L. Rev. 2019, 61, 33. [Google Scholar]
- Krakowski, I.; Kim, J.; Cai, Z.R.; Daneshjou, R.; Lapins, J.; Eriksson, H.; Lykou, A.; Linos, E. Human-AI interaction in skin cancer diagnosis: A systematic review and meta-analysis. Npj Digit. Med. 2024, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- von Itzstein, M.S.; Hullings, M.; Mayo, H.; Beg, M.S.; Williams, E.L.; Gerber, D.E. Application of Information Technology to Clinical Trial Evaluation and Enrollment: A Review. JAMA Oncol. 2021, 7, 1559. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Su, G.H.; Ma, D.; Xiao, Y.; Shao, Z.M.; Jiang, Y.Z. Technological advances in cancer immunity: From immunogenomics to single-cell analysis and artificial intelligence. Signal Transduct. Target. Ther. 2021, 6, 312. [Google Scholar] [CrossRef] [PubMed]
- Barisoni, L.; Lafata, K.J.; Hewitt, S.M.; Madabhushi, A.; Balis, U.G.J. Digital pathology and computational image analysis in nephropathology. Nat. Rev. Nephrol. 2020, 16, 669–685. [Google Scholar] [CrossRef]
- Ho, D.; Quake, S.R.; McCabe, E.R.; Chng, W.J.; Chow, E.K.; Ding, X.; Gelb, B.D.; Ginsburg, G.S.; Hassenstab, J.; Ho, C.M.; et al. Enabling Technologies for Personalized and Precision Medicine. Trends Biotechnol. 2020, 38, 497–518. [Google Scholar] [CrossRef]
- Li, Z.; Koban, K.C.; Schenck, T.L.; Giunta, R.E.; Li, Q.; Sun, Y. Artificial Intelligence in Dermatology Image Analysis: Current Developments and Future Trends. J. Clin. Med. 2022, 11, 6826. [Google Scholar] [CrossRef]
- Khozeimeh, F.; Alizadehsani, R.; Roshanzamir, M.; Khosravi, A.; Layegh, P.; Nahavandi, S. An expert system for selecting wart treatment method. Comput. Biol. Med. 2017, 81, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Han, S.S.; Park, I.; Eun Chang, S.; Lim, W.; Kim, M.S.; Park, G.H.; Chae, J.B.; Huh, C.H.; Na, J.I. Augmented Intelligence Dermatology: Deep Neural Networks Empower Medical Professionals in Diagnosing Skin Cancer and Predicting Treatment Options for 134 Skin Disorders. J. Investig. Dermatol. 2020, 140, 1753–1761. [Google Scholar] [CrossRef]
- Avram, M.R.; Watkins, S.A. Robotic Follicular Unit Extraction in Hair Transplantation. Dermatol. Surg. 2014, 40, 1319–1327. [Google Scholar] [CrossRef]
- Cianci, S.; Arcieri, M.; Vizzielli, G.; Martinelli, C.; Granese, R.; La Verde, M.; Fagotti, A.; Fanfani, F.; Scambia, G.; Ercoli, A. Robotic Pelvic Exenteration for Gynecologic Malignancies, Anatomic Landmarks, and Surgical Steps: A Systematic Review. Front. Surg. 2021, 8, 79015. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Codella, N.C.; Dusza, S.W.; Gutman, D.A.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, M.E.; DeFazio, J.L.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018, 78, 270–277.e1. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Liopyris, K.; Dusza, S.W.; Codella, N.C.; Gutman, D.A.; Helba, B.; Kalloo, A.; Halpern, A.C.; Soyer, H.P.; Curiel-Lewandrowski, C.; et al. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol. 2020, 82, 622–627. [Google Scholar] [CrossRef]
- Maron, R.C.; Haggenmüller, S.; von Kalle, C.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hauschild, A.; French, L.E.; Schlaak, M.; Ghoreschi, K.; et al. Robustness of convolutional neural networks in recognition of pigmented skin lesions. Eur. J. Cancer 2021, 145, 81–91. [Google Scholar] [CrossRef]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; et al. Melanoma recognition by a deep learning convolutional neural network—Performance in different melanoma subtypes and localisations. Eur. J. Cancer 2020, 127, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Yang, S.; Kim, W.; Jung, J.; Chung, K.Y.; Lee, S.W.; Oh, B. Acral melanoma detection using a convolutional neural network for dermoscopy images. PLoS ONE 2018, 13, e0193321. [Google Scholar] [CrossRef]
- Lee, S.; Chu, Y.; Yoo, S.; Choi, S.; Choe, S.; Koh, S.; Chung, K.; Xing, L.; Oh, B.; Yang, S. Augmented decision-making for acral lentiginous melanoma detection using deep convolutional neural networks. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1842–1850. [Google Scholar] [CrossRef]
- Pham, T.C.; Luong, C.M.; Hoang, V.D.; Doucet, A. AI outperformed every dermatologist in dermoscopic melanoma diagnosis, using an optimized deep-CNN architecture with custom mini-batch logic and loss function. Sci. Rep. 2021, 11, 17485. [Google Scholar] [CrossRef] [PubMed]
- Shetty, B.; Fernandes, R.; Rodrigues, A.P.; Chengoden, R.; Bhattacharya, S.; Lakshmanna, K. Skin lesion classification of dermoscopic images using machine learning and convolutional neural network. Sci. Rep. 2022, 12, 18134. [Google Scholar] [CrossRef]
- Olayah, F.; Senan, E.M.; Ahmed, I.A.; Awaji, B. AI Techniques of Dermoscopy Image Analysis for the Early Detection of Skin Lesions Based on Combined CNN Features. Diagnostics 2023, 13, 1314. [Google Scholar] [CrossRef]
- De Guzman, L.C.; Maglaque, R.P.C.; Torres, V.M.B.; Zapido, S.P.A.; Cordel, M.O. Design and Evaluation of a Multi-model, Multi-level Artificial Neural Network for Eczema Skin Lesion Detection. In Proceedings of the 2015 3rd International Conference on Artificial Intelligence, Modelling and Simulation (AIMS), Kota Kinabalu, Malaysia, 2–4 December 2015; pp. 42–47. [Google Scholar] [CrossRef]
- Ngoo, A.; Finnane, A.; McMeniman, E.; Soyer, H.P.; Janda, M. Fighting Melanoma with Smartphones: A Snapshot of Where We are a Decade after App Stores Opened Their Doors. Int. J. Med. Inform. 2018, 118, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar] [CrossRef]
- Ech-Cherif, A.; Misbhauddin, M.; Ech-Cherif, M. Deep Neural Network Based Mobile Dermoscopy Application for Triaging Skin Cancer Detection. In Proceedings of the 2019 2nd International Conference on Computer Applications & Information Security (ICCAIS), Riyadh, Saudi Arabia, 1–3 May 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional Neural Networks Using MobileNet for Skin Lesion Classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10–12 July 2019; pp. 242–247. [Google Scholar] [CrossRef]
- Velasco, J. A Smartphone-Based Skin Disease Classification Using MobileNet CNN. Int. J. Adv. Trends Comput. Sci. Eng. 2019, 8, 2632–2637. [Google Scholar] [CrossRef]
- SkinVision. Skin Cancer Melanoma Detection App. Available online: https://www.skinvision.com (accessed on 8 November 2024).
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. arXiv 2016, arXiv:1611.07004. [Google Scholar] [CrossRef]
- Maier, T.; Kulichova, D.; Schotten, K.; Astrid, R.; Ruzicka, T.; Berking, C.; Udrea, A. Accuracy of a smartphone application using fractal image analysis of pigmented moles compared to clinical diagnosis and histological result. J. Eur. Acad. Dermatol. Venereol. 2014, 29, 663–667. [Google Scholar] [CrossRef]
- Thissen, M.; Udrea, A.; Hacking, M.; von Braunmuehl, T.; Ruzicka, T. mHealth App for Risk Assessment of Pigmented and Nonpigmented Skin Lesions—A Study on Sensitivity and Specificity in Detecting Malignancy. Telemed. E-Health 2017, 23, 948–954. [Google Scholar] [CrossRef]
- Sun, M.; Kentley, J.; Mehta, P.; Dusza, S.; Halpern, A.; Rotemberg, V. Accuracy of commercially available smartphone applications for the detection of melanoma. Br. J. Dermatol. 2022, 186, 744–746. [Google Scholar] [CrossRef]
- Koka, S.S.A.; Burkhart, C.G. Artificial Intelligence in Dermatology: Current Uses, Shortfalls, and Potential Opportunities for Further Implementation in Diagnostics and Care. Open Dermatol. J. 2023, 17, e187437222304140. [Google Scholar] [CrossRef]
- Kania, B.; Montecinos, K.; Goldberg, D.J. Artificial intelligence in cosmetic dermatology. J. Cosmet. Dermatol. 2024, 23, 3305–3311. [Google Scholar] [CrossRef]
- VisualDx. Aysa. 2025. Available online: https://www.visualdx.com/blog/visualdx-launches-aysa-for-consumers-to-check-skin-conditions-using-ai/ (accessed on 17 May 2025).
- Marri, S.S.; Albadri, W.; Hyder, M.S.; Janagond, A.B.; Inamadar, A.C. Efficacy of an Artificial Intelligence App (Aysa) in Dermatological Diagnosis: Cross-Sectional Analysis. JMIR Dermatol. 2024, 7, e48811. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Yu, Z.; Primiero, C.; Vico-Alonso, C.; Wang, Z.; Yang, L.; Tschandl, P.; Hu, M.; Ju, L.; Tan, G.; et al. A Multimodal Vision Foundation Model for Clinical Dermatology. arXiv 2024, arXiv:2410.15038. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Primiero, C.; Betz-Stablein, B.; Nguyen, T.D.; Gal, Y.; Bowling, A.; Haskett, M.; Sashindranath, M.; Bonnington, P.; Mar, V.; et al. Multi-task AI models in dermatology: Overcoming critical clinical translation challenges for enhanced skin lesion diagnosis. J. Eur. Acad. Dermatol. Venereol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Mevorach, L.; Farcomeni, A.; Pellacani, G.; Cantisani, C. A Comparison of Skin Lesions’ Diagnoses Between AI-Based Image Classification, an Expert Dermatologist, and a Non-Expert. Diagnostics 2025, 15, 1115. [Google Scholar] [CrossRef]
- Riaz, S.; Naeem, A.; Malik, H.; Naqvi, R.A.; Loh, W.K. Federated and Transfer Learning Methods for the Classification of Melanoma and Nonmelanoma Skin Cancers: A Prospective Study. Sensors 2023, 23, 8457. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Schmitt, M.; Krieghoff-Henning, E.; Hekler, A.; Maron, R.C.; Wies, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; et al. Federated Learning for Decentralized Artificial Intelligence in Melanoma Diagnostics. JAMA Dermatol. 2024, 160, 303–311. [Google Scholar] [CrossRef]
- Cheslerean-Boghiu, T.; Fleischmann, M.E.; Willem, T.; Lasser, T. Transformer-based interpretable multi-modal data fusion for skin lesion classification. arXiv 2023, arXiv:2304.14505. [Google Scholar] [CrossRef]
- Ou, C.; Zhou, S.; Yang, R.; Jiang, W.; He, H.; Gan, W.; Chen, W.; Qin, X.; Luo, W.; Pi, X.; et al. A deep learning based multimodal fusion model for skin lesion diagnosis using smartphone collected clinical images and metadata. Front. Surg. 2022, 9, 1029991. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.H.; Shah, S.T.H.; Khaled, R.; Buccoliero, A.; Shah, S.B.H.; Di Terlizzi, A.; Di Benedetto, G.; Deriu, M.A. Explainable AI-Based Skin Cancer Detection Using CNN, Particle Swarm Optimization and Machine Learning. J. Imaging 2024, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, F.; Ali, H.; El Hajj, N.; Shah, Z. Artificial intelligence-based methods for fusion of electronic health records and imaging data. Sci. Rep. 2022, 12, 17981. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, L.; Maron, R.C.; Hekler, A.; Haggenmüller, S.; Wies, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; Sergon, M.; et al. Prospective multicenter study using artificial intelligence to improve dermoscopic melanoma diagnosis in patient care. Commun. Med. 2024, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.A.; Cowen, E.A.; Kurtansky, N.R.; Weber, J.; Dauscher, M.; DeFazio, J.; Deng, L.; Dusza, S.W.; Haliasos, H.; Halpern, A.C.; et al. Prospective validation of dermoscopy-based open-source artificial intelligence for melanoma diagnosis (PROVE-AI study). Npj Digit. Med. 2023, 6, 127. [Google Scholar] [CrossRef]
- Abbas, Q.; Daadaa, Y.; Rashid, U.; Ibrahim, M. Assist-Dermo: A Lightweight Separable Vision Transformer Model for Multiclass Skin Lesion Classification. Diagnostics 2023, 13, 2531. [Google Scholar] [CrossRef]
- Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 2023, 15, 2179. [Google Scholar] [CrossRef]
- Ayas, S. Multiclass skin lesion classification in dermoscopic images using swin transformer model. Neural Comput. Appl. 2022, 35, 6713–6722. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin Lesions Classification Into Eight Classes for ISIC 2019 Using Deep Convolutional Neural Network and Transfer Learning. IEEE Access 2020, 8, 114822–114832. [Google Scholar] [CrossRef]
- Liu, X.; Yu, Z.; Tan, L.; Yan, Y.; Shi, G. Enhancing Skin Lesion Diagnosis with Ensemble Learning. arXiv 2024, arXiv:2409.04381. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. arXiv 2014, arXiv:1409.4842. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2018, arXiv:1801.04381. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Ray, A.; Sarkar, S.; Schwenker, F.; Sarkar, R. Decoding skin cancer classification: Perspectives, insights, and advances through researchers’ lens. Sci. Rep. 2024, 14, 30542. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, C.; Combalia, M.; Podlipnik, S.; Codella, N.C.F.; Rotemberg, V.; Halpern, A.C.; Reiter, O.; Carrera, C.; Barreiro, A.; Helba, B.; et al. BCN20000: Dermoscopic Lesions in the Wild. Sci. Data 2024, 11, 641. [Google Scholar] [CrossRef]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Med. Image Anal. 2022, 75, 102305. [Google Scholar] [CrossRef]
- Alipour, N.; Burke, T.; Courtney, J. Skin Type Diversity in Skin Lesion Datasets: A Review. Curr. Dermatol. Rep. 2024, 13, 198–210. [Google Scholar] [CrossRef]
- Gomolin, A.; Netchiporouk, E.; Gniadecki, R.; Litvinov, I.V. Artificial Intelligence Applications in Dermatology: Where Do We Stand? Front. Med. 2020, 7, 100. [Google Scholar] [CrossRef]
- Patel, S.; Wang, J.V.; Motaparthi, K.; Lee, J.B. Artificial intelligence in dermatology for the clinician. Clin. Dermatol. 2021, 39, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.K.; Park, C.; Henao, R.; Kheterpal, M. Deep Learning in Dermatology: A Systematic Review of Current Approaches, Outcomes, and Limitations. JID Innov. 2023, 3, 100150. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Achar, U.; Hakim, B.; Achar, A. Artificial Intelligence in Dermatology: A Systematized Review. Int. J. Dermatol. Venereol. 2024, 8, 33–39. [Google Scholar] [CrossRef]
- Martínez-Vargas, E.; Mora-Jiménez, J.; Arguedas-Chacón, S.; Hernández-López, J.; Zavaleta-Monestel, E. The Emerging Role of Artificial Intelligence in Dermatology: A Systematic Review of Its Clinical Applications. Dermato 2025, 5, 9. [Google Scholar] [CrossRef]
- Nahm, W.J.; Sohail, N.; Burshtein, J.; Goldust, M.; Tsoukas, M. Artificial Intelligence in Dermatology: A Comprehensive Review of Approved Applications, Clinical Implementation, and Future Directions. Int. J. Dermatol. 2025. Online ahead of print. [Google Scholar] [CrossRef]

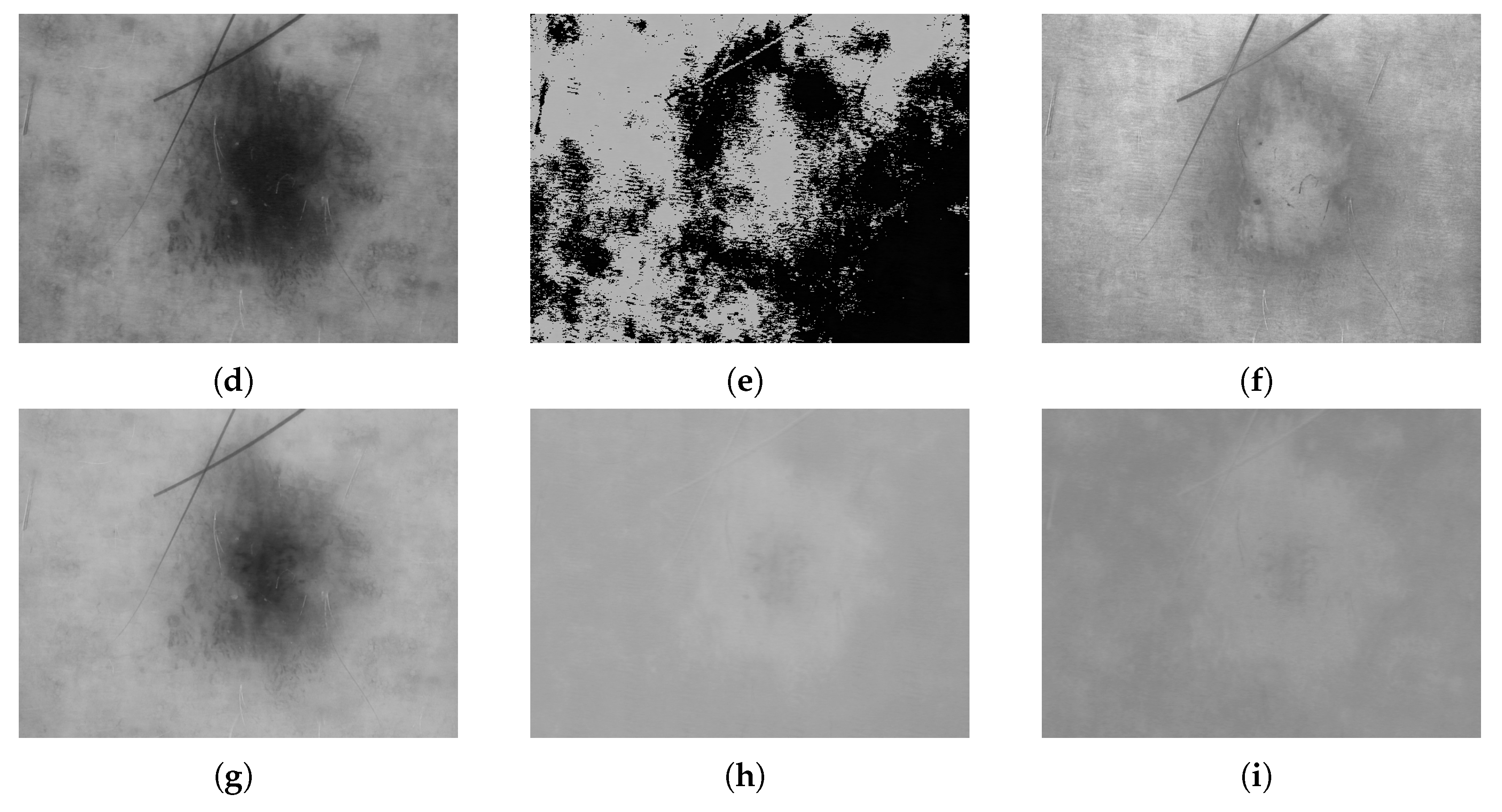

| Type of ML | Examples of Solutions |
|---|---|
| Supervised | |
| Unsupervised | |
| Reinforcement |
| Method | Input | Key Innovation | Application in Dermatology |
|---|---|---|---|
| StyleGAN2 | Random hidden vector + mapping network | AdaIN for style control at different network levels | Generation of synthetic images of melanocytic nevi to augment the dermatoscopic set [60] |
| CycleGAN | Dermatoscopic images ↔ clinical images | Loss of cyclic consistency | Translation of clinical images into dermatoscopic style (and vice versa) to unify domains and augment training data [61] |
| Tags/Year | Artificial Intelligence AND Dermatology | Machine Learning AND Dermatology | Deep Learning AND Dermatology | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PubMed | Scopus | WoS | PubMed | Scopus | WoS | PubMed | Scopus | WoS | |
| 2009 | 8 | 6 | 2 | 0 | 5 | 3 | 0 | 0 | 0 |
| 2010 | 6 | 8 | 2 | 3 | 6 | 0 | 0 | 0 | 0 |
| 2011 | 7 | 10 | 2 | 1 | 11 | 2 | 1 | 0 | 0 |
| 2012 | 6 | 10 | 1 | 1 | 8 | 3 | 0 | 0 | 0 |
| 2013 | 8 | 10 | 0 | 1 | 3 | 1 | 0 | 0 | 0 |
| 2014 | 26 | 8 | 0 | 9 | 8 | 1 | 0 | 0 | 0 |
| 2015 | 13 | 21 | 2 | 12 | 13 | 10 | 0 | 1 | 0 |
| 2016 | 26 | 30 | 0 | 11 | 18 | 3 | 0 | 5 | 0 |
| 2017 | 41 | 33 | 2 | 23 | 32 | 9 | 4 | 23 | 3 |
| 2018 | 77 | 41 | 14 | 53 | 49 | 12 | 26 | 57 | 9 |
| 2019 | 135 | 50 | 20 | 95 | 95 | 26 | 44 | 150 | 19 |
| 2020 | 235 | 71 | 53 | 152 | 137 | 27 | 88 | 207 | 35 |
| 2021 | 330 | 99 | 70 | 196 | 149 | 42 | 125 | 301 | 41 |
| 2022 | 310 | 102 | 73 | 191 | 199 | 42 | 118 | 385 | 38 |
| 2023 | 408 | 195 | 115 | 247 | 329 | 63 | 157 | 654 | 69 |
| 2024 | 644 | 309 | 224 | 320 | 288 | 85 | 144 | 526 | 87 |
| Tag/Year | Artificial Intelligence | Machine Learning | Deep Learning | Dermatology | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PubMed | Scopus | WoS | PubMed | Scopus | WoS | PubMed | Scopus | WoS | PubMed | Scopus | WoS | |
| 2009 | 4640 | 11,150 | 1420 | 604 | 4622 | 2742 | 106 | 127 | 44 | 7040 | 1097 | 1159 |
| 2010 | 4473 | 12,518 | 1399 | 718 | 4669 | 2729 | 107 | 162 | 74 | 7358 | 1145 | 1099 |
| 2011 | 5280 | 14,737 | 1397 | 1148 | 4685 | 3075 | 118 | 186 | 100 | 8281 | 1293 | 1336 |
| 2012 | 5549 | 15,503 | 1440 | 1498 | 4941 | 3453 | 146 | 239 | 136 | 8536 | 1309 | 1462 |
| 2013 | 6811 | 14,740 | 1575 | 1931 | 5723 | 4388 | 207 | 331 | 227 | 10,504 | 1586 | 1635 |
| 2014 | 6894 | 16,064 | 1863 | 2426 | 7336 | 5943 | 265 | 679 | 481 | 14,094 | 1649 | 1556 |
| 2015 | 6773 | 18,332 | 2148 | 3292 | 9251 | 8044 | 384 | 1250 | 1200 | 16,145 | 1849 | 1701 |
| 2016 | 6804 | 21,627 | 2615 | 3921 | 12,048 | 10,508 | 643 | 2872 | 2588 | 17,987 | 1904 | 1727 |
| 2017 | 8240 | 21,430 | 3602 | 5284 | 17,052 | 14,691 | 1353 | 8134 | 6370 | 19,731 | 2284 | 1929 |
| 2018 | 11,245 | 23,154 | 6393 | 8331 | 25,896 | 23,017 | 3085 | 17,088 | 13,618 | 20,790 | 2495 | 2070 |
| 2019 | 16,422 | 23,082 | 11,370 | 12,676 | 45,281 | 34,673 | 5613 | 32,987 | 23,602 | 22,452 | 2840 | 2494 |
| 2020 | 22,692 | 30,020 | 16,731 | 18,240 | 56,636 | 44,872 | 9328 | 46,750 | 31,789 | 27,515 | 3580 | 3029 |
| 2021 | 31,476 | 34,080 | 23,899 | 25,664 | 71,723 | 59,819 | 14,522 | 63,368 | 45,096 | 29613 | 4250 | 3223 |
| 2022 | 39,132 | 36,190 | 30,871 | 31,426 | 88,916 | 71,351 | 19,640 | 82,530 | 57,790 | 28,317 | 4374 | 3332 |
| 2023 | 38,695 | 47,355 | 36,708 | 33,865 | 109,766 | 74,386 | 21,222 | 99,458 | 59,014 | 27,131 | 4891 | 3254 |
| 2024 | 50,970 | 60,469 | 54,470 | 41,594 | 130,263 | 90,995 | 23,112 | 99,742 | 69,072 | 27,912 | 5183 | 3788 |
| Architecture | Source | Dataset | Accuracy [%] | AUC | Sensitivity [%] | Specificity [%] |
|---|---|---|---|---|---|---|
| Separable Vision Transformer | [213] (2023) | PH2 + ISBI-2017 + HAM10000 + ISIC (9 classes) | 95.6 | 0.95 | 96.7 | 95.00 |
| DSCC_Net | [214] (2023) | ISIC–2020 + HAM10000 + DermIS (4 classes) | 94.17 | 0.9943 | 93.76 | — |
| Swin Transformer | [215] (2022) | ISIC-2019 (8 classes) | 97.20 | — | 82.30 | 97.90 |
| GoogleNet-TL | [216] (2020) | ISIC-2019 (8 classes) | 94.92 | — | 79.80 | 97.00 |
| SkinNet | [217] (2024) | HAM10000 (7 classes) | 86.70 | 0.96 | — | — |
| Dataset | Number of Images | Number of Classes | Evaluation Measure | Main Limitations |
|---|---|---|---|---|
| ISIC (2019) | 25,331 train + 8239 test | 8 (+OOD) | Balanced accuracy | Class imbalance; limited phototype diversity; partial lack of histopathology |
| HAM10000 | 10,015 | 7 | Balanced accuracy | Class imbalance; no images outside the pigmented lesion range; ∼50% without histopathological confirmation |
| BCN20000 | 18,946 (train) | 8 (+OOD) | Balanced accuracy | Class imbalance; one site (geographic/skin bias); limited phototype data |
| Method/Architecture | Advantages | Disadvantages | Recommended Application |
|---|---|---|---|
| ResNet-50 (transfer) |
|
|
|
| VGG-16 |
|
|
|
| GoogLeNet (transfer learning) |
|
|
|
| DenseNet-121 |
|
|
|
| EfficientNet-B0 |
|
|
|
| DSCC_Net |
|
|
|
| SkinNet (ensemble learning) |
|
|
|
| Swin Transformer |
|
|
|
| Separable Vision Transformer |
|
|
|
| UNet |
|
|
|
| Attention UNet |
|
|
|
| StyleGAN2 (augmentation) |
|
|
|
| Authors | Year | Scope | Main Contributions | Limitations |
|---|---|---|---|---|
| Gomolin et al. [227] | 2020 | Narrative review of AI applications in dermatology | Overview of AI use in melanoma, ulcers, inflammatory dermatoses; identifies barriers to clinical use (e.g., black-box models) | Non-systematic; early 2020 state-of-the-art; no quantitative synthesis |
| Patel et al. [228] | 2021 | Educational overview for clinicians on AI in dermatology | Summarises diagnostic performance of AI (∼67–99% accuracy); stresses improved access and faster diagnosis | Narrative form; limited in regulation, bias, and external validation |
| Jeong et al. [229] | 2022 | Systematic review of CNNs in dermatology imaging | Comprehensive summary of CNN-based classification; dataset comparison; discusses regulatory paths | Focuses only on CNN image models; no clinical outcomes or ethical analysis |
| Biswas et al. [230] | 2025 | Review of diagnostic performance of AI across skin diseases | Reports high sensitivity/accuracy for melanoma, psoriasis, acne, tinea; stresses health access benefits | Lacks technical/regulatory depth; semi-systematic review only |
| Martinez-Vargas et al. [231] | 2025 | Systematic review of clinical outcomes with AI in dermatology | Shows reduced missed melanoma cases (58.8% to 4.1%), improved triage; identifies heterogeneity and bias risks | Limited external validation; scope excludes technical/ethical discussions |
| Nahm et al. [232] | 2025 | Review of FDA/regulated AI tools in dermatology | Identifies 15 approved AI tools; highlights clinical integration in screening and education | Only covers approved systems; omits experimental and technical details |
| This work | 2025 | Broad review: AI methods, clinical use, ethics, regulation, validation, bibliometrics | Combines technical, legal, and ethical dimensions; bibliometric analysis 2009–2024; regulatory gaps; architecture/dataset comparison; recommendations for standardisation and explainability | Very broad scope; limited depth in specific subdomains; only English-language sources |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zbrzezny, A.M.; Krzywicki, T. Artificial Intelligence in Dermatology: A Review of Methods, Clinical Applications, and Perspectives. Appl. Sci. 2025, 15, 7856. https://doi.org/10.3390/app15147856
Zbrzezny AM, Krzywicki T. Artificial Intelligence in Dermatology: A Review of Methods, Clinical Applications, and Perspectives. Applied Sciences. 2025; 15(14):7856. https://doi.org/10.3390/app15147856
Chicago/Turabian StyleZbrzezny, Agnieszka M., and Tomasz Krzywicki. 2025. "Artificial Intelligence in Dermatology: A Review of Methods, Clinical Applications, and Perspectives" Applied Sciences 15, no. 14: 7856. https://doi.org/10.3390/app15147856
APA StyleZbrzezny, A. M., & Krzywicki, T. (2025). Artificial Intelligence in Dermatology: A Review of Methods, Clinical Applications, and Perspectives. Applied Sciences, 15(14), 7856. https://doi.org/10.3390/app15147856