A U-Shaped Architecture Based on Hybrid CNN and Mamba for Medical Image Segmentation
Abstract
1. Introduction
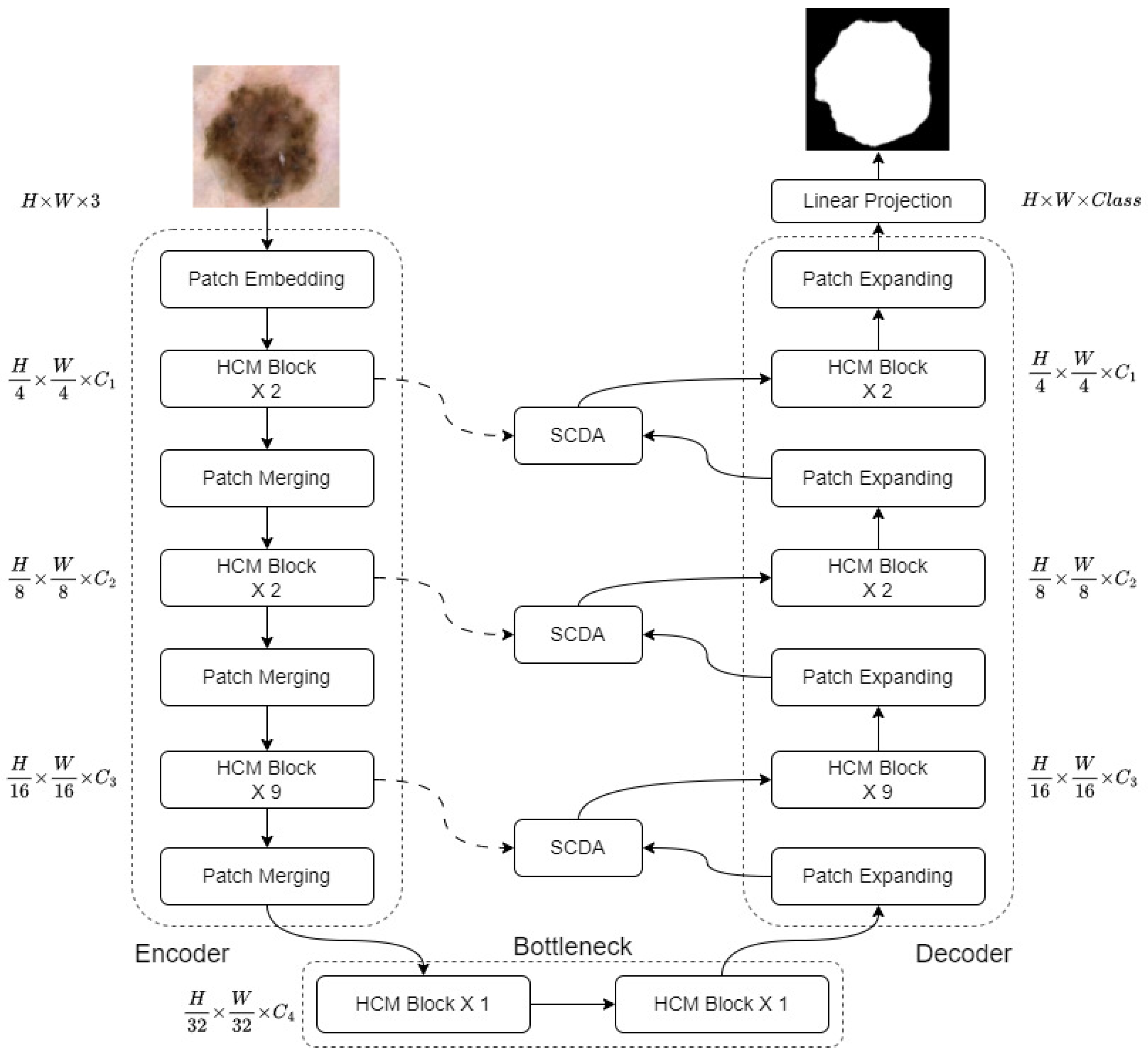
- We propose HCMUNet, a novel U-shaped architecture that integrates a hybrid CNN-Mamba network within a dual-branch encoder-decoder framework. This design facilitates efficient extraction of both local and global features while maintaining low computational complexity.
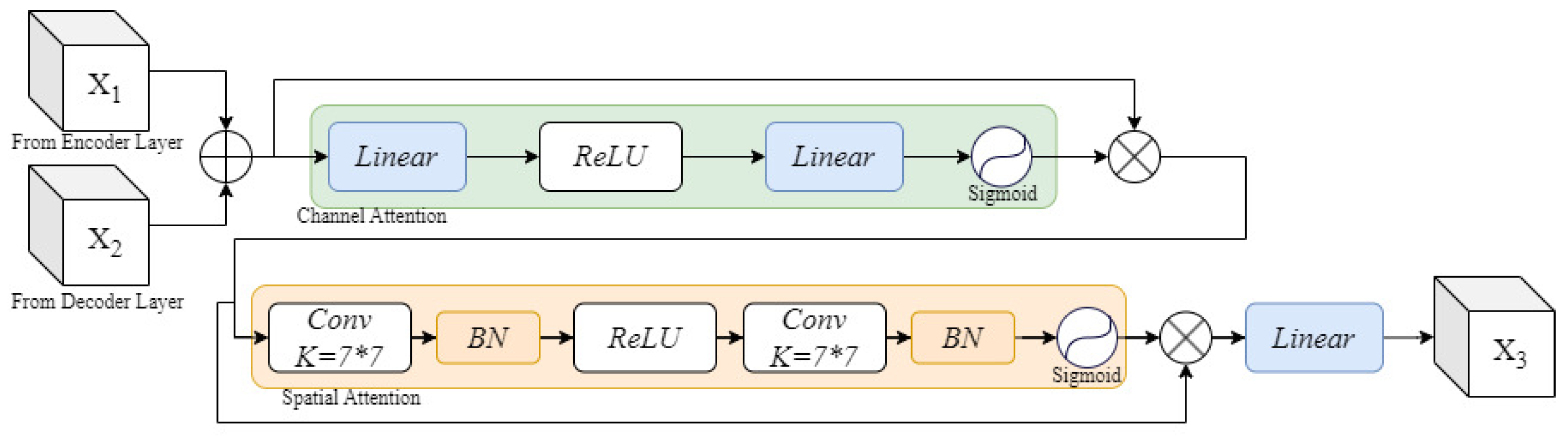
- We introduce Skip Connection Dual Attention (SCDA), which enhances conventional skip connections by incorporating both channel and spatial attention. This mechanism strengthens cross-dimensional feature fusion and improves the recovery of spatial information lost during down-sampling.
- We validate HCMUNet on three benchmark datasets: ISIC 2018, Synapse, and ACDC. The experimental results indicate that HCMUNet achieves high segmentation performance and exhibits strong generalization capability across diverse medical image segmentation tasks.
2. Related Work
2.1. U-Shaped Model for Medical Image Segmentation
2.2. Attention Mechanism
3. Method
3.1. Framework Overview
| Algorithm 1 Pseudo code of HCMUNet. |
| Require: Image , Ground truth , T |
| Ensure: , |
|
3.2. Hybrid Convolutional Mamba Block
3.3. Skip Connection Dual Attention
3.4. Loss Function
4. Experimental Results and Analysis
4.1. Dataset
4.2. Implementation Details
4.3. Evaluation Metrics
4.4. Experimental Results
4.5. Ablation Study
4.6. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the 9th International Conference on Learning Representations, Vienna, Austria, 3–7 May 2021. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assiste Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Rao, Y.; Zhao, W.; Tang, Y.; Zhou, J.; Lim, S.; Lu, J. Hornet:Efficient high-order spatial interactions with recursive gated convolutions. In Proceedings of the 36th International Conference on Neural Information Processing Systems, New Orleans, LA, USA, 28 November–9 December 2022; pp. 10353–10366. [Google Scholar]
- Wu, R.; Liang, P.; Huang, X.; Shi, L.; Gu, Y.; Zhu, H.; Chang, Q. Mhorunet: High-order spatial interaction unet for skin lesion segmentation. Biomed. Signal Process. Control 2024, 88, 105517. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Li, X.; Fan, D.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 548–558. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF international Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Gu, A.; Goel, K.; Ré, C. Efficiently Modeling Long Sequences with Structured State Spaces. In Proceedings of the 10th International Conference on Learning Representations, Virtually, 25–29 April 2022. [Google Scholar]
- Gu, A.; Dao, T. Mamba: Linear-time sequence modeling with selective state spaces. In Proceedings of the 12th International Conference on Learning Representations, Vienna, Austria, 7–11 May 2024. [Google Scholar]
- Zhu, L.; Liao, B.; Zhang, Q.; Wang, X.; Liu, W.; Wang, X. Vision mamba: Efficient visual representation learning with bidirectional state space model. In Proceedings of the 41st International Conference on Machine Learning, Vienna, Austria, 21–27 July 2024; pp. 62429–62442. [Google Scholar]
- Liu, Y.; Tian, Y.; Zhao, Y.; Yu, H.; Xie, L.; Wang, Y.; Ye, Q.; Jiao, J.; Liu, Y. Vmamba: Visual state space model. In Proceedings of the Conference on Neural Information Processing Systems, Vancouver, ON, Canada, 10–15 December 2024; Volume 37, pp. 103031–103063. [Google Scholar]
- Ma, J.; Li, F.; Wang, B. U-mamba: Enhancing long-range dependency for biomedical image segmentation. arXiv 2024, arXiv:2401.04722. [Google Scholar]
- Ruan, J.; Li, J.; Xiang, S. Vm-unet: Vision mamba unet for medical image segmentation. arXiv 2024, arXiv:2402.02491. [Google Scholar]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2018, arXiv:1902.03368. [Google Scholar]
- Harrigr. Segmentation Outside the Cranial Vault Challenge. Synapse 2015. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 3431–3440. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. ResUNet++: An Advanced Architecture for Medical Image Segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. In Proceedings of the Medical Imaging with Deep Learning, Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Peng, Y.; Sonka, M.; Chen, D.Z.; Sonka, M. U-Net v2: Rethinking the skip connections of U-Net for medical image segmentation. In Proceedings of the IEEE 22nd International Symposium on Biomedical Imaging, Houston, TX, USA, 14–17 April 2025; pp. 1–5. [Google Scholar]
- Xing, Z.; Ye, T.; Yang, Y.; Liu, G.; Zhu, L. Segmamba: Long-range sequential modeling mamba for 3d medical image segmentation. In Proceedings of the 27th International Conference On Medical lmage Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 578–588. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 18–28. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In Proceedings of the International MICCAI Brainlesion Workshop, Virtual Event, 27 September 2021; pp. 272–284. [Google Scholar]
- Liu, J.; Yang, H.; Zhou, H.; Xi, Y.; Yu, L.; Li, C.; Liang, Y.; Shi, G.; Yu, Y.; Zhang, S.; et al. Swin-umamba: Mamba-based unet with imagenet-based pretraining. In Proceedings of the 27th International Conference on Medical lmage Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 615–625. [Google Scholar]
- Fan, C.; Yu, H.; Huang, Y.; Wang, L.; Yang, Z.; Jia, X. SliceMamba with Neural Architecture Search for Medical Image Segmentation. IEEE J. Biomed. Health Inform. 2025, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the 17th European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Wu, R.; Liu, Y.; Liang, P.; Chang, Q. H-vmunet: High-order vision mamba unet for medical image segmentation. Neurocomputing 2025, 624, 129447. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.; Kweon, I. Cbam: Convolutional block attention module. In Proceedings of the 15th European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Park, J.; Woo, S.; Lee, J.; Kweon, I. Bam: Bottleneck attention module. In Proceedings of the British Machine Vision Conference, Newcastle, UK, 3–6 September 2018; p. 147. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- Xu, W.; Wan, Y.; Zhao, D. SFA: Efficient Attention Mechanism for Superior CNN Performance. Neural Process. Lett. 2025, 57, 38. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified Linear Units Improve Restricted Boltzmann Machines. In Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Elfwing, S.; Uchibe, E.; Doya, K. Sigmoid-Weighted Linear Units for Neural Network Function Approximation in Reinforcement Learning. Neural Netw. 2018, 107, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, X.; Ke, Z.; Zhang, W.; Lau, R. Biformer: Vision transformer with bi-level routing attention. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 10323–10333. [Google Scholar]
- Lan, L.; Cai, P.; Jiang, L.; Liu, X.; Li, Y.; Zhang, Y. BRAU-Net++: U-Shaped Hybrid CNN-Transformer Network for Medical Image Segmentation. arXiv 2024, arXiv:2401.00722. [Google Scholar]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Unified Focal Loss: Generalising Dice and Cross Entropy-Based Losses to Handle Class Imbalanced Medical Image Segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef] [PubMed]
- Murguía, M.; Villaseñor, J.L. Estimating the Effect of the Similarity Coefficient and the Cluster Algorithm on Biogeographic Classifications. Ann. Bot. Fennici 2003, 40, 415–421. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Karimi, D.; Salcudean, S.E. Reducing the Hausdorff Distance in Medical Image Segmentation with Convolutional Neural Networks. IEEE Trans. Med. Imaging 2019, 39, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Yang, Y.B. Sa-net: Shuffle attention for deep convolutional neural networks. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, Toronto, ON, Canada, 6–11 June 2021; pp. 2235–2239. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtually, 19–25 June 2021; pp. 574–584. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the 24th International Conference on Medical Image Computing and Computer-Assisted Intervention, Virtually, 27 September–1 October 2021; pp. 14–24. [Google Scholar]
- Wang, H.; Cao, P.; Wang, J.; Zaiane, O.R. Uctransnet: Rethinking the skip connections in u-net from a channel-wise perspective with transformer. In Proceedings of the 36th AAAI Conference on Artificial Intelligence, Virtually, 22 February–1 March 2022; pp. 2441–2449. [Google Scholar]
- Xu, G.; Zhang, X.; He, X.; Wu, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. In Proceedings of the 6th Chinese Conference on Pattern Recognition and Computer Vision, Xiamen, China, 13–15 October 2023; pp. 42–53. [Google Scholar]
- Heidari, M.; Kazerouni, A.; Soltany, M.; Azad, R.; Aghdam, E.; Cohen-Adad, J.; Merhof, D. Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 6202–6212. [Google Scholar]
- Rahman, M.; Munir, M.; Marculescu, R. Emcad: Efficient multi-scale convolutional attention decoding for medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 19–21 January 2024; pp. 11769–11779. [Google Scholar]
- Rahman, M.; Marculescu, R. Medical image segmentation via cascaded attention decoding. In Proceedings of the IEEE/CVF Winter Conference on Applications Of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 6222–6231. [Google Scholar]


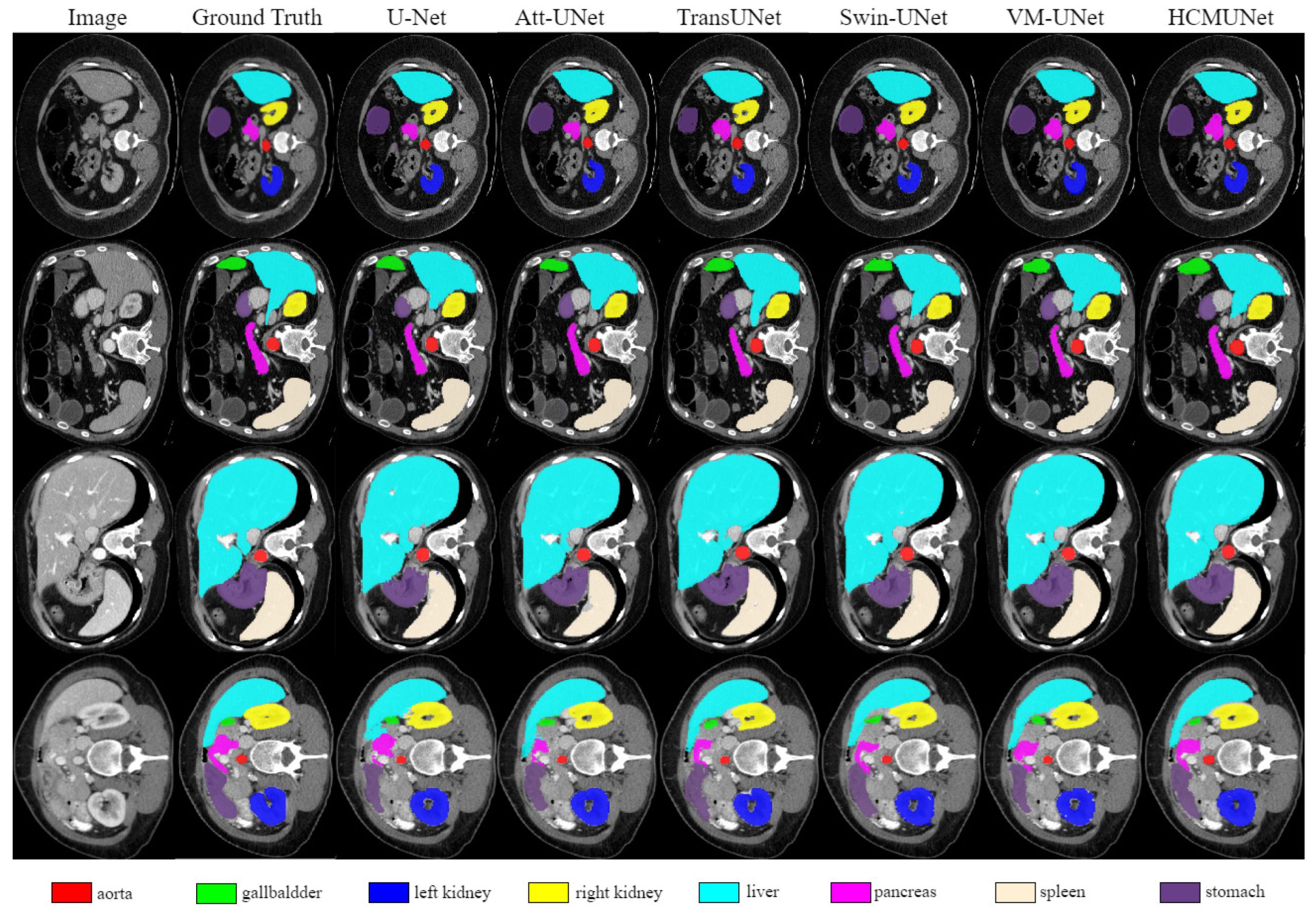

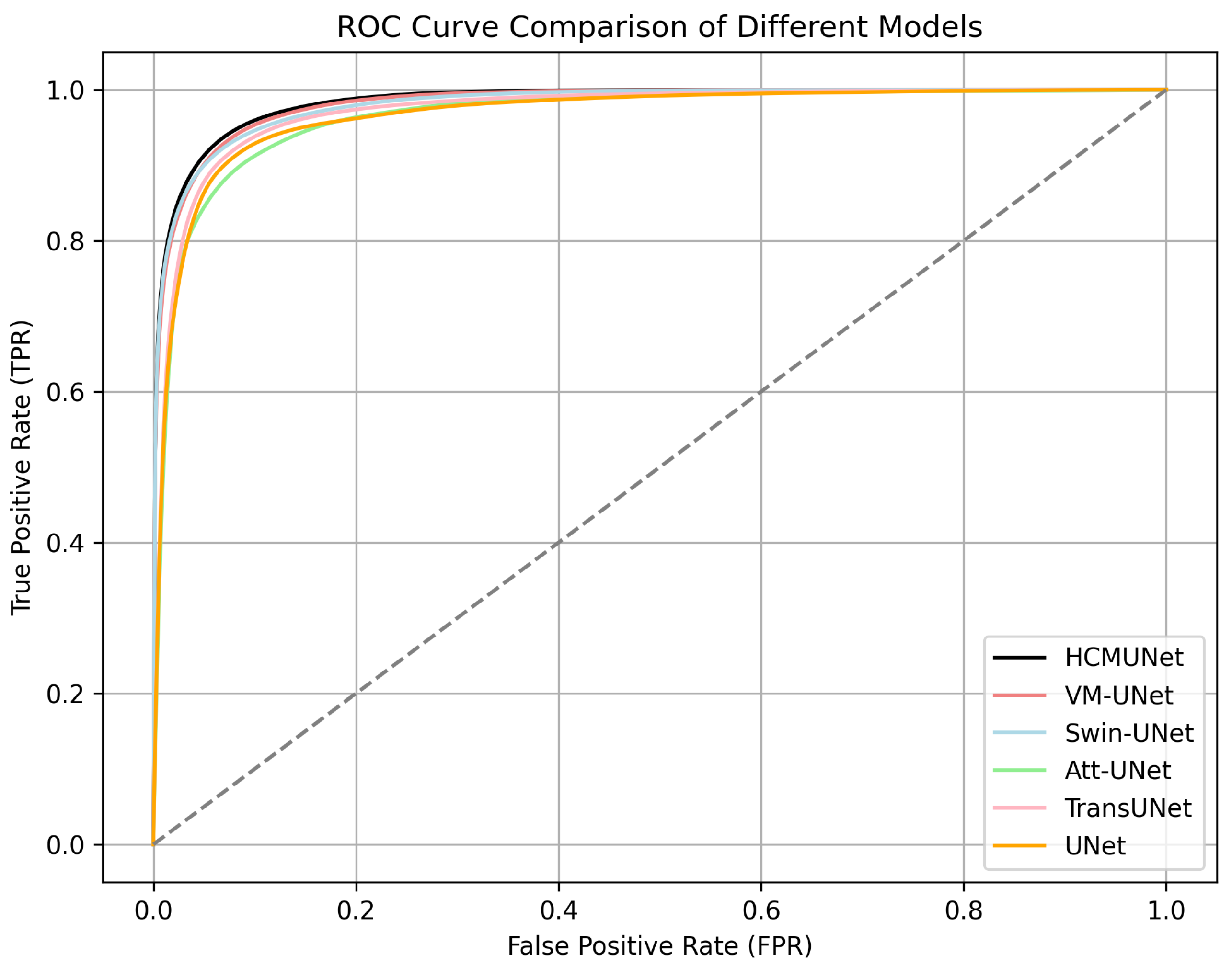
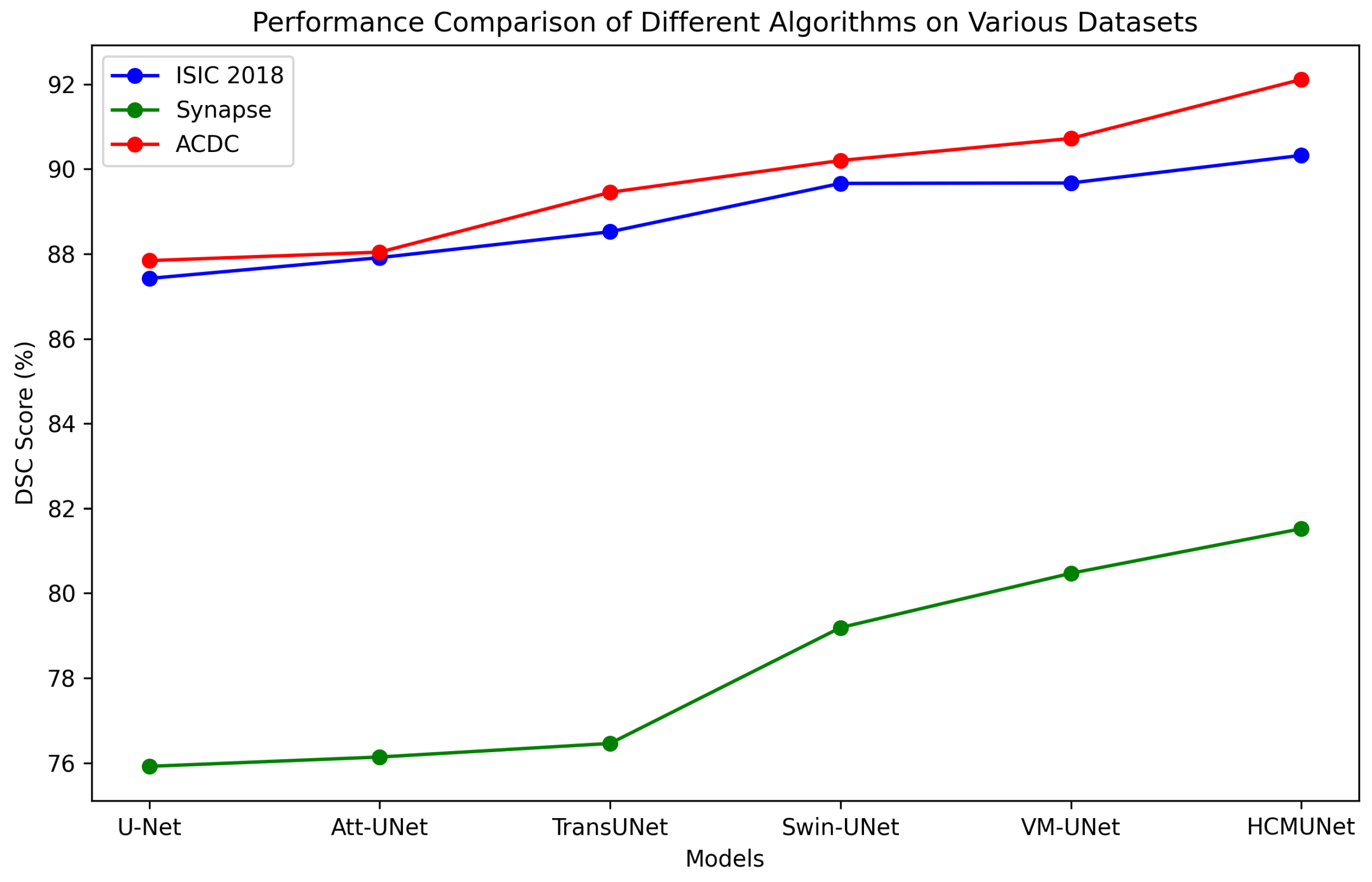
| Model | mIoU (%) | DSC (%) | Acc (%) | Spe (%) | Sen (%) |
|---|---|---|---|---|---|
| U-Net [2] | 77.64 ± 0.73 | 87.42 ± 0.87 | 93.88 ± 0.95 | 96.32 ± 1.20 | 87.71 ± 1.44 |
| U-Net++ [17] | 78.30 ± 0.90 | 87.63 ± 0.71 | 93.76 ± 0.78 | 95.19 ± 1.07 | 88.10 ± 1.23 |
| Att-UNet [19] | 78.95 ± 1.17 | 87.91 ± 1.01 | 93.21 ± 1.15 | 96.23 ± 1.35 | 87.60 ± 1.53 |
| SANet [42] | 79.47 ± 1.29 | 88.42 ± 1.11 | 94.29 ± 0.69 | 95.57 ± 1.42 | 89.46 ± 1.37 |
| TransUNet [43] | 79.71 ± 1.20 | 88.52 ± 1.27 | 94.57 ± 1.41 | 96.05 ± 1.30 | 89.14 ± 1.19 |
| TransFuse [44] | 80.66 ± 1.25 | 89.33 ± 1.28 | 93.66 ± 0.67 | 93.73 ± 1.17 | 90.78 ± 1.45 |
| Swin-UNet [26] | 80.71 ± 1.13 | 89.66 ± 1.04 | 94.19 ± 0.78 | 95.41 ± 1.22 | 90.31 ± 1.10 |
| VM-UNet [12] | 81.27 ± 0.79 | 89.67 ± 0.68 | 94.83 ± 0.63 | 96.13 ± 1.01 | 90.79 ± 0.98 |
| HCMUNet (Ours) | 82.19 ± 0.62 | 90.32 ± 0.53 | 94.71 ± 0.70 | 96.19 ± 0.78 | 91.21 ± 0.80 |
| Model | DSC | HD95 | Aorta | Gallbladder | Kidney (L) | Kidney (R) | Liver | Pancreas | Spleen | Stomach |
|---|---|---|---|---|---|---|---|---|---|---|
| U-Net [2] | 75.92 ± 2.71 | 37.55 ± 4.80 | 87.39 ± 2.87 | 67.52 ± 3.11 | 78.72 ± 2.04 | 68.87 ± 3.19 | 92.45 ± 1.26 | 51.51 ± 3.60 | 86.09 ± 3.67 | 74.82 ± 2.08 |
| Att-UNet [19] | 76.14 ± 3.32 | 33.51 ± 3.17 | 88.61 ± 2.12 | 66.40 ± 2.44 | 77.12 ± 1.49 | 71.07 ± 1.41 | 91.78 ± 2.43 | 55.01 ± 3.61 | 85.66 ± 2.72 | 73.47 ± 2.46 |
| TransUNet [43] | 76.46 ± 1.80 | 29.32 ± 4.27 | 86.55 ± 1.69 | 61.65 ± 3.40 | 79.41 ± 2.41 | 76.30 ± 2.95 | 93.13 ± 1.33 | 55.30 ± 3.29 | 84.63 ± 3.43 | 74.72 ± 1.87 |
| UCTransNet [45] | 78.24 ± 1.77 | 26.35 ± 1.38 | 86.52 ± 3.13 | 65.23 ± 2.05 | 80.69 ± 1.61 | 73.19 ± 2.58 | 93.05 ± 1.62 | 57.07 ± 2.65 | 87.55 ± 1.05 | 77.28 ± 2.48 |
| LeViT-UNet [46] | 78.82 ± 3.99 | 18.89 ± 3.99 | 85.23 ± 3.73 | 66.32 ± 1.39 | 82.68 ± 3.33 | 78.13 ± 3.37 | 93.61 ± 1.80 | 60.95 ± 2.50 | 89.00 ± 1.56 | 74.62 ± 3.68 |
| Swin-UNet [26] | 79.19 ± 2.07 | 22.07 ± 2.67 | 84.72 ± 2.61 | 66.60 ± 3.40 | 82.82 ± 1.19 | 79.41 ± 0.68 | 93.94 ± 0.75 | 59.49 ± 3.67 | 89.21 ± 2.31 | 77.36 ± 1.22 |
| HiFormer [47] | 80.55 ± 2.19 | 15.63 ± 3.41 | 87.07 ± 2.24 | 66.67 ± 2.27 | 83.92 ± 2.24 | 81.09 ± 2.91 | 94.09 ± 1.84 | 60.17 ± 2.72 | 90.76 ± 1.74 | 80.61 ± 2.31 |
| VM-UNet [12] | 80.47 ± 1.71 | 18.91 ± 2.42 | 86.84 ± 1.59 | 68.43 ± 4.13 | 85.04 ± 2.16 | 81.12 ± 2.67 | 94.11 ± 0.48 | 59.49 ± 1.78 | 87.77 ± 3.16 | 80.97 ± 2.33 |
| EMCAD [48] | 81.16 ± 2.01 | 16.72 ± 4.44 | 85.24 ± 1.80 | 67.24 ± 4.64 | 87.62 ± 0.30 | 81.38 ± 0.91 | 94.67 ± 1.77 | 61.02 ± 3.96 | 91.81 ± 2.35 | 80.30 ± 1.93 |
| HCMUNet (Ours) | 81.52 ± 1.14 | 17.83 ± 1.47 | 88.06 ± 1.63 | 69.60 ± 2.30 | 87.04 ± 0.67 | 82.35 ± 1.48 | 95.10 ± 3.71 | 59.24 ± 3.62 | 90.63 ± 1.54 | 80.76 ± 2.30 |
| Model | DSC | RV | MYO | LV |
|---|---|---|---|---|
| U-Net [2] | 87.84 ± 1.38 | 86.51 ± 1.32 | 84.66 ± 5.65 | 92.36 ± 3.92 |
| Att-UNet [19] | 88.04 ± 2.02 | 86.70 ± 2.33 | 84.59 ± 8.96 | 92.83 ± 1.57 |
| TransUNet [43] | 89.45 ± 1.13 | 87.85 ± 2.22 | 86.09 ± 2.23 | 94.39 ± 1.43 |
| nnUNet [22] | 90.14 ± 1.83 | 88.58 ± 1.44 | 89.72 ± 1.18 | 92.13 ± 2.24 |
| Swin-UNet [26] | 90.20 ± 0.78 | 87.44 ± 2.46 | 88.20 ± 3.40 | 94.97 ± 1.26 |
| LeViT-UNet [46] | 90.42 ± 1.15 | 88.85 ± 1.52 | 88.29 ± 2.04 | 94.12 ± 1.88 |
| HiFormer [47] | 90.69 ± 0.85 | 90.06 ± 1.30 | 89.00 ± 1.29 | 93.00 ± 1.07 |
| VM-UNet [12] | 90.72 ± 0.77 | 90.61 ± 1.67 | 89.40 ± 1.88 | 92.14 ± 0.76 |
| EMCAD [48] | 91.30 ± 0.35 | 90.17 ± 1.88 | 89.16 ± 1.24 | 94.55 ± 1.59 |
| TransCASCADE [49] | 91.59 ± 0.21 | 90.12 ± 2.66 | 90.14 ± 0.68 | 94.51 ± 2.55 |
| HCMUNet (Ours) | 92.11 ± 0.26 | 91.50 ± 0.78 | 90.20 ± 0.56 | 94.61 ± 1.12 |
| Dataset | Components | Evaluation Metrics | |||
|---|---|---|---|---|---|
| Mamba | Conv | SCDA | Paras (M) | DSC (%) | |
| ISIC 2018 | ✓ | 44.27 | 89.67 ± 0.68 | ||
| ✓ | ✓ | 25.43 | 90.16 ± 0.59 | ||
| ✓ | ✓ | 55.11 | 90.03 ± 0.73 | ||
| ✓ | ✓ | 67.17 | 85.22 ± 1.21 | ||
| ✓ | ✓ | ✓ | 36.27 | 90.32 ± 0.53 | |
| Synapse | ✓ | 44.27 | 80.47 ± 1.71 | ||
| ✓ | ✓ | 25.43 | 80.92 ± 1.44 | ||
| ✓ | ✓ | 55.11 | 80.67 ± 2.02 | ||
| ✓ | ✓ | 67.17 | 74.48 ± 3.13 | ||
| ✓ | ✓ | ✓ | 36.27 | 81.52 ± 1.14 | |
| ACDC | ✓ | 44.27 | 90.72 ± 0.77 | ||
| ✓ | ✓ | 25.43 | 91.34 ± 0.55 | ||
| ✓ | ✓ | 55.11 | 90.88 ± 0.97 | ||
| ✓ | ✓ | 67.17 | 87.05 ± 1.35 | ||
| ✓ | ✓ | ✓ | 36.27 | 92.11 ± 0.26 | |
| Dataset | Different Data Volumes from the Dataset | |||
|---|---|---|---|---|
| 25% | 50% | 75% | 100% | |
| ISIC 2018 | 87.15 ± 1.39 | 88.68 ± 0.71 | 89.73 ± 0.68 | 90.32 ± 0.53 |
| Synapse | 75.35 ± 3.41 | 79.46 ± 1.85 | 81.08 ± 1.45 | 81.52 ± 1.14 |
| ACDC | 88.95 ± 1.67 | 90.36 ± 0.74 | 91.68 ± 0.45 | 92.11 ± 0.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Du, Y.; Sui, D. A U-Shaped Architecture Based on Hybrid CNN and Mamba for Medical Image Segmentation. Appl. Sci. 2025, 15, 7821. https://doi.org/10.3390/app15147821
Ma X, Du Y, Sui D. A U-Shaped Architecture Based on Hybrid CNN and Mamba for Medical Image Segmentation. Applied Sciences. 2025; 15(14):7821. https://doi.org/10.3390/app15147821
Chicago/Turabian StyleMa, Xiaoxuan, Yingao Du, and Dong Sui. 2025. "A U-Shaped Architecture Based on Hybrid CNN and Mamba for Medical Image Segmentation" Applied Sciences 15, no. 14: 7821. https://doi.org/10.3390/app15147821
APA StyleMa, X., Du, Y., & Sui, D. (2025). A U-Shaped Architecture Based on Hybrid CNN and Mamba for Medical Image Segmentation. Applied Sciences, 15(14), 7821. https://doi.org/10.3390/app15147821





