Truncation Artifact Reduction in Stationary Inverse-Geometry Digital Tomosynthesis Using Deep Convolutional Generative Adversarial Network
Abstract
1. Introduction
2. Materials and Methods
2.1. s-IGDT System
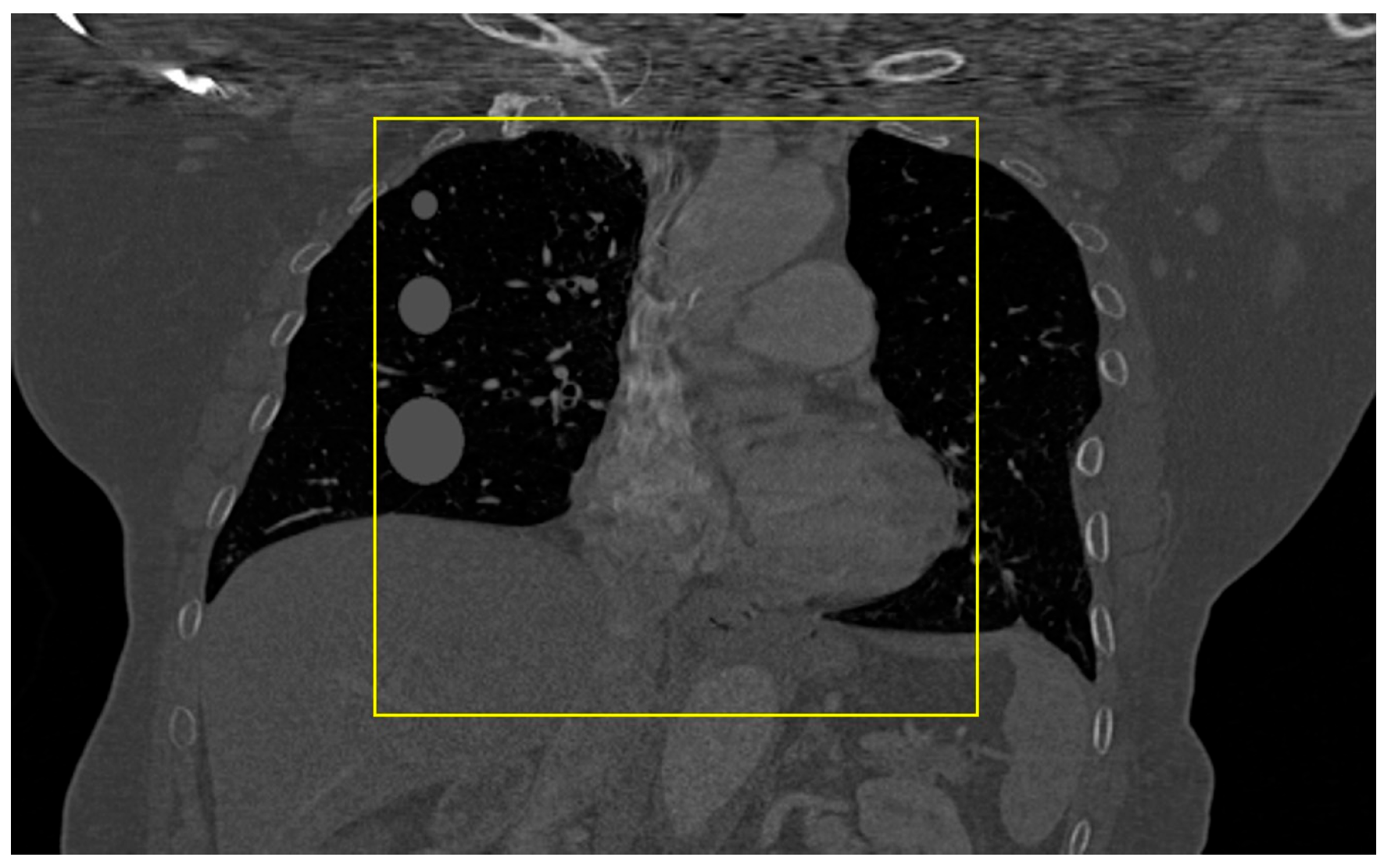
2.2. Data Acquisition
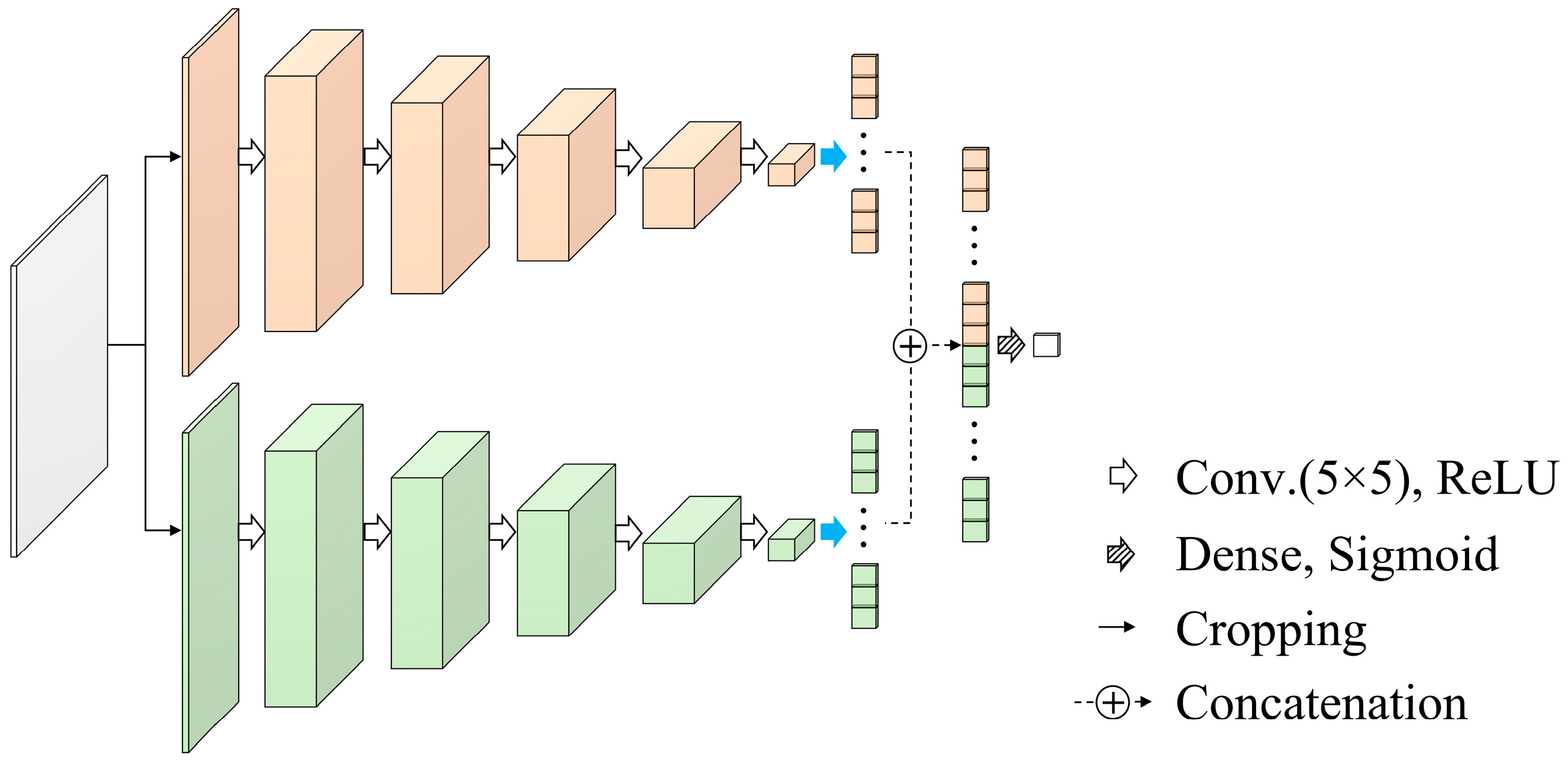
2.3. Network Architecture
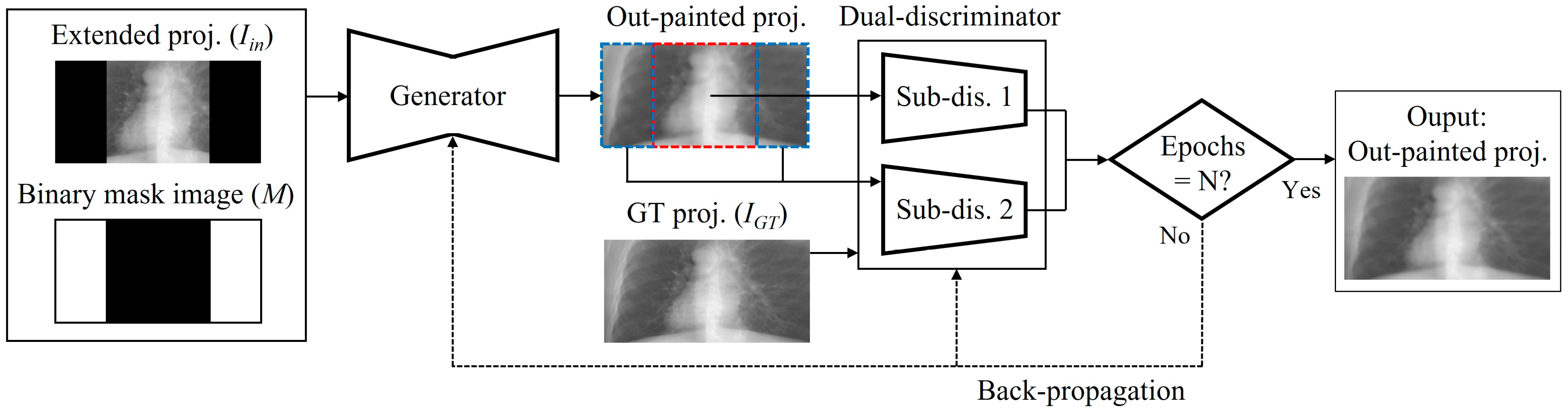
2.4. Network Training
2.5. Quantitative Evaluation and Comparison
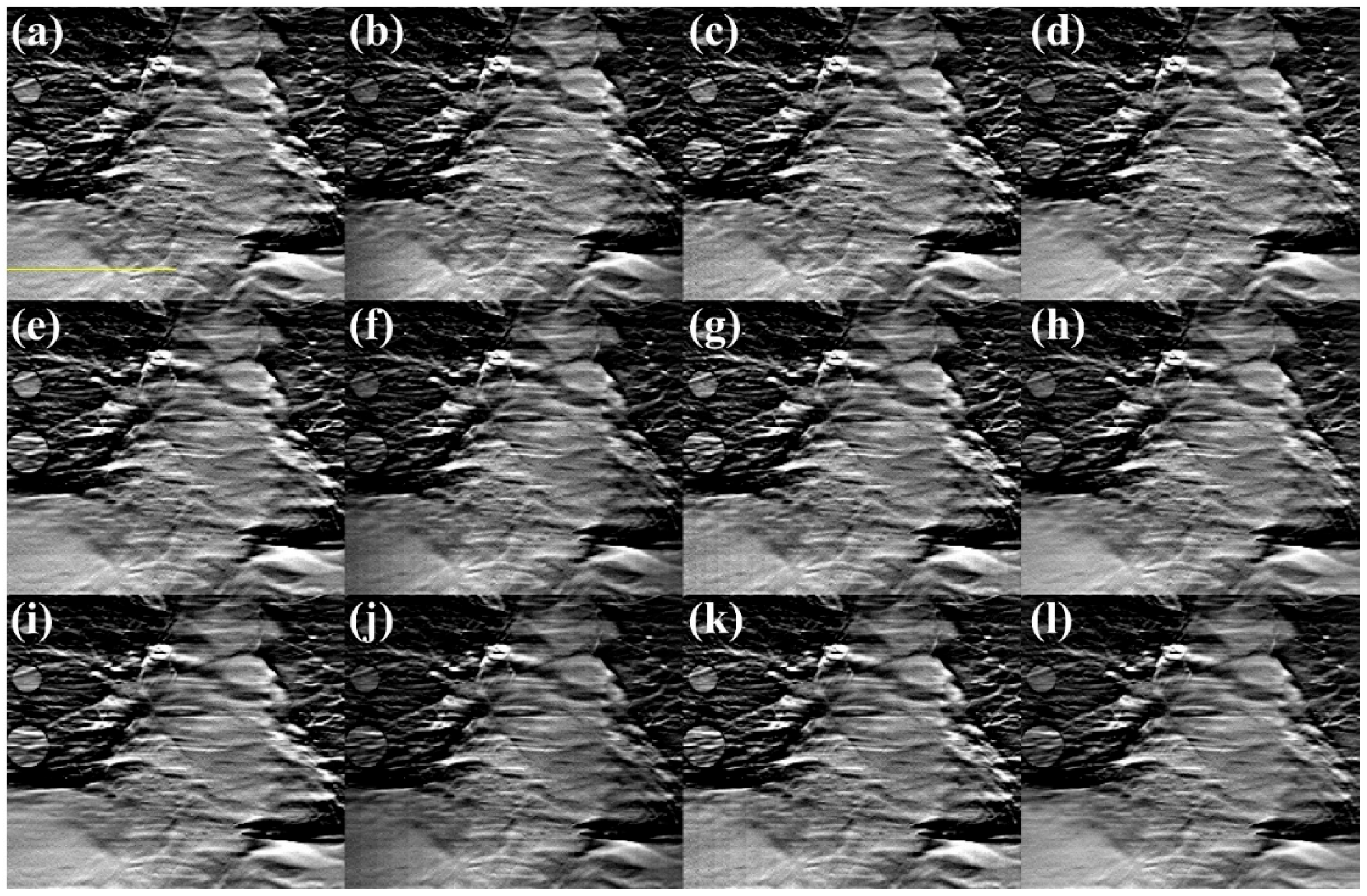
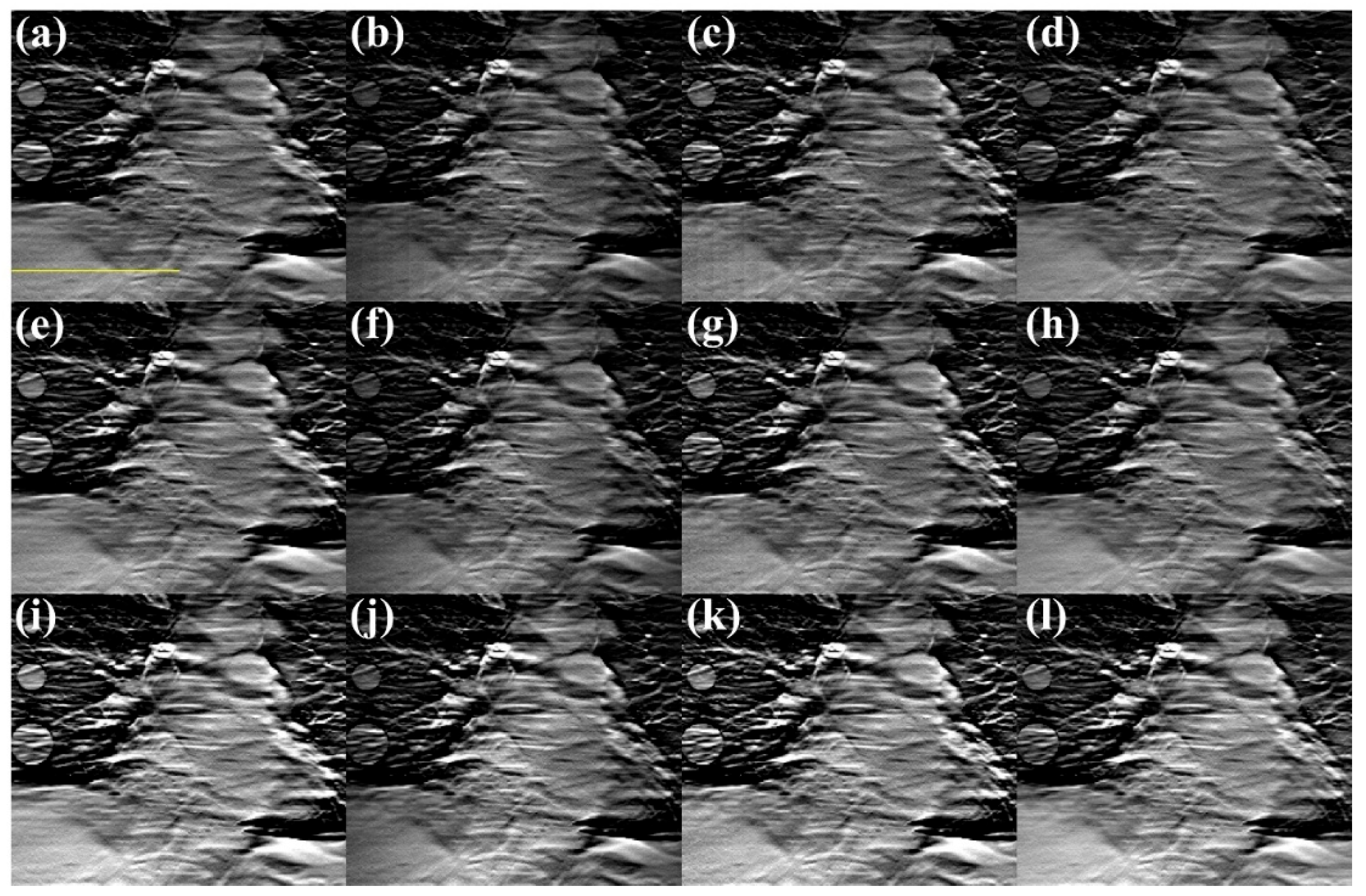
3. Results
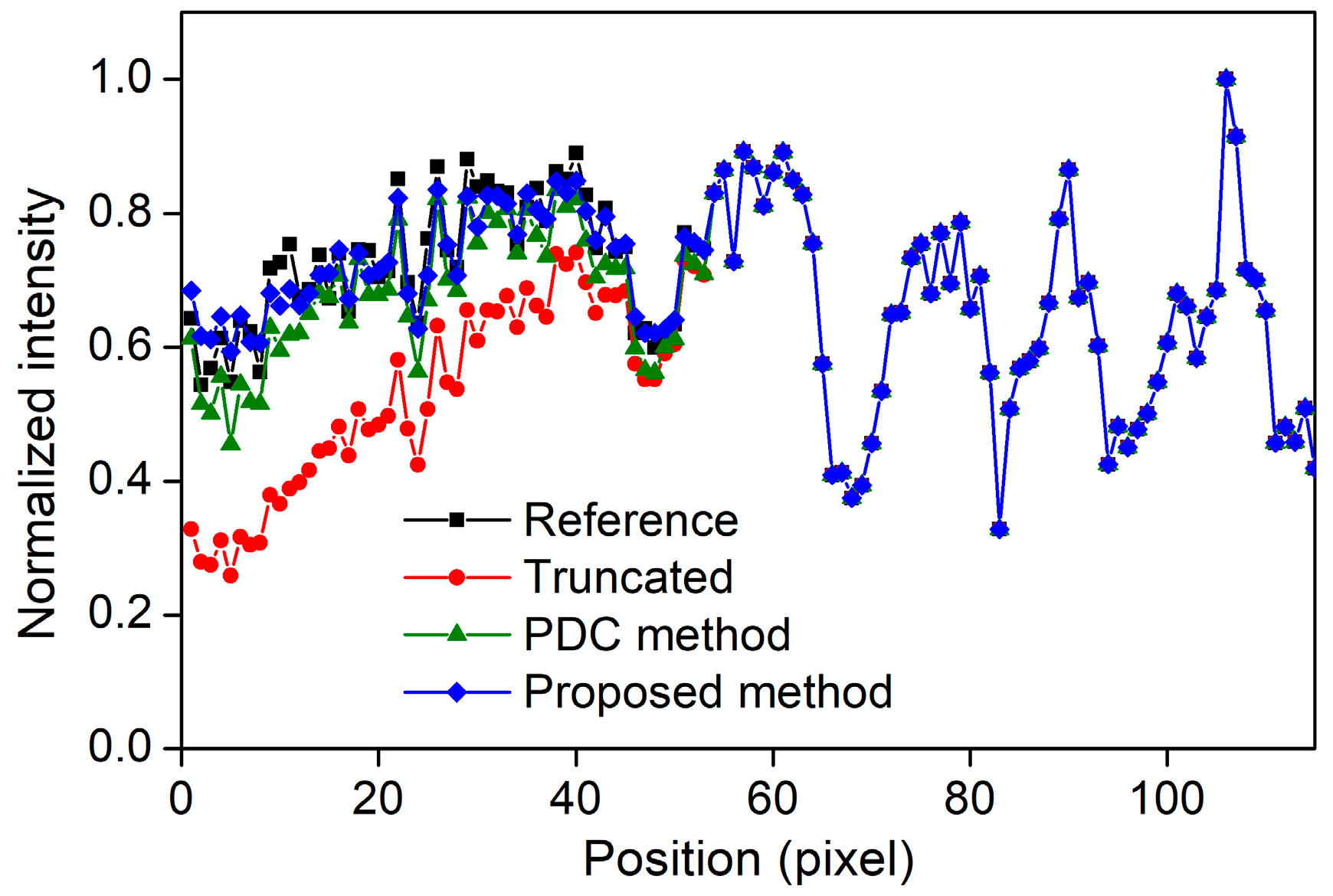
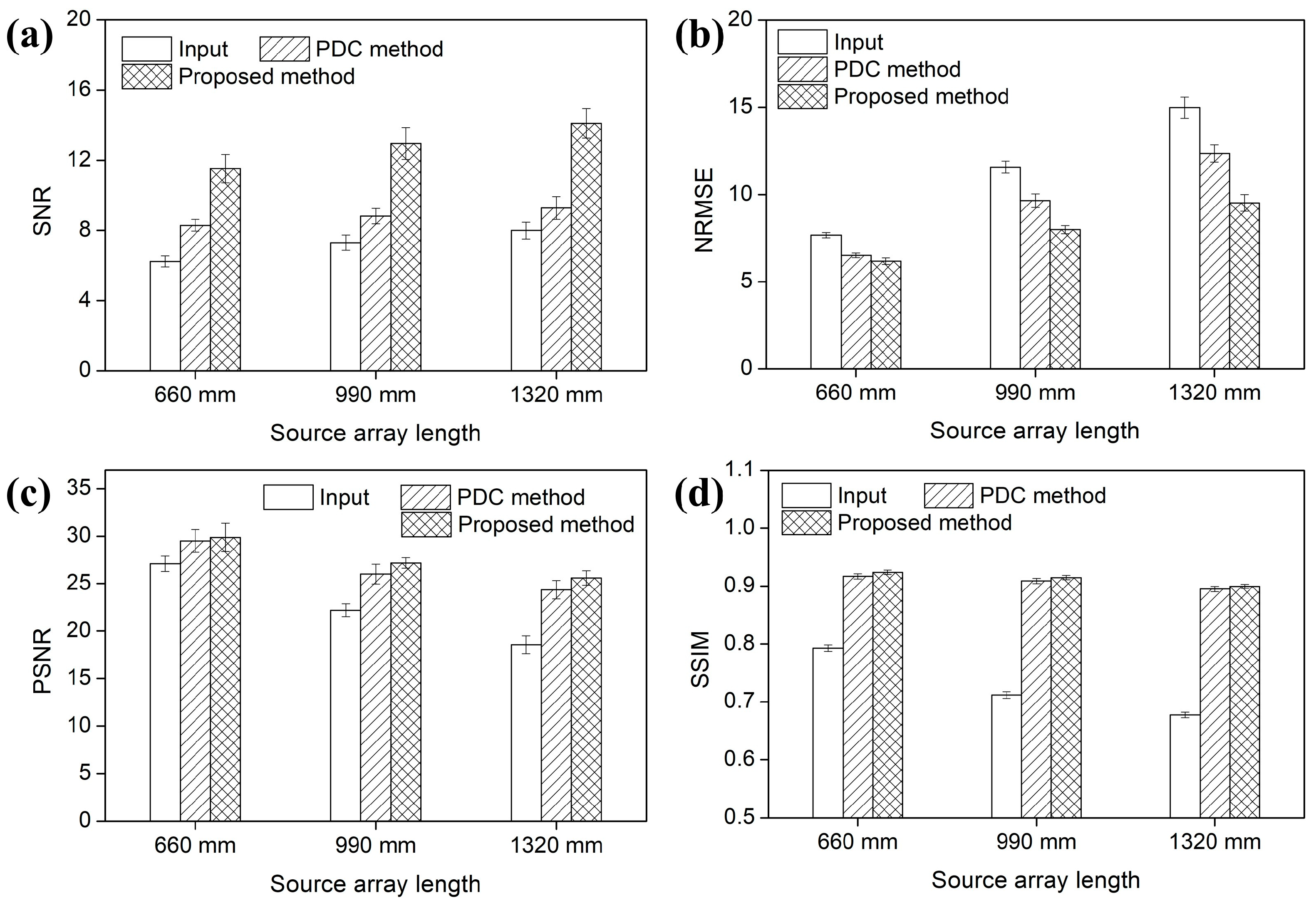
3.1. Lengths of the Source Array
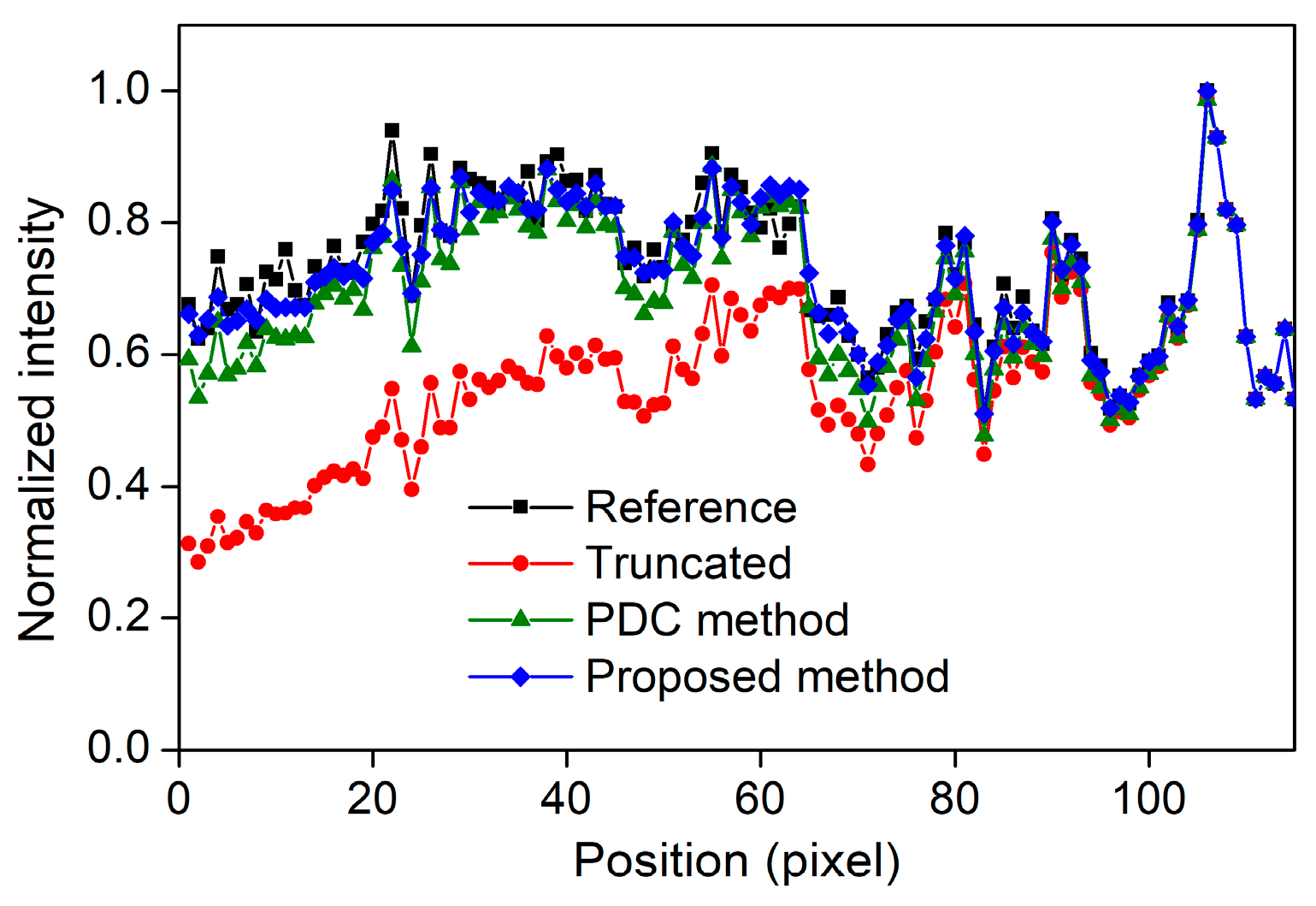
3.2. Numbers of the Focal Spots
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petropoulos, A.E.; Skiadopoulos, S.G.; Messaris, G.A.T.; Karahaliou, A.N.; Costaridou, L.I. Contrast and depth resolution of breast lessions in a digital breast tomosynthesis system. Phys. Med. 2016, 32, 277. [Google Scholar] [CrossRef]
- Chou, S.-H.S.; Kicska, G.A.; Pipavath, S.N.; Reddy, G.P. Digital Tomosynthesis of the Chest: Current and Emerging Applications. Radiographics 2014, 34, 359–372. [Google Scholar] [CrossRef]
- Kim, B.; Yim, D.; Lee, S. Development of a truncation artifact reduction method in stationary inverse-geometry X-ray laminography for non-destructive testing. Nucl. Eng. Technol. 2021, 53, 1626–1633. [Google Scholar] [CrossRef]
- Qian, X.; Rajaram, R.; Calderon-Colon, X.; Yang, G.; Phan, T.; Lalush, D.S.; Lu, J.; Zhou, O. Design and characterization of a spatially distributed multibeam field emission X-ray source for stationary digital breast tomosynthesis. Med. Phys. 2009, 36, 4389–4399. [Google Scholar] [CrossRef]
- Yang, G.; Rajarm, R.; Cao, G.; Sultana, S.; Liu, Z.; Lalush, D.; Lu, J.; Zhou, O. Stationary digital breast tomosynthesis system with a multi-beam field emission x-ray source array. In Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 16–21 February 2008; Volume 6913, pp. 441–450. [Google Scholar] [CrossRef]
- Kim, B.; Yim, D.; Lee, S. Reduction of truncation artifact in stationary inverse-geometry digital tomosynthesis using convolutional neural network. In Proceedings of the SPIE Medical Imaging, Houston, TX, USA, 15–20 February 2020; Volume 11312, pp. 1111–1116. [Google Scholar] [CrossRef]
- Thapa, D.; Billingsley, A.; Luo, Y.; Inscoe, C.; Zhou, O.; Lu, J.; Lee, Y. Orthogonal tomosynthesis for whole body skeletal imaging enabled by carbon nanotube x-ray source array. In Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 20 February–28 March 2022; Volume 12031, pp. 1018–1024. [Google Scholar] [CrossRef]
- Lopez Montes, A.; McSkimming, T.; Zbijewski, W.; Siewerdsen, J.H.; Delnooz, C.; Skeats, A.; Gonzales, B.; Sisniega, A. Stationary X-ray tomography for hemorrhagic stroke imaging: Sampling and resolution properties. In Proceedings of the 7th International Conference on Image Formation in X-ray Computed Tomography, Baltimore, MD, USA, 12–16 June 2022; Volume 12304, pp. 151–157. [Google Scholar] [CrossRef]
- Brock, A.; Donahue, J.; Simonyan, K. Large Scale GAN Training for High Fidelity Natural Image Synthesis. arXiv 2019, arXiv:1809.11096. [Google Scholar]
- Bergmann, U.; Jetchev, N.; Vollgraf, R. Learning Texture Manifolds with the Periodic Spatial GAN. arXiv 2017, arXiv:1705.06566. [Google Scholar]
- Yu, J.; Lin, Z.; Yang, J.; Shen, X.; Lu, X.; Huang, T.S. Generative image inpainting with contextual attention. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; Volume 2018, pp. 5505–5514. [Google Scholar] [CrossRef]
- Gomi, T.; Sakai, R.; Hara, H.; Watanabe, Y.; Mizukami, S. Usefulness of a Metal Artifact Reduction Algorithm in Digital Tomosynthesis Using a Combination of Hybrid Generative Adversarial Networks. Diagnostics 2021, 11, 1629. [Google Scholar] [CrossRef]
- Gomi, T.; Kijima, Y.; Kobayashi, T.; Koibuchi, Y. Evaluation of a Generative Adversarial Network to Improve Image Quality and Reduce Radiation-Dose during Digital Breast Tomosynthesis. Diagnostics 2022, 12, 495. [Google Scholar] [CrossRef]
- Nazeri, K.; Ng, E.; Joseph, T.; Qureshi, F.Z.; Ebrahimi, M. EdgeConnect: Generative Image Inpainting with Adversarial Edge Learning. arXiv 2019, arXiv:1901.00212. [Google Scholar]
- Podgorsak, A.R.; Bhurwani, M.M.S.; Ionita, C.N. CT artifact correction for sparse and truncated projection data using generative adversarial networks. Med. Phys. 2021, 48, 615–626. [Google Scholar] [CrossRef]
- Armato, S.G., III; Hadjiiski, L.M.; Tourassi, G.D.; Drukker, K.; Giger, M.L.; Li, F.; Redmond, G.; Farahani, K.; Kirby, J.S.; Clarke, L.P. LUNGx Challenge for computerized lung nodule classification: Reflections and lessons learned. J. Med. Imaging 2015, 2, 020103. [Google Scholar] [CrossRef]
- Zeng, G.L.; Gullberg, G.T. Unmatched projector/backprojector pairs in an iterative reconstruction algorithm. IEEE Trans. Med. Imaging 2000, 19, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2016, arXiv:1511.06434. [Google Scholar]
- Chen, X.; Duan, Y.; Houthooft, R.; Schulman, J.; Sutskever, I.; Abbeel, P. InfoGAN: Interpretable Representation Learning by Information Maximizing Generative Adversarial Nets. Adv. Neural. Inf. Process. Syst. 2016, 29, 2180–2188. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Volume 70, pp. 214–223. [Google Scholar]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. arXiv 2016, arXiv:1511.07122. [Google Scholar]
- Lucic, M.; Kurach, K.; Michalski, M.; Gelly, S.; Bousquet, O. Are gans created equal? a large-scale study. Adv. Neural. Inf. Process. Syst. 2018, 31, 698–707. [Google Scholar]
- Zhang, Z.; Deng, C.; Shen, Y.; Williamson, D.S.; Sha, Y.; Zhang, Y.; Song, H.; Li, X. On Loss Functions and Recurrency Training for GAN-based Speech Enhancement Systems. arXiv 2020, arXiv:2007.14974. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simonceli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Hore, A.; Ziou, D. Image quality metrics: PSNR versus SSIM. In Proceedings of the International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar]
- Park, J.; Hwang, D.; Kim, K.Y.; Kang, S.K.; Kim, Y.K.; Lee, J.S. Computed tomography super-resolution using deep convolutional neural network. Phys. Med. Biol. 2018, 63, 145011. [Google Scholar] [CrossRef]
- Pathak, D.; Krahenbuhl, P.; Donahue, J.; Darrell, T.; Efros, A.A. Context encoders: Feature learning by inpainting. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; Volume 2016, pp. 2536–2544. [Google Scholar]
- Sun, H.; Fan, R.; Li, C.; Lu, Z.; Xie, K.; Ni, X.; Yang, J. Imaging study of pseudo-CT synthesized from cone-beam CT based on 3D CycleGAN in radiotherapy. Front. Oncol. 2021, 11, 603844. [Google Scholar] [CrossRef]
- Li, J.; Liang, X.; Wei, Y.; Xu, T.; Feng, J.; Yan, S. Perceptual generative adversarial networks for small object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; Volume 2017, pp. 1222–1230. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, C.; Wang, A.; Yang, L.; Hao, Y. CT super-resolution using multiple dense residual block based GAN. Signal Image Video Process. 2021, 15, 725–733. [Google Scholar] [CrossRef]
- Bulat, A.; Yang, J.; Tzimiropoulos, G. To learn image super-resolution, use a GAN to learn how to do image degradation first. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 185–200. [Google Scholar]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In Proceedings of the Computer Vision-ECCV 2016 Part II, Amsterdam, The Netherlands, 11–14 October 2016; Volume 14, pp. 694–711. [Google Scholar] [CrossRef]
- Gholizadeh-Ansari, M.; Alirezaie, J.; Babyn, P. Deep Learning for Low-Dose CT Denoising Using Perceptual Loss and Edge Detection Layer. J. Digit. Imaging 2020, 33, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Primidis, T.G.; Wells, S.G.; Soloviev, V.Y.; Welsch, C.P. 3D chest tomosynthesis using a stationary flat panel source array and a stationary detector: A Monte Carlo proof of concept. Biomed. Phys. Eng. Express 2022, 8, 015006. [Google Scholar] [CrossRef]
- Billingsley, A.; Inscoe, C.; Lu, J.; Zhou, O.; Lee, Y.Z. Second generation stationary chest tomosynthesis with faster scan time and wider angular span. Med. Phys. 2024, 52, 542–552. [Google Scholar] [CrossRef]






| Parameters | |
|---|---|
| Type of the source array | Linear |
| Length of the source array | 660 mm, 990 mm, 1320 mm * |
| Number of the focal spots | 21 *, 41, 81 |
| Detector size | 170 × 170 mm2 |
| Source-to-center of object distance | 1207 mm |
| Source-to-detector distance | 1348.5 mm |
| Layer | Kernel Size | Dilation Rate | Activation Function | Output Channel |
|---|---|---|---|---|
| Conv. | 5 × 5 | - | ReLU | 64 |
| Conv. | 4 × 4 | - | ReLU | 128 |
| Conv. | 4 × 4 | - | ReLU | 256 |
| Conv. | 4 × 4 | - | ReLU | 512 |
| Conv. | 4 × 4 | - | ReLU | 512 |
| Dilated Conv. | 4 × 4 | 4 | ReLU | 512 |
| Dilated Conv. | 4 × 4 | 8 | ReLU | 512 |
| Dilated Conv. | 4 × 4 | 16 | ReLU | 512 |
| Dilated Conv. | 4 × 4 | 32 | ReLU | 512 |
| Conv. | 4 × 4 | - | ReLU | 512 |
| Conv. | 4 × 4 | - | ReLU | 512 |
| Deconv. | 4 × 4 | - | ReLU | 256 |
| Deconv. | 4 × 4 | - | ReLU | 128 |
| Conv. | 4 × 4 | - | ReLU | 128 |
| Conv. | 4 × 4 | - | ReLU | 64 |
| Conv. | 4 × 4 | - | Sigmoid | 1 |
| Layer | Kernel Size | Activation Function | Output Channel | |
|---|---|---|---|---|
| Cropping | - | - | 2 | |
| Sub- discriminator 1/2 | Conv./Conv. | 5 × 5/5 × 5 | ReLU/ReLU | 32/32 |
| Conv./Conv. | 5 × 5/5 × 5 | ReLU/ReLU | 64/64 | |
| Conv./Conv. | 5 × 5/5 × 5 | ReLU/ReLU | 64/64 | |
| Conv./Conv. | 5 × 5/5 × 5 | ReLU/ReLU | 64/64 | |
| Conv./Conv. | 5 × 5/5 × 5 | ReLU/ReLU | 64/64 | |
| Flatten/Flatten | - | ReLU/ReLU | 1024/1024 | |
| Conca. | - | - | 2048 | |
| Dense | - | Sigmoid | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.; Lee, S. Truncation Artifact Reduction in Stationary Inverse-Geometry Digital Tomosynthesis Using Deep Convolutional Generative Adversarial Network. Appl. Sci. 2025, 15, 7699. https://doi.org/10.3390/app15147699
Kim B, Lee S. Truncation Artifact Reduction in Stationary Inverse-Geometry Digital Tomosynthesis Using Deep Convolutional Generative Adversarial Network. Applied Sciences. 2025; 15(14):7699. https://doi.org/10.3390/app15147699
Chicago/Turabian StyleKim, Burnyoung, and Seungwan Lee. 2025. "Truncation Artifact Reduction in Stationary Inverse-Geometry Digital Tomosynthesis Using Deep Convolutional Generative Adversarial Network" Applied Sciences 15, no. 14: 7699. https://doi.org/10.3390/app15147699
APA StyleKim, B., & Lee, S. (2025). Truncation Artifact Reduction in Stationary Inverse-Geometry Digital Tomosynthesis Using Deep Convolutional Generative Adversarial Network. Applied Sciences, 15(14), 7699. https://doi.org/10.3390/app15147699






