1. Introduction
Sleep is an essential physiological process for brain and body restoration. During sleep, significant changes occur in brain, muscle, ocular, cardiac, and respiratory activity [
1]. Heart rate and respiration decrease, muscles enter a state of paralysis, and the brain processes daily experiences, storing them in memory [
2]. A healthy sleep cycle, characterized by an adequate duration, quality, consistency, and regularity, improves cognitive development, consolidates learning, and supports physical and emotional well-being [
3]. It also strengthens the immune system and regulates functions such as metabolism, insulin sensitivity, and glucose tolerance, helping to prevent metabolic diseases such as diabetes and obesity [
4]. In contrast, sleep deprivation has widespread negative effects. It can lead to a decrease in body temperature, disrupted growth hormone release, diminished immune function, drowsiness, a lack of concentration, and memory loss [
4]. It is also associated with increased appetite, mood disturbances, slower reaction times, cognitive impairment, and the development of neurodegenerative diseases such as Parkinson’s and Alzheimer’s [
5]. Beyond these vital functions, sleep disorders have a huge impact on quality of life.
According to the American Academy of Sleep Medicine (AASM), sleep disorders affect a third of the global population [
3,
6]. The most common include insomnia, hypersomnia, parasomnia, sleep-related breathing disorders, narcolepsy, circadian rhythm disorders, and abnormal movements during sleep [
6,
7]. An example is obstructive sleep apnea (OSA), which disrupts the oxygen supply of the body and, if left untreated, can lead to hypertension, cardiovascular diseases, and other serious conditions [
8]. These disorders not only affect individual health, but also have societal consequences. They are related to increased medical and psychiatric morbidity, traffic accidents, reduced work productivity, lower quality of life, and even higher mortality rates [
9].
Given the importance of sleep for human health, it is fundamental to understand its behavior and define patterns or stages that facilitate the early diagnosis of related disorders. The accurate classification of these stages can improve medical interventions. In fact, this task is typically performed manually by experts, but it is time-consuming and labor-intensive, requiring detailed visual analysis. In addition, measurements are often invasive, performed in clinical settings or at the patient’s home under expert supervision. Recent technological developments aim to reduce invasiveness through non-contact or wearable alternatives. For example, optical fiber-based systems embedded in mattresses have been investigated to monitor heart rate and respiration with promising precision [
10], and similar efforts include textile-integrated sensors in garments designed for overnight monitoring [
11]. These approaches offer greater comfort and practicality for home environments and also facilitate the acquisition of biomedical signals during sleep, making them highly useful for classification-related applications.
In relation to sleep stage classification, various mathematical models have been explored to automate the identification of sleep stages. Advances in deep learning have shown promising results, achieving high precision in multiclass classification tasks [
12]. Beyond neurological disorders, these methods have also been applied in medical engineering, such as in the evaluation of muscle strength using wearable sensors with temporal convolutional and transformer-based models [
13], and in breast cancer imaging [
14], supporting more accurate screening and diagnosis. These developments illustrate the growing impact of deep learning across clinical domains and its potential to improve diagnostic precision and therapeutic decision-making.
To assess sleep quality, neurophysiologists collect various physiological data from patients during sleep, including respiration, heart rate, body temperature, and movements [
2]. These parameters provide insights into the sleep duration, onset latency, interruptions, efficiency, and other key aspects. The clinical standard for recording biological signals during sleep is polysomnography (PSG). PSG recordings typically include electroencephalography (EEG) to measure cortical electrical activity through scalp electrodes, electrooculography (EOG) used to track eye movements, electromyography (EMG) to measure muscle activity, and electrocardiography (ECG) to monitor cardiovascular activity. Additional PSG recordings may include body temperature, respiratory effort, motion annotations, and other physiological parameters. By integrating these signals, PSG enables an extensive assessment of neural and physical patterns during sleep, resulting in multidimensional datasets.
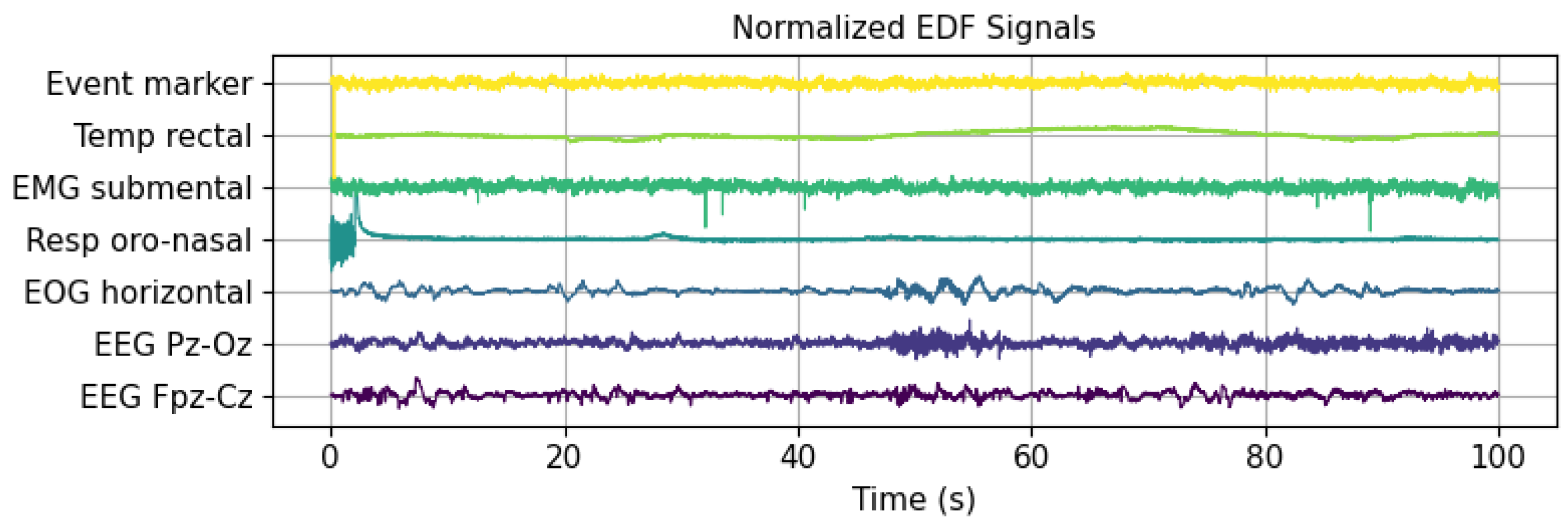
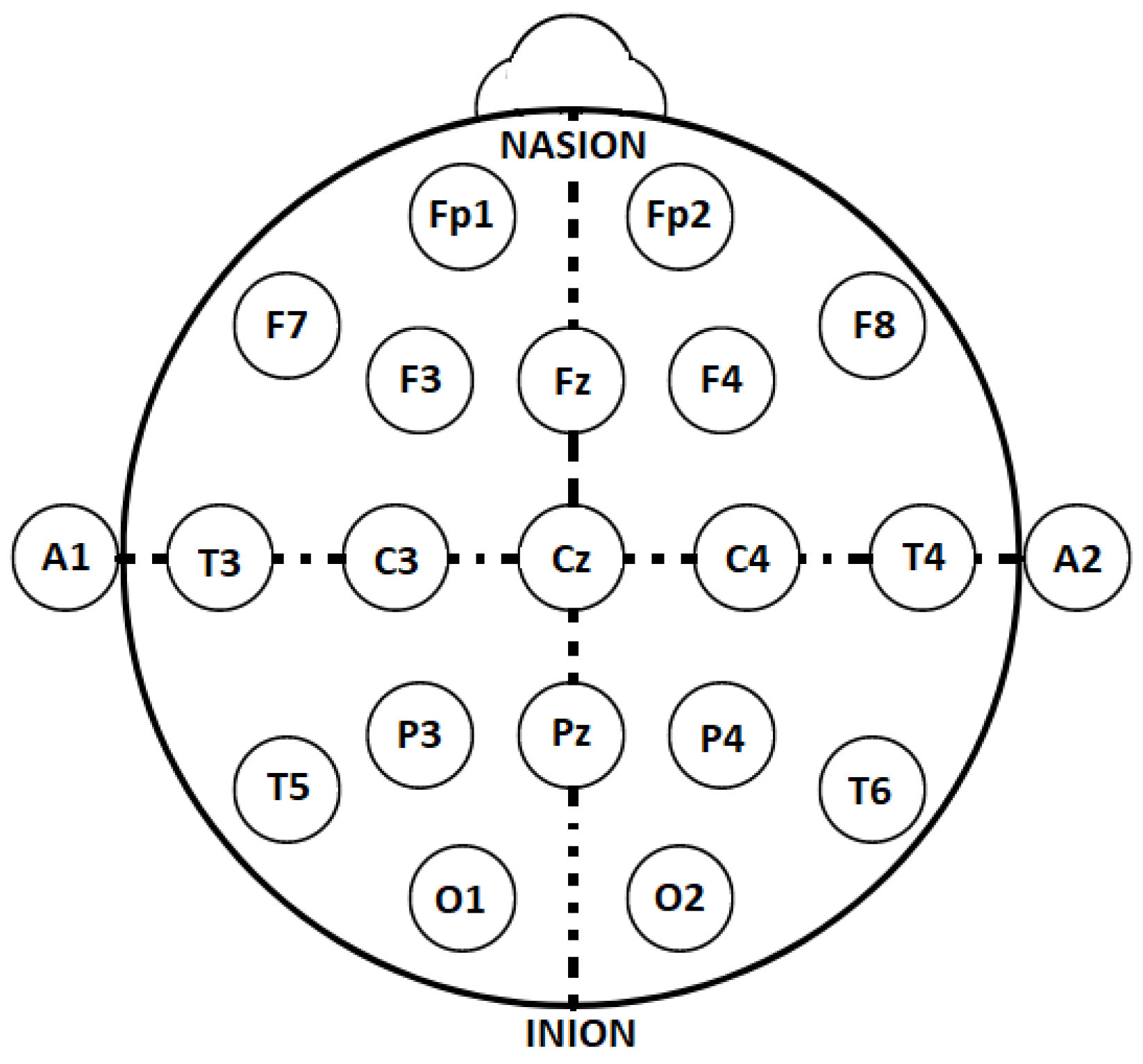
Figure 1 illustrates the first 100 s of several signals from a PSG recording in the publicly available Sleep EDF Expanded database, utilized in this research. This database includes various signals, such as event markers, temperature, EMG, respiration, EOG, and two EEG channels. The latter were recorded at different cortical locations after the international 10–20 electrode placement standard for EEG (
Figure 2). The 10–20 placement protocol divides the cortex into areas based on the main brain lobes, assigning each a combination of letters and numbers. Fp, F, C, P, T, O, and A correspond to the frontal polar, frontal central, parietal, temporal, occipital, and auricular regions, respectively [
15]. Even and odd numbers refer to the right and left brain hemispheres, respectively, while the letter z designates the central region of each lobe.
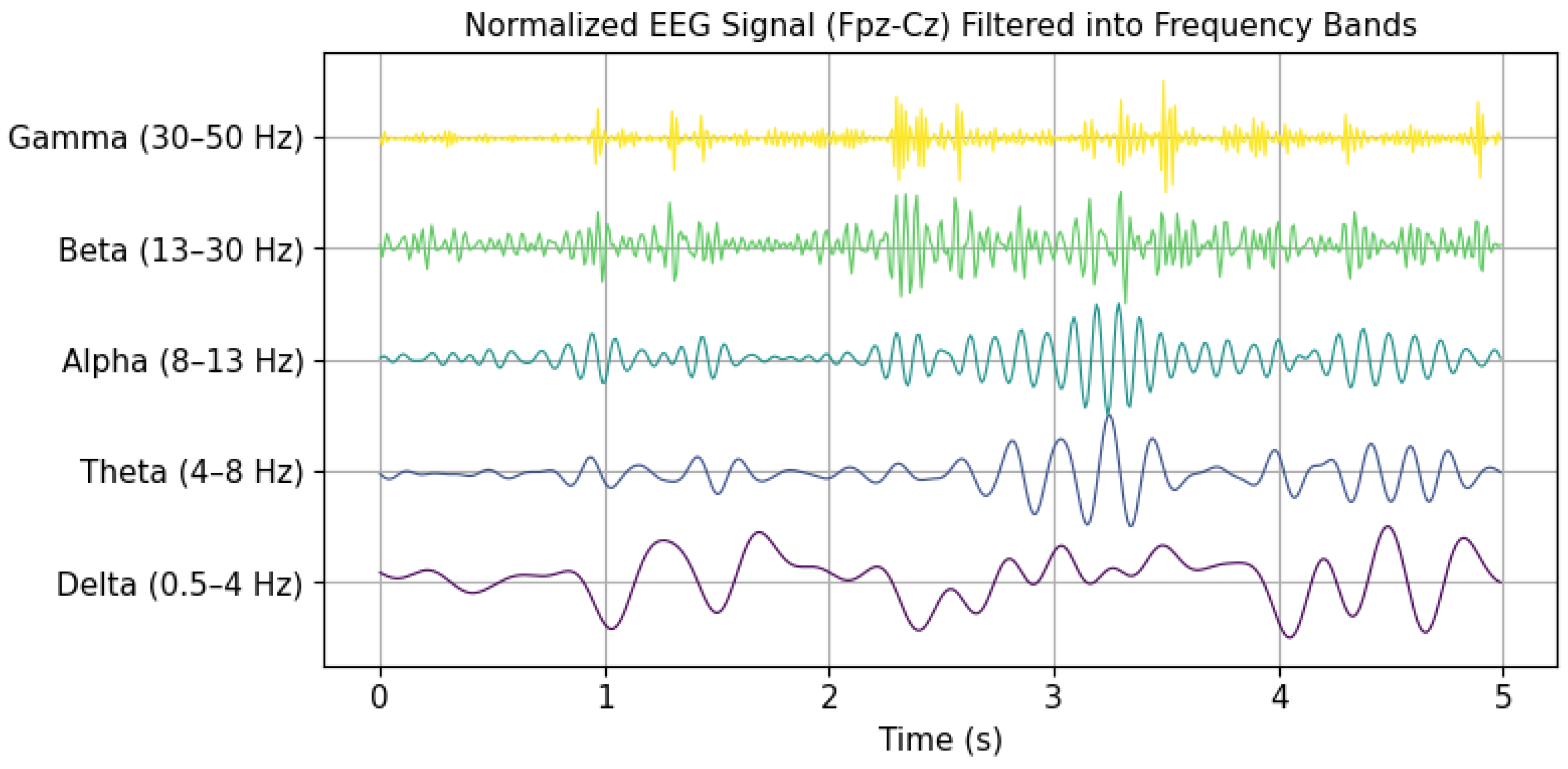
Information for the analysis of brain activity during sleep is obtained through EEG recordings. These signals are characterized by their non-linear, nonstationary, and often random nature, with voltage amplitudes ranging from 5
V to
V [
16]. EEG signals are classified into five main wave types, varying in frequency from 0.5 to 128 Hz, and are identified by Greek letters: gamma (30–128 Hz), beta (13–30 Hz), alpha (8–13 Hz), theta (4–8 Hz), and delta (0.5–4 Hz) [
12].
Figure 3 presents a temporal representation of brain waves over a 5 s interval. These waves were extracted from a signal within the Sleep EDF Expanded database. In this case, gamma wave activity above 50 Hz was discarded due to the 100 Hz sampling frequency, in accordance with the Nyquist criterion. Electrical patterns captured by EEG, such as variations in amplitude, frequency, and waveform morphology, are key indicators in identifying and characterizing different stages of sleep.
The first set of rules for categorizing sleep stages was introduced by Rechtchaffen and Kales in 1968, known as the R&K rules [
17]. These rules define a sleep cycle with a duration between 90 and 110 min, comprising wakefulness (W), rapid eye movement (REM, hereafter referred to as R) sleep, and non-REM (NREM) sleep stages. The NREM phase consists of four distinct stages: S1, S2, S3, and S4. Sleep stages are typically assessed in 30 s intervals. In
Table 1, each sleep stage with its physiological characteristics is presented [
17].
In 2007, the AASM revised the classification system to improve its clinical applicability. The update merged states S3 and S4 into a unified stage, designated N3, while stages S1 and S2 were changed to N1 and N2, respectively. Furthermore, the wakefulness stage was renamed wake [
18,
19].
This research presents a deep neural network framework for the classification of sleep stages using EEG data. Following the AASM guidelines, the classification targets the five main sleep stages: W, R, N1, N2, and N3. To address class imbalance, random undersampling at the epoch level is applied. The proposed architecture integrates convolutional neural networks (CNN) and bidirectional long short-term memory (BiLSTM) networks, further enhanced by an attention mechanism. This approach aims to eliminate the need for hand-crafted feature design and to enable the processing of extensive amounts of EEG data.
The methodology follows a structured pipeline consisting of five key steps: data acquisition and preprocessing, data splitting, class balancing, model training, and performance evaluation. Initially, EEG signals are obtained from PSG data along with the corresponding annotations from hypnogram (HYP) files. The signals then undergo preprocessing steps. Subsequently, the dataset is randomly partitioned into training, validation, and testing subsets. To address class imbalance, this work proposes an epoch-level random undersampling (E-LRUS) technique, applied to the training and validation sets. In the model training phase, deep learning architectures, specifically CNNs in combination with BiLSTM, are implemented to learn features directly from the EEG signals. In addition, an attention mechanism is integrated into the architecture to assign varying importance to different time steps. Finally, the trained models are evaluated using standard machine learning metrics to assess the classification performance.
Some contributions of this study include (1) the application of E-LRUS to address class imbalance across the training and validation sets; (2) the use of a hybrid deep learning architecture combining convolutional neural networks and bidirectional long short-term memory layers, enabling the extraction of both spatial and temporal features from raw EEG data; (3) the incorporation of an attention-based strategy that enhances the model’s ability to prioritize the most informative segments within each EEG sequence; (4) the development of a full deep learning pipeline that avoids manual feature engineering, supporting scalability and the efficient processing of large EEG datasets; and (5) the use of the publicly available Sleep-EDF Expanded dataset, promoting reproducibility and enabling direct benchmarking against existing approaches.
The remainder of this paper is structured as follows:
Section 1 introduces the study;
Section 2 discusses related works;
Section 3 explains the developed methodology;
Section 4 includes the data, experimental findings, and analysis; and, finally,
Section 5 summarizes the conclusions and outlines possible future research.
2. Related Works
Classifying sleep stages using PSG recordings has been the objective of considerable research. Several works focus on EEG signals, as they provide valuable frequency-based insights into brain function in all stages of sleep. These signals, being one-dimensional, also offer the advantage of reducing the computational complexity compared to multimodal approaches, making them especially attractive for both clinical and computational applications. Most recent approaches leverage these EEG signals through machine learning or deep learning techniques, often reporting standard performance metrics such as accuracy or F1-scores on epoch-based datasets.
Many studies have used feature extraction in combination with classical classification algorithms. A notable early study by Doroshenkov et al. [
20] leveraged the temporal structure of EEG signals, incorporating a hidden Markov model (HMM) to model state transitions in sleep stages. This model was enhanced by extracting features by applying a fast Fourier transform (FFT) using a Hamming window, which led to accuracy of 61.08%. Another study, conducted by Vural and Yildiz [
21], assessed various EEG-derived features for the classification of the sleep stage. Their methodology included temporal features such as the mean value, standard deviation, and Hjorth parameter, along with frequency-based attributes such as the relative spectral power, spectral centroid, and spectral bandwidth. In addition, they incorporated time–frequency characteristics by combining both domains. Using principal component analysis (PCA) as a dimension reduction method, they achieved accuracy of 69.98%. Focusing on efficient classification, Huang et al. [
22] proposed a low-computational-cost system designed for implementation in portable sleep monitoring devices. Their method used power spectral density (PSD) features obtained through a short-time Fourier transform (STFT) with a window overlapped in 0.5 s. To optimize feature selection, they applied prototype space feature extraction (PSFE) with fuzzy C-Means (FCM) clustering, alongside reduction methods including linear discriminant analysis (LDA) and PCA. Then, a support vector machine (SVM) was employed in the classification using a radial basis function (RBF) kernel, which produced accuracy of 70.92%. Other researchers have explored alternative classification strategies. Sanders et al. [
23] focused on EEG-based spectral analysis, extracting features such as cross-frequency coupling (CFC) and spectral power across the delta, theta, alpha, beta, and gamma bands. Their approach used a continuous wavelet transform (CWT) with a Morlet wavelet to analyze phase–amplitude coupling, and classification was performed using LDA with cross-validation, resulting in accuracy of 75%. To further improve the classification accuracy, Nateghi et al. [
24] introduced a novel approach using a Kalman time-varying autoregressive filter (TVAR) for feature extraction. Their classification process was divided into two steps: first, distinguishing between the stages W, REM, and non-REM, followed by refining the classification of the NREM substages (N1, N2, and N3). Using LightGBM and XGBoost classifiers, their system achieved accuracy that exceeded 77% with an average F1-score of 69%. Clustering-based methodologies have also been explored. Shuyuan et al. [
25] proposed an improved K-Means clustering algorithm for EEG data segmentation. Unlike traditional K-Means, their method selected cluster centers based on the point density, addressing issues like initialization sensitivity and outliers. The cluster centroids were refined using the “Three Sigma Rule” during iterations, leading to classification accuracy of 75% in five sleep stages. Herrera [
26], who used self-organizing maps (SOM) to create a structured EEG feature space, presented an alternative representation-based technique. This method was complemented by a feature selection process using mutual information (MI) criteria, which identified the most relevant variables for classification. Using an SVM with an RBF kernel and hyperparameter tuning via cross-validation, this approach yielded a classification accuracy score of 70.43%.
Recent advances in deep learning have significantly improved the sleep stage classification performance. Unlike traditional methods that rely on manually crafted features, deep learning models can automatically extract relevant patterns from raw EEG signals [
27]. One of the first CNN-based approaches was introduced by Tsinalis et al. [
5], who designed a convolutional neural network for the classification of sleep stages using single-channel EEG data. Their architecture consisted of a pair of convolutional layers that incorporated a pooling function, connected to dense layers, and a softmax classifier. They employed rectified linear activation functions and used a cross-entropy loss criterion. The EEG signals were preprocessed using wavelet analysis, and class balancing was achieved through stochastic gradient descent with weighted sampling. Their model achieved accuracy of 74%. More advanced CNN architectures have been developed since then. Sors et al. [
7] proposed a CNN model that classified 30 s EEG epochs while incorporating contextual information from neighboring epochs. The training process used the ADAM optimizer with a data distribution of training (50%), validation (20%), and testing (30%). Their system demonstrated high accuracy of 87%. Hybrid deep learning models have also gained traction in sleep stage classification. Yang et al. [
28] developed a nine-layer CNN-BiLSTM architecture that combined convolutional layers for feature extraction, BiLSTM layers for temporal modeling, and residual connections. Given the unbalanced distribution of data between categories of sleep, they implemented adaptive learning rate adjustments to avoid overfitting in the CNN layers. In addition, gradient clipping was applied in the BiLSTM layers to stabilize the training. The model achieved accuracy of 84.1%. Another hybrid model, SleepEEGNet, was proposed by Mousavi et al. [
29]. The architecture combined CNN layers with BiLSTM layers that functioned as both encoders and decoders. Their system reached 84.26% precision, an F1-score of 79.66%, and a Cohen’s kappa coefficient of 0.79. Similarly, Toma et al. [
30] introduced a multichannel CNN-BiLSTM model that integrated EEG (Fpz-Cz, Pz-Oz), EOG, and EMG signals. Their two-stage design leveraged pretrained dual-channel modules combined via transfer learning. The architecture reported accuracy of 91.44% with an F1-score of 88.69% using the Sleep-EDF 20 dataset, as well as accuracy of 90.21% over the extended Sleep-EDF 78 version.
Despite these improvements, certain challenges remain in classifying sleep stages, including the difficulty in accurately identifying stage N1, which is often underrepresented in datasets due to its short duration within the sleep cycle, and issues related to class imbalance. However, the ongoing development of deep learning methodologies and feature engineering strategies has contributed to increased classification accuracy. The use of models of hybrid neural networks and advanced preprocessing techniques suggests promising directions for the construction of improved models for the detection of sleep stages.
4. Results and Discussion
In the context of implementing deep learning approaches for multiclass classification tasks, each decision significantly impacts the performance. Selecting a suitable sleep dataset for research, ensuring a computational environment capable of processing large amounts of raw data, and preprocessing signals are essential steps in this workflow. In addition, aspects related to the neural network, such as the parameter setup, architecture, training process, and performance evaluation metrics, are fundamental considerations that must be addressed and can be experimentally adjusted to optimize model performance. These considerations provide the foundation for optimizing the effectiveness of the model when evaluating sleep stage classification using EEG signals. To validate these considerations, the following experiments are presented.
4.1. Dataset
In this study, the Sleep-EDF Expanded Database is considered, which is widely recognized in EEG-based sleep research [
72]. This dataset contains 197 full PSG recordings in EDF format, with multiple sessions from 78 subjects (healthy and with sleep disorders) between 25 and 101 years of age. These data were collected during approximately 20 consecutive hours of sleep monitoring in a home environment [
33]. The recordings comprise several physiological signals: two EEG signals, EOG, submental EMG, event makers, respiration, and body temperature, all stored in EDF format with a 16-bit resolution. Each EDF file is accompanied by metadata, including patient information, recording settings, signal specifications, and administrative details. The EEG channels (Fpz-Cz and Pz-Oz) follow the 10–20 international system, covering the frontal and occipital regions, as suggested by the AASM. EEG and EOG signals were sampled at 100 Hz, while the remaining signals were sampled at 1 Hz [
33]. The recordings are divided into 30 s epochs and each labeled according to the R&K guidelines as “W”, “1”, “2”, “3”, “4”, “R”, “movement”, and unlabeled segments.
4.2. Experimental Environment
Deploying deep learning architectures with raw EEG data requires significant computational resources. All experiments were carried out on a device equipped with a 13th-gen Intel Core i9-13900K processor (3.00 GHz), an Nvidia RTX 4070 GPU, and 16 GB RAM, running Windows 11 Pro 64-bit. The development environment included Python 3.9.0, using frameworks like PyTorch, NumPy, Scikit-learn, and PyEDFlib. Visualization was performed using Matplotlib V. 3.10.0 and Seaborn V. 0.13.2. This configuration provided the computational power necessary to carry out all the experiments presented in order to achieve the presented results.
4.3. Data Preprocessing
To ensure the consistency and relevance of the input data, several preprocessing steps were applied to the EEG recordings prior to model training. First, the Fpz-Cz EEG channel was extracted from each EDF file to adapt the input to a single-channel architecture, minimizing the complexity while preserving critical information for sleep stage classification. To improve the data quality and address class imbalance, only epochs with valid sleep stage annotations were retained. Epochs labeled “movement” or left unannotated were discarded entirely because they introduced noise and lacked ground truth. Furthermore, epochs labeled stage W were selectively retained only during transitional periods around the onset of sleep and awakening; this was done to avoid the overrepresentation of wakefulness and to balance the distribution between classes. According to the AASM guidelines, stages 3 and 4 were combined into the N3 category. The final class labels were then numerically encoded as follows: N1 = 0, N2 = 1, N3 = 2, R = 3, and W = 4. After class filtering and relabeling, all EEG signals were normalized to the range [0, 1] using min–max normalization. No additional artifact rejection or filtering was applied, following an end-to-end modeling strategy where the model learned directly from the raw normalized signal.
Regarding data organization, complete EDF files were stored per patient. The dataset was divided by assigning entire EDF files: 60% for training, 20% for validation, and 20% for testing. This prevented the split of files across subsets, ensuring full recordings per patient and a subject-independent evaluation. To address class imbalance, the E-LRUS technique was applied to the training and validation sets, promoting balanced learning and better model generalization. This preprocessing pipeline is designed to maintain data integrity, reduce bias, and enable robust subject-level evaluation, ensuring that the model is trained and tested under realistic and representative input conditions.
4.4. Proposed Neural Network Architecture
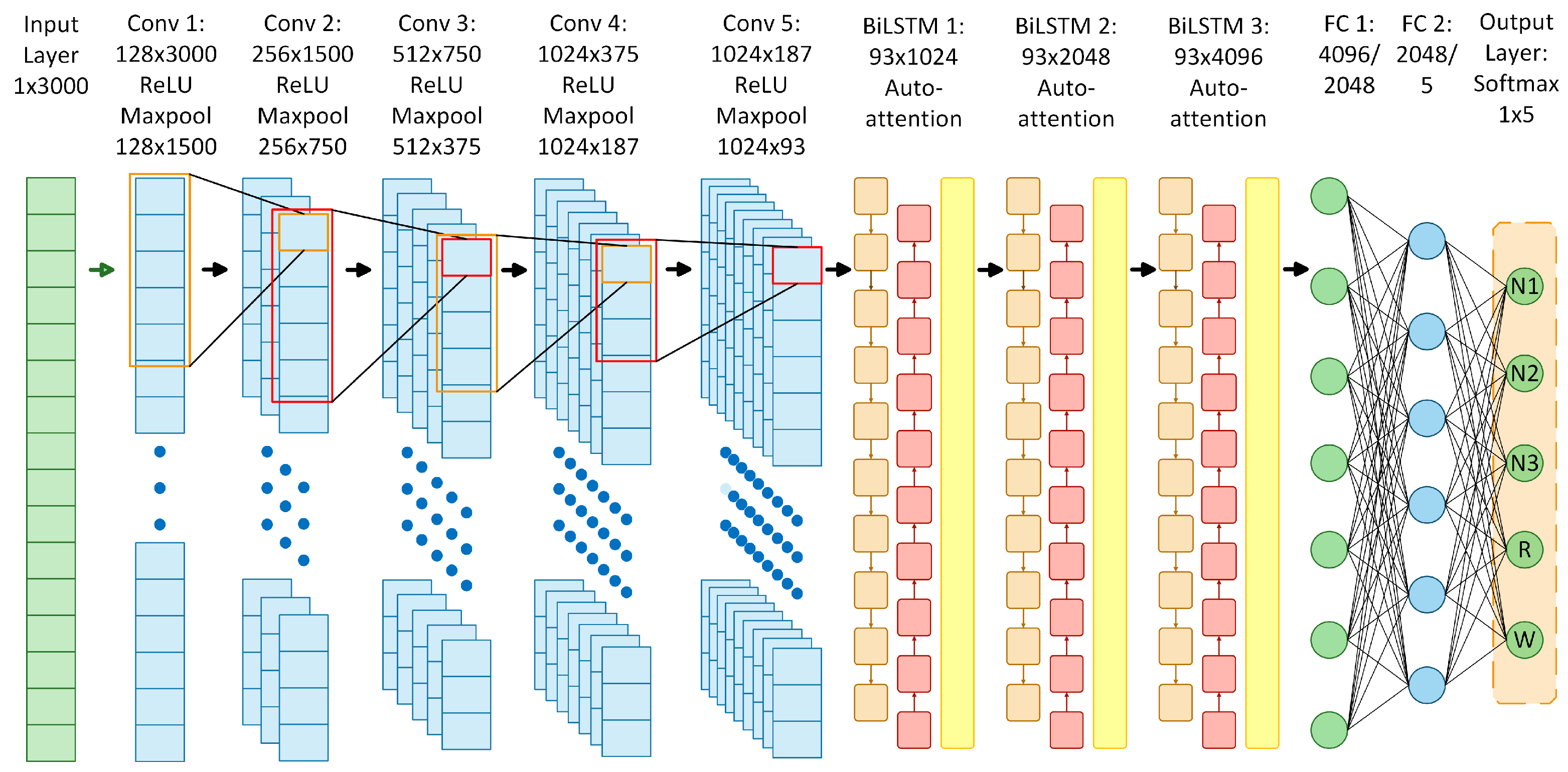
The proposed neural network architecture was designed to perform sleep stage classification in EEG recordings using a combination of CNNs, BiLSTM, and self-attention mechanisms. It consists of five main components.
1D Convolutional Layers: Five layers with increasing filters (from 128 to 1024), each with ReLU activation, maximum pooling operations, and batch normalization. The choice of five layers adds depth to capture complex EEG features, while max pooling enables hierarchical extraction and dimensionality reduction, allowing the efficient processing of long sequences.
Bidirectional LSTM Layers: Three BiLSTM layers with 512, 1024, and 2048 memory cells, respectively. The use of three layers helps to capture complex long-term temporal dependencies in both directions, supporting the modeling of the sequential nature of sleep stages.
Self-Attention Layers: Placed after each BiLSTM, these dynamically weight the hidden states over time. This allows the model to focus on the most relevant temporal features, complementing BiLSTM’s ability to capture dependencies and improving the discrimination of transient or subtle sleep stage patterns.
Fully Connected Layers: Two dense layers with 2048 and 5 outputs, respectively, with ReLU activation. The first dense layer reduces the high-dimensional LSTM output to 2048 units for feature consolidation, while the second generates classification scores for the five sleep stages.
Softmax Output Layer: A final softmax output layer that maps the outputs to probabilities for each of the five sleep stages: N1, N2, N3, R, and W.
The number of layers and units was empirically selected based on the validation performance in multiple configurations. Although increasing the complexity of the model generally improved the performance, the upper limit was constrained by the available computational resources. To efficiently process long EEG sequences and handle large volumes of data, the model leverages hierarchical convolutional feature extraction and max pooling to reduce the dimensionality. This reduction, combined with dense layers, enables scalability to extended recordings despite computational limitations. In addition, attention mechanisms were incorporated to explore their potential impact on the capture of important temporal features in EEG signals and to selectively emphasize informative EEG segments relevant to each stage of sleep. The scheme of the proposed model architecture is shown in
Figure 11, which illustrates the arrangement of the layers involved in the learning process. This architecture yielded the best classification performance among several alternatives and was used in subsequent experiments.
4.5. Parameter Configuration
After completing data preprocessing and defining the architecture of the neural network, the model training process was carried out. The loss function used was the categorical cross-entropy, handling non-numeric classes probabilistically. The SGD, RMSprop, and ADAM optimizers were tested, with the latter yielding the best results. Since ADAM adjusts the learning rate automatically, it was not specified manually. In the context of regularization techniques, various dropout values between 0 and 1 were tested, and a value of 0.7 led to the best model performance. Furthermore, weight decay was applied using a penalty coefficient of . The signals were divided into 30 s sequences. For EEG signals sampled at 100 Hz, this is equal to 3000 samples per sequence. During the application of E-LRUS, the training set was balanced to 537 epochs per class, based on the least represented class, N1. Several batch sizes (16, 32, 64, and 128) were tested, with the best results achieved with 64 samples per batch. The learning cycles were set to a total of 100 epochs, with early stopping configured at 30 epochs. These parameters were fine-tuned across training sessions and corresponded to those that led to the best-performing model.
4.6. Network Training
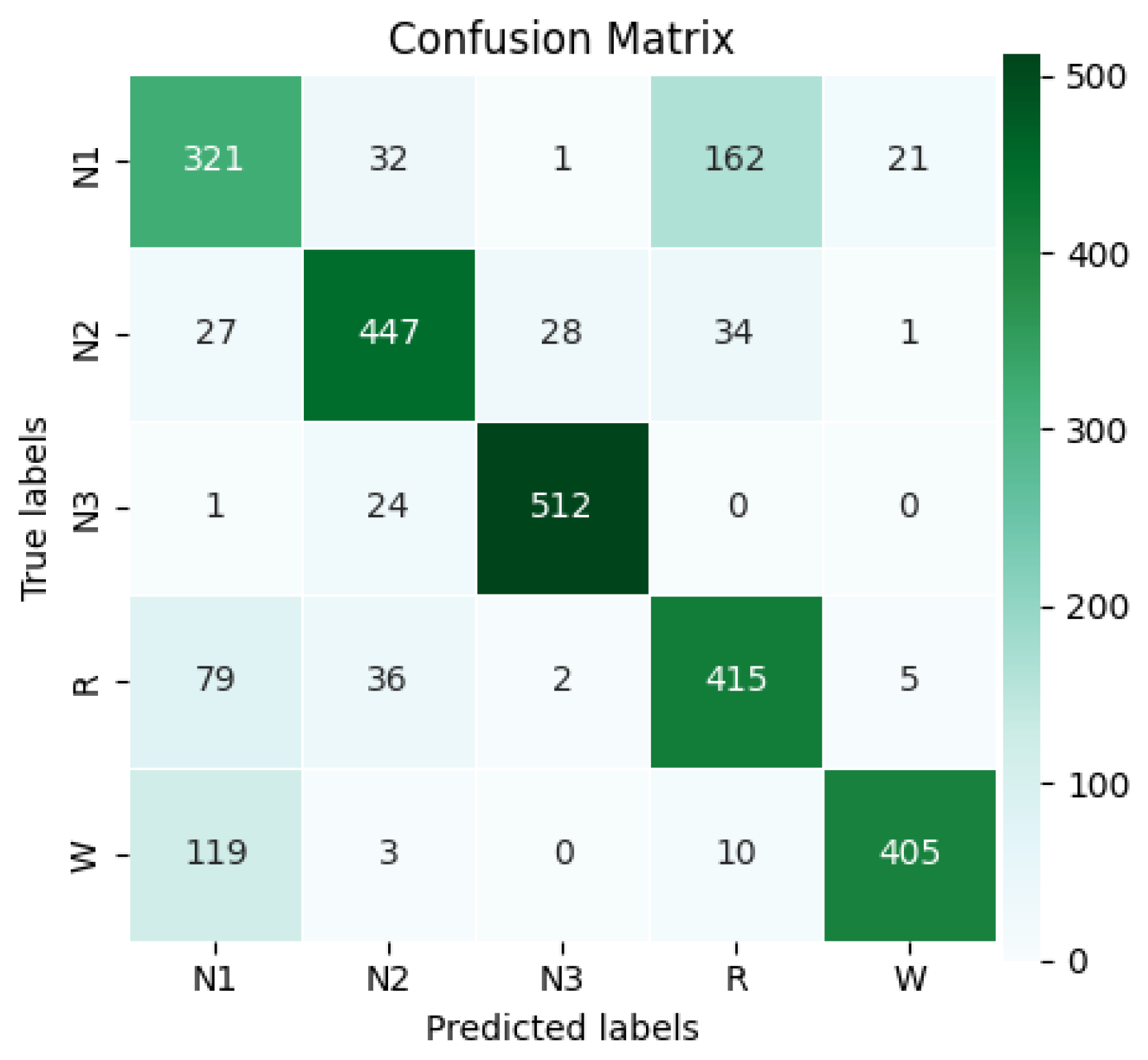
After each training iteration, the model evaluation on the validation data was performed by selecting the epoch where the validation loss reached its lowest point. The model was trained for up to 77 epochs, but early stopping with a patience value of 30 epochs was triggered at epoch 47, where the best loss was recorded. Due to the complexity of the network and the large EEG data, training required high-performance hardware and optimizations such as early stopping and batch tuning. The layer-wise computational cost analysis in the Methodology section highlights performance impacts, helping to identify bottlenecks and architectural optimization to ensure a balance of accuracy and efficiency. After completing the training session, the confusion matrix of the classification results was obtained, as shown in
Figure 12.
This matrix represents 2685 predictions made by the model, corresponding to the processing of 8,055,000 samples, with 3000 samples per example and 537 examples per class. Class N1 had the fewest correct predictions, with a total of 321, representing the poorest performance. The accuracy for classes N2, R, and W was similar, achieving 447, 415, and 405 correct predictions, respectively. However, class N3 performed the best, with 512 correct predictions. The model achieved overall accuracy of 78.21%. Using the confusion matrix, the
,
,
, and
were calculated for each class. The results of the calculation of the metrics per class, including the precision, recall, F1-score, specificity, NPV, and Jaccard index, are shown in
Table 3. The classification results for each class are detailed below.
N1: Class N1 was the most challenging for the model. The precision was 58.68% and recall was 59.78%, with an F1-score of 59.23%, indicating that the model correctly identified this class just over half the time, showing balanced but modest performance. The specificity was 89.48%, and the NPV was 89.90%, showing that the model is more reliable in rejecting this class than confirming it. The Jaccard index was 42.07%, reflecting weak agreement between the predictions and true labels.
N2: Class N2 showed solid and balanced performance. The precision was 82.47% and recall was 83.24%, with an F1-score of 82.85%, reflecting good accuracy and detection and overall consistent results. The specificity was 95.58% and the NPV was 95.80%, showing effective distinction from other classes. The Jaccard index was 70.73%, indicating strong classification consistency.
N3: Class N3 achieved the best overall performance. The precision was 94.29% and recall was 95.34%, with an F1-score of 94.81%, demonstrating excellent accuracy and sensitivity and outstanding balanced performance. The specificity was 98.56%, and the NPV was 98.83%. The Jaccard index was 90.14%, showing strong agreement between the predictions and actual values.
R: For class R, the precision was 66.83% and the recall reached 77.28%, with an F1-score of 71.68%, indicating moderate precision with fairly good sensitivity and acceptable overall performance. The specificity was 90.41% and the NPV was 94.09%, reflecting the good detection of negatives. The Jaccard index was 55.85%, indicating moderate agreement between the predicted and true labels.
W: Class W achieved 93.75% precision and 75.42% recall, with an F1-score of 83.59%, showing high precision and a good detection rate. The specificity was 98.74% and the NPV was 94.14%, demonstrating the model’s ability to avoid false positives. The Jaccard index was 71.81%, indicating good overall performance.
After computing the individual metrics per class, the macro-average for each metric was obtained. Additionally, Cohen’s kappa coefficient was calculated, resulting in a value of 0.7277, which indicates strong concordance between the predicted and actual class labels, with minimal influence of chance. The performance model metrics calculated in the validation step are presented in
Table 4.
Based on the previous results, the accuracy of 78.21% suggests that the system successfully classified most of the cases. The 79.20% precision indicates a strong ability to correctly predict positive cases, with a low false positive rate. The recall of 78.21% demonstrates the consistency of the model in detecting actual positive cases. The F1-score of 78.43% shows the harmony between precision and sensitivity. The high specificity and NPV, both at 94.55%, indicate strong performance in correctly identifying negative cases. Meanwhile, the Jaccard index of 66.12% suggests that the model can still improve in terms of achieving an exact match between the predictions and the class labels. Finally, the Cohen’s kappa value of 72.77% shows substantial alignment between the predicted outcomes and actual labels, largely excluding chance. These validation results show solid and consistent performance, suggesting that the model was effectively trained. They provide a reliable basis for the final evaluation.
4.7. Model Evaluation
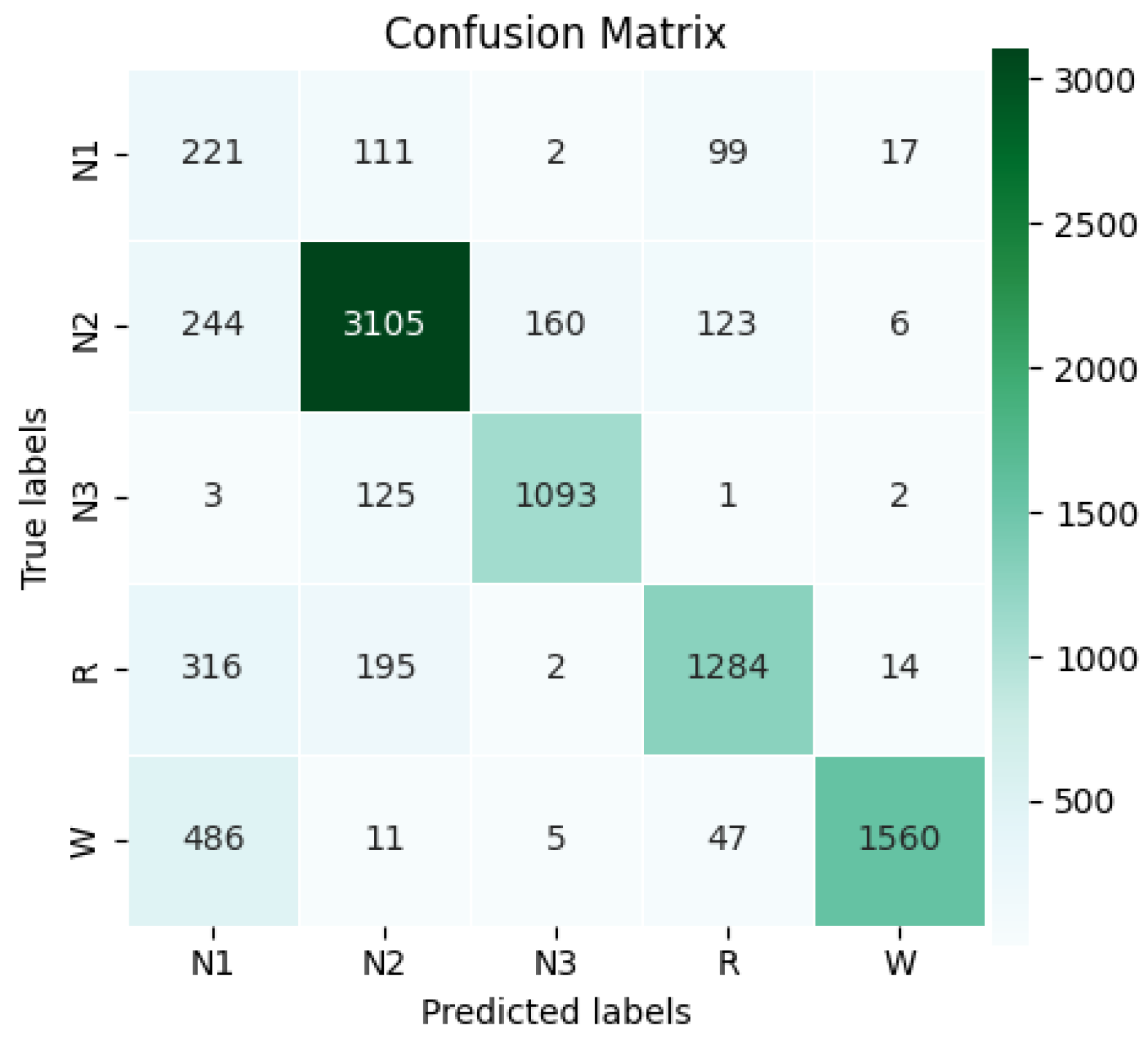
Once the best model was trained, its performance was evaluated in the test set. The confusion matrix shown in
Figure 13 summarizes the classification results by class. Since this set was neither preprocessed nor balanced, the natural class distribution was maintained, allowing the model’s performance to be observed in a real-data context. In total, 9232 test examples were used, equivalent to 276,960 s of recordings (27,696,000 samples). The class distribution was as follows: N1 = 450, N2 = 3638, N3 = 1224, R = 1811, and W = 2109. This distribution is typical in sleep analysis, where N1 is often underrepresented and N2 is usually the most frequent stage, reflecting the natural prevalence of certain sleep cycle patterns.
According to the confusion matrix, the model correctly classified 221 N1 examples, 3105 N2, 1093 N3, 1284 R, and 1560 W. With these results, the overall accuracy of 78.67% was calculated for the test set, demonstrating the model’s ability to correctly classify most examples not used during training. Detailed class-wise metrics are shown in
Table 5. A summary of the performance metrics per class is presented below.
N1: Class N1 remained the hardest to predict. The precision was 17.40% and recall 49.11%, with an F1-score of 25.70%, revealing limited performance in this class but with some success in detection. The specificity was 88.06% and the NPV 97.12%, indicating more reliable rejection than detection. The Jaccard index was 14.74%, reflecting poor overlap between the predictions and the true labels.
N2: Class N2 maintained strong metrics. The precision was 87.54% and recall 85.35%, with an F1-score of 86.43%, indicating effective classification with balanced accuracy and coverage. The specificity was 92.10% and the NPV 90.62%, showing the reliable identification of non-N2 cases. The Jaccard index was 76.10%, confirming the consistent predictions.
N3: Class N3 produced the most reliable results. The precision was 86.61% and recall 89.30%, with an F1-score of 87.93%, reflecting both accurate predictions and strong sensitivity. The specificity reached 97.89% and the NPV 98.36%, with a Jaccard index of 78.46%, showing a high degree of overlap with the true labels.
R: For class R, the precision was 82.63% and recall 70.90%, with an F1-score of 76.32%, showing decent performance with slightly lower coverage. The specificity was 96.36% and the NPV 93.14%, indicating the effective rejection of incorrect cases. The Jaccard index was 61.70%, reflecting moderate agreement.
W: Class W achieved 97.56% precision and 73.97% recall, with an F1-score of 84.14%, suggesting high accuracy and good but improvable detection. The specificity was 99.45% and the NPV 92.81%, highlighting an excellent ability to avoid false positives. The Jaccard index was 72.63%, supporting its solid overall performance.
These results reveal varying performance across different classes, with N1 being the most difficult to classify and N3 performing the best. The poor performance of N1 in several metrics can be attributed to its underrepresentation in the data, a common challenge in unbalanced classification problems. In contrast, classes N2, N3, R, and W showed consistent and balanced performance. These differences underscore the importance of addressing class imbalance to improve the accuracy and balance in predictions. After calculating the metrics for each class, macro-average metrics were derived, and Cohen’s kappa coefficient was calculated for the test set matrix, yielding a value of 73.39%, confirming the model’s consistency and the validity of its predictions. The results of the overall test metrics are presented in
Table 6. In the following, a comparative analysis between the test results and validation results is provided.
Overall accuracy: The accuracy on the test set was 78.67%, indicating relatively accurate classification in general. This result is satisfactory, especially given the significant class imbalance in this multiclass classification problem. The close match with the accuracy of 78.21% in the validation set suggests good consistency between model training and generalization to new data.
Precision: The precision of 74.35% indicates an acceptable rate of correct predictions for the positive class. However, compared to the precision of 79.20% on the validation set, it can be inferred that the underrepresentation of class N1 limited the performance in the test set.
Recall: The recall of 73.73% indicates that the model successfully classified most of the positive samples. This is consistent with the recall of 78.21% from the validation set, indicating that the model’s capacity to identify positive classes showed minimal variation between the two sets.
F1-Score: The F1-score of 72.10% indicates a reasonable equilibrium between the precision and recall metrics. The drop in the F1-score from the validation set, where it was 78.43%, is mainly due to the significant decrease in precision for class N1 in the test set.
Specificity: The specificity of 94.77% suggests excellent performance in identifying negative samples, consistent with the specificity of 94.55% from the validation set, reflecting a low false negative rate.
NPV: The NPV of 94.41% indicates the effectiveness of the model in accurately identifying negative classes, corroborated by the NPV of 94.55% observed in the validation set.
Jaccard Index: The Jaccard index of 60.73% indicates acceptable similarity between the model outputs and the actual labels, although there is room for improvement compared to the Jaccard index on the validation set of 66.12%. The difference is mainly due to the class imbalance for N1.
Cohen’s Kappa: Finally, the Cohen’s Kappa score of 73.39% indicates a high degree of consistency between the predictions and the true labels, which implies that the model’s decisions are not coincidental. This value, slightly higher than the 72.77% from the validation set, suggests consistent performance in generating reliable agreements.
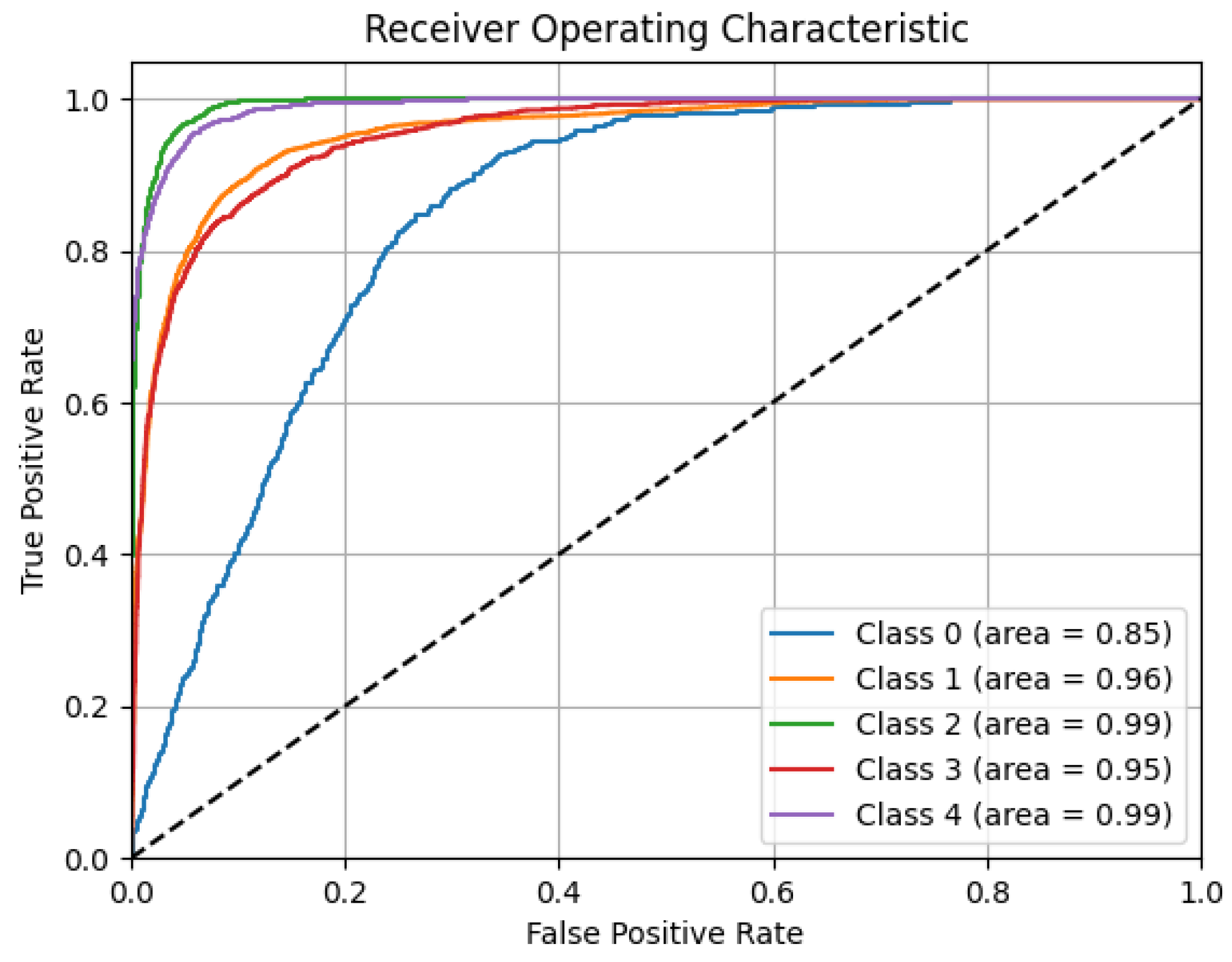
The results show the model’s overall strong performance, effectively managing class imbalances and excelling in identifying negative classes. Although there is room for improvement in classifying less frequent categories, the model maintains a good balance between precision and recall, minimizes random predictions, and generalizes well across datasets. To reinforce these outcomes,
Figure 14 presents the overall ROC curve of the model, which reflects the model’s ability to differentiate among classes using different decision thresholds.
The area under curve (AUC) values reflect high performance for nearly all classes: W and N3 both achieve an outstanding AUC of 0.99, while R and N2 also produce excellent results, with AUCs of 0.95 and 0.96, respectively. Although the N1 class has a lower AUC of 0.85, it still indicates an acceptable discriminative ability, especially considering its low representation in the data distribution. The ROC curves illustrate the model’s strong ability to distinguish between sleep stages, maintaining high true positive rates while minimizing false positives, even with unbalanced class distributions.
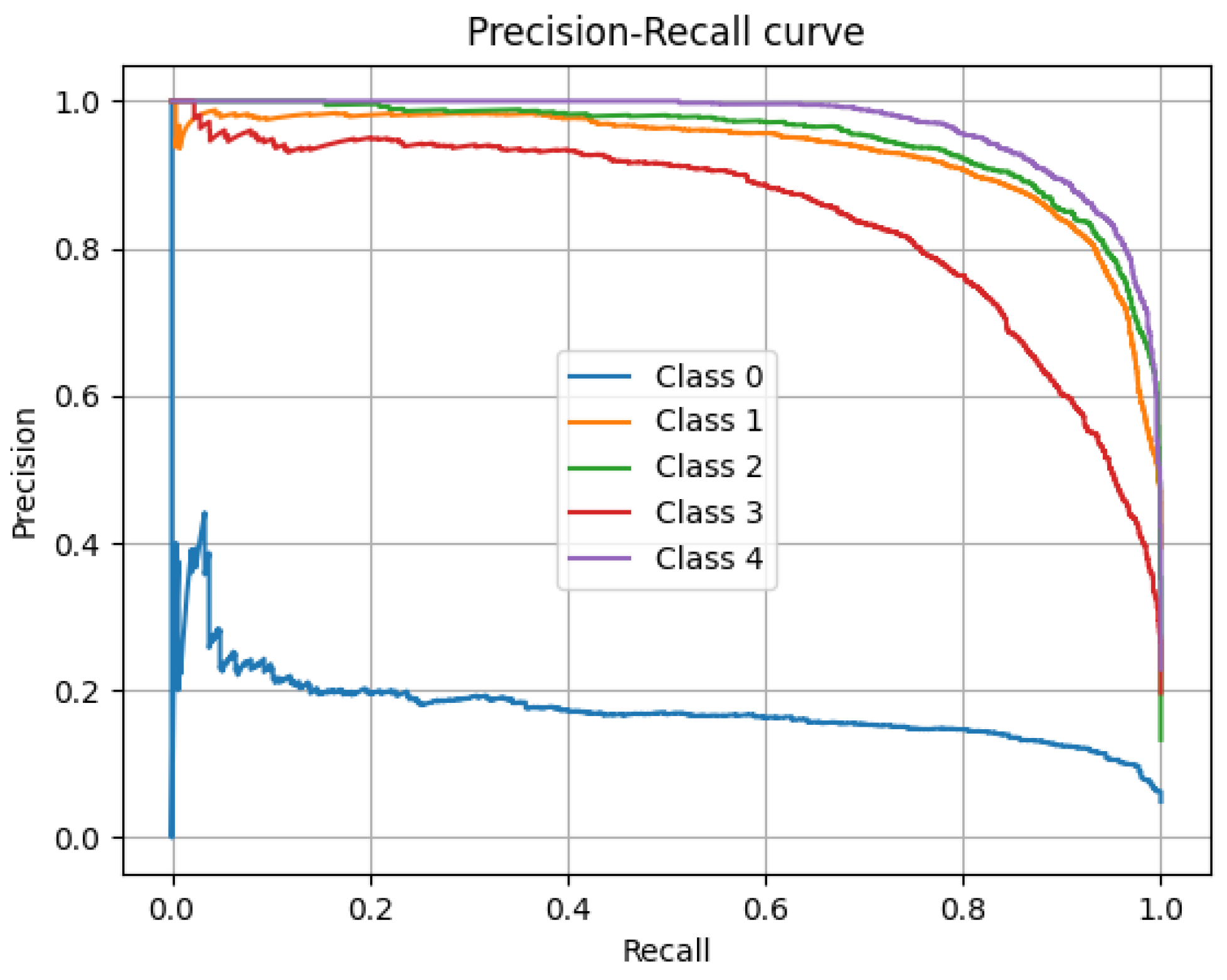
Complementarily, the model’s overall precision–recall curve, particularly valuable in evaluating performance under class imbalances, is presented in
Figure 15. The curves for classes 1 through 4 (N2, N3, R, and W) remain high, indicating a favorable equilibrium between recall and the correctness of positive predictions. In particular, N3 and W exhibit the most stable and elevated curves, maintaining high precision levels even with increasing recall. In contrast, the precision of class N1 decreases as the recall increases, reflecting the difficulty of the model in identifying this class without generating false positives. Although the model exhibits strong results across most stages, improvements are still needed to handle less frequent categories.
The per-class metrics and the analyses conducted confirm the model’s classification capabilities across various sleep stages, despite variations due to class imbalance. Although N1 performs poorly relative to more prevalent stages, such as N2 and N3, the model manages the dominant classes well, with a good balance between precision and recall. Global metrics such as the F1-score and Jaccard index indicate its solid classification capacity, which could be further improved by addressing the dataset imbalance. The ROC and precision–recall curves reinforce the evidence of the model’s reliable discriminative power. Despite its limitations with minority classes, the model demonstrates strong generalization and overall robustness. The next section will provide a detailed discussion of these results, their implications, and comparisons with previous studies to guide future model development.
4.8. Discussion
When the process moves from validation to testing, the model still filters out negative examples reliably, showing that its nonstage rule holds up in real data. There is a small but expected drop in capturing true positives for the rare stages when faced with the natural imbalance of the test set; this does not negate our results but points to areas for fine-tuning (for example, through rebalancing or class-aware loss). The overall agreement remains similar, meaning that most errors happen in the least common classes, while the major stages remain solidly identified. These findings show that the model is generally robust and suggest simple steps to strengthen its ability to identify underrepresented patterns without degrading its strong performance on the more frequent ones.
To validate the effectiveness of the proposed model, we compare its accuracy with that of leading sleep stage classifiers (
Table 7). The traditional methods—K-Means (75%) [
25], SVM (70.4%) [
26], CFC + LDA (75%) [
23], and LGBM + XGBoost (77%) [
24]—achieve reasonable results but lack the ability to capture subtle time-dependent features (e.g., sleep spindles, K-complexes). They also rely on manual feature extraction, which demands expert knowledge, limits scalability, and hinders practical deployment. Deep learning models extract features directly from raw data, streamlining processing and improving the scalability and generalization to large datasets. In Sleep-EDF Expanded, pure CNNs achieve 74% accuracy [
5]. CNN-BiLSTM models improve the performance, reaching 84.1% on ISRUC-Sleep [
28] and 84.26% on Sleep-EDF 13 and 18 [
29], all using single-channel EEGs. Meanwhile, a multichannel approach on Sleep-EDF 20 and 78 achieves 91.44% accuracy [
30]. Hybrid models combining CNNs and LSTM achieved the best performance in capturing EEG dynamics. However, based on the computational cost formulas outlined in the Methodology section, pure CNNs may be significantly more efficient, as their structure favors parallel processing. This highlights the importance of weighing accuracy against resource demands in real-world applications.
The proposed CNN–BiLSTM–Attention model achieves an overall accuracy rate of 78.67%, slightly below the top-performing models [
28,
29,
30]. However, our approach includes methodological elements that aim to improve the generalization and control bias. In particular, we introduce E-LRUS, which subsamples entire 30 s segments to preserve temporal coherence, unlike conventional oversampling or synthetic methods. This enabled the class N3, which was the second most underrepresented after N1, to achieve the highest performance (precision: 86.61%, recall: 89.30%, F1-score: 87.93%), outperforming more frequent classes like N2 and W. E-LRUS thus facilitates the robust detection of underrepresented classes without skewing toward dominant ones. In contrast, several state-of-the-art studies lack explicit balancing or preprocessing, possibly leading to bias or overfitting despite their higher accuracy.
The method demonstrates solid performance in most sleep stages, with N1 remaining the most challenging to classify. The model effectively distinguishes N1 from N3 due to clear spectral and temporal differences. However, it shows moderate difficulty in separating N1 from N2 during validation and more substantial confusion with W and R, particularly in the test set. These errors likely result from overlapping transient EEG patterns and the inherently variable and transitional nature of N1. Despite class balancing, its low intraclass consistency and the absence of distinct biomarkers continue to limit the classification performance. Further improvements could involve adapting the complexity of the model by adjusting the depth of the network or exploring alternative architectures and training schemes. Repeating E-LRUS sampling with varied random seeds and merging datasets could also increase N1’s representation without adding artificial variability, although care is needed to avoid memorization. Enhancing the N1 classification in these ways would significantly improve the model’s generalizability and clinical applicability.
5. Conclusions
The proposed hybrid architecture, which combines CNNs, BiLSTM layers, and attention mechanisms, demonstrated solid performance by extracting meaningful features from raw EEG data. It achieved overall accuracy of 78.67% and an F1-score of 72.10%, reflecting balanced performance across sleep stages. To improve its generalization and reduce bias, the model incorporates the E-LRUS procedure, which subsamples entire 30 s segments to preserve temporal coherence. This strategy contributed to strong performance in N3, the second most underrepresented stage, with an F1-score of 87.93%, outperforming more frequent stages such as N2 and W. By balancing the training data without altering its natural structure, the model mitigated overfitting and enabled robust detection between stages. This synergy between the architecture and data handling supports a reproducible end-to-end solution for biomedical signal classification.
Future work should address class imbalance through targeted data augmentation and class-aware training, while also integrating time- and frequency-domain processing within deeper or residual network architectures to capture more intricate patterns with fewer parameters. Additional improvements may include refined architectures, multiscale temporal features, temporal context enhancement, and specialized loss functions designed to better detect the underrepresented stage N1. Since model performance depends on factors such as the dataset size, diversity, class imbalance, and design choices, continued efforts could focus on hybrid architectures, advanced data enhancement, and balanced training strategies to improve the reliability, generalizability, and clinical applicability. Incorporating complementary modalities such as EMG or EOG, developing interpretability tools for clinicians, and exploring lightweight model variants suitable for portable devices will further support real-world clinical adoption. Extending this framework to other EEG-based diagnostic tasks could broaden its impact in sleep medicine.