Machine Learning for Chronic Kidney Disease Detection from Planar and SPECT Scintigraphy: A Scoping Review
Abstract
:1. Introduction
- Aassess ML applications for renal SPECT and planar scintigraphy;
- Identify gaps and challenges in ML applications for CKD detection and prognosis;
- Explore related research efforts in ML applications to other scintigraphy domains;
- Set grounds for potential future research pathways in the medical image analysis for SPECT and planar scintigraphy images.
- RQ1: What ML methods are currently utilized for detecting, predicting, and diagnosing CKD using PLANAR and SPECT images in renal scintigraphy?
- RQ2: What ML methods are currently utilized for processing PLANAR and SPECT images across various scintigraphy domains?
- RQ3: What are the challenges and limitations associated with applying ML methods to PLANAR and SPECT images in scintigraphy?
2. Background
3. Methodology
3.1. Search Strategy
3.1.1. Preliminary Screening:Addressing the RQ1
- Not related to planar scintigraphy or SPECT image modalities;
- Not related to the CKD prediction;
- Not related to this scoping review;
- Not written in English language.
3.1.2. Broader Screening: Addressing the RQ2 and RQ3
- Not related to any research question;
- Considers other image modalities;
- Considers dual or multi-modal approach;
- Not written in the English language.
4. Results
4.1. Preliminary Screening; Addressing the RQ1
4.2. Broader Screening; Addressing the RQ2 & RQ3
4.3. Categorization of the Results
4.3.1. Year of Publishing
4.3.2. Type of Research
4.3.3. Research Methods
5. Discussion
5.1. Key Insights
5.2. Data, Data Sources, and Data Utilization
5.3. Research Methods
5.4. Diagnosis of Renal Pathologies
5.5. Advancing Future Research
5.6. Towards Robust and Trustworthy Research
5.7. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AB | Adaptive Boosting |
| AE | Autoencoders |
| AI | Artificial intelligence |
| BC | Bagging Classifier |
| BT | Boosted Tree |
| CAD | Coronary artery disease |
| CAM | Class activation mapping |
| CKD | Chronic kidney disease |
| CNN | Convolutional Neural Network |
| CT | Computed tomography |
| DM | Diffusion Maps |
| DT | Decision tree |
| EHR | Electronic health record |
| ESRD | End-stage renal disease |
| FNN | Feed-forward NN |
| GAN | Generative Adversarial Network |
| GB | Gradient Boosting |
| GFR | Glomerular filtration rate |
| GO | Growth Optimizer |
| KNN | K-Nearest Neighbors |
| LDA | Linear Discriminant Analysis |
| LIME | Local interpretable model-agnostic explanations |
| LR | Logistic Regression |
| ML | Machine learning |
| MLP | Multilayer Perceptron |
| MRI | Magnetic resonance imaging |
| NB | Naive Bayes |
| NN | Neural network |
| PCA | Principal component analysis |
| PET | Positron emission tomography |
| RF | Random Forest |
| SGD | Stochastic Gradient Descent |
| SHAP | Shapley additive explanations |
| SMOTE | Synthetic minority oversampling technique |
| SPECT | Single photon emission computed tomography |
| SR | Scoping review |
| SVM | Support vector machine |
| UE | Ultrasound elastography |
| VGG | Visual geometry group |
| XGB | Extreme Gradient Boosting |
References
- World Health Statistics 2019: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2019.
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef] [PubMed]
- Lei, N.; Zhang, X.; Wei, M.; Lao, B.; Xu, X.; Zhang, M.; Chen, H.; Xu, Y.; Xia, B.; Zhang, D.; et al. Machine learning algorithms’ accuracy in predicting kidney disease progression: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2022, 22, 205. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Lerman, L.O. Prediction of chronic kidney disease progression by magnetic resonance imaging: Where are we? Am. J. Nephrol. 2019, 49, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Fried, J.G.; Morgan, M.A. Renal Imaging: Core Curriculum 2019. Am. J. Kidney Dis. 2019, 73, 552–565. [Google Scholar] [CrossRef]
- Database of Dynamic Renal Scintigraphy. Available online: https://dynamicrenalstudy.org/pages/about-project.html (accessed on 17 May 2025).
- Dietz, M.; Jacquet-Francillon, N.; Sadr, A.B.; Collette, B.; Mure, P.Y.; Demède, D.; Pina-Jomir, G.; Moreau-Triby, C.; Grégoire, B.; Mouriquand, P.; et al. Ultrafast cadmium-zinc-telluride-based renal single-photon emission computed tomography: Clinical validation. Pediatr. Radiol. 2023, 53, 1911–1918. [Google Scholar] [CrossRef]
- Alnazer, I.; Bourdon, P.; Urruty, T.; Falou, O.; Khalil, M.; Shahin, A.; Fernandez-Maloigne, C. Recent advances in medical image processing for the evaluation of chronic kidney disease. Med. Image Anal. 2021, 69, 101960. [Google Scholar] [CrossRef]
- Cheplygina, V.; De Bruijne, M.; Pluim, J.P. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 2019, 54, 280–296. [Google Scholar] [CrossRef]
- Decuyper, M.; Maebe, J.; Van Holen, R.; Vandenberghe, S. Artificial intelligence with deep learning in nuclear medicine and radiology. EJNMMI Phys. 2021, 8, 81. [Google Scholar] [CrossRef]
- Magherini, R.; Mussi, E.; Volpe, Y.; Furferi, R.; Buonamici, F.; Servi, M. Machine learning for renal pathologies: An updated survey. Sensors 2022, 22, 4989. [Google Scholar] [CrossRef]
- Rubini, L.; Soundarapandian, P.; Eswaran, P. Chronic Kidney Disease. UCI Machine Learning Repository. 2015. Available online: https://archive.ics.uci.edu/dataset/336/chronic+kidney+disease (accessed on 23 February 2025).
- Sanmarchi, F.; Fanconi, C.; Golinelli, D.; Gori, D.; Hernandez-Boussard, T.; Capodici, A. Predict, diagnose, and treat chronic kidney disease with machine learning: A systematic literature review. J. Nephrol. 2023, 36, 1101–1117. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, Z.; Yuan, E.; Lv, X.; Zhang, Y.; Tan, Y.; Xia, C.; Tang, J.; Huang, J.; Li, Z. Imaging-based deep learning in kidney diseases: Recent progress and future prospects. Insights Imaging 2024, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Rebouças Filho, P.P.; da Silva, S.P.P.; Almeida, J.S.; Ohata, E.F.; Alves, S.S.A.; Silva, F.d.S.H. An Approach to Classify Chronic Kidney Diseases Using Scintigraphy Images. In Proceedings of the XXXII Conference on Graphics, Patterns and Images (SIBGRAPI 2019), Rio de Janeiro, Brazil, 28–31 October 2019; pp. 156–159. [Google Scholar]
- Ardakani, A.A.; Hekmat, S.; Abolghasemi, J.; Reiazi, R. Scintigraphic texture analysis for assessment of renal allograft function. Pol. J. Radiol. 2018, 83, e1. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. PLoS Med. 2021, 18, e1003583. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Alexandria, A.R.D.; Ferreira, M.C.; Ohata, E.F.; Cavalcante, T.D.S.; Mota, F.A.X.D.; Nogueira, I.C.; Albuquerque, V.H.C.; Gondim, V.J.T.; Neto, E.C. Automated Classification of Dynamic Renal Scintigraphy Exams to Determine the Stage of Chronic Kidney Disease: An Investigation. In Proceedings of the 2021 3rd International Conference on Research and Academic Community Services (ICRACOS 2021), Surabaya, Indonesia, 9–10 October 2021; pp. 305–310. [Google Scholar] [CrossRef]
- Kikuchi, A.; Wada, N.; Kawakami, T.; Nakajima, K.; Yoneyama, H. A myocardial extraction method using deep learning for 99mTc myocardial perfusion SPECT images: A basic study to reduce the effects of extra-myocardial activity. Comput. Biol. Med. 2022, 141, 105164. [Google Scholar] [CrossRef]
- Zhu, F.B.; Li, L.X.; Zhao, J.Y.; Zhao, C.; Tang, S.J.; Nan, J.F.; Li, Y.T.; Zhao, Z.Q.; Shi, J.Z.; Chen, Z.H.; et al. A new method incorporating deep learning with shape priors for left ventricular segmentation in myocardial perfusion SPECT images. Comput. Biol. Med. 2023, 160, 106954. [Google Scholar] [CrossRef]
- Papandrianos, N.I.; Feleki, A.; Moustakidis, S.; Papageorgiou, E.; Apostolopoulos, I.D.; Apostolopoulos, D.J. An Explainable Classification Method of SPECT Myocardial Perfusion Images in Nuclear Cardiology Using Deep Learning and Grad-CAM. Appl. Sci. 2022, 12, 7592. [Google Scholar] [CrossRef]
- Wen, H.; Wei, Q.; Huang, J.L.; Tsai, S.C.; Wang, C.Y.; Chiang, K.F.; Deng, Y.; Cui, X.; Gao, R.; Zhou, W.; et al. Analysis on SPECT myocardial perfusion imaging with a tool derived from dynamic programming to deep learning. Optik 2021, 240, 166842. [Google Scholar] [CrossRef]
- Papandrianos, N.; Papageorgiou, E. Automatic Diagnosis of Coronary Artery Disease in SPECT Myocardial Perfusion Imaging Employing Deep Learning. Appl. Sci. 2021, 11, 6362. [Google Scholar] [CrossRef]
- Berkaya, S.K.; Sivrikoz, I.A.; Gunal, S. Classification models for SPECT myocardial perfusion imaging. Comput. Biol. Med. 2020, 123, 103893. [Google Scholar] [CrossRef]
- Chen, J.J.; Su, T.Y.; Chen, W.S.; Chang, Y.H.; Lu, H.H.S. Convolutional Neural Network in the Evaluation of Myocardial Ischemia from CZT SPECT Myocardial Perfusion Imaging: Comparison to Automated Quantification. Appl. Sci. 2021, 11, 514. [Google Scholar] [CrossRef]
- Abdi, M.E.H.; Naili, Q.; Habbache, M.; Said, B.; Boumenir, A.; Douibi, T.; Djermane, D.; Berrani, S.A. Effectively Detecting Left Bundle Branch Block False Defects in Myocardial Perfusion Imaging (MPI) with a Convolutional Neural Network (CNN); Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Papathanasiou, N.D.; Papandrianos, N.; Papageorgiou, E.; Apostolopoulos, D.J. Innovative Attention-Based Explainable Feature-Fusion VGG19 Network for Characterising Myocardial Perfusion Imaging SPECT Polar Maps in Patients with Suspected Coronary Artery Disease. Appl. Sci. 2023, 13, 8839. [Google Scholar] [CrossRef]
- Amini, M.; Pursamimi, M.; Hajianfar, G.; Salimi, Y.; Saberi, A.; Mehri-Kakavand, G.; Nazari, M.; Ghorbani, M.; Shalbaf, A.; Shiri, I.; et al. Machine learning-based diagnosis and risk classification of coronary artery disease using myocardial perfusion imaging SPECT: A radiomics study. Sci. Rep. 2023, 13, 14920. [Google Scholar] [CrossRef] [PubMed]
- Sabouri, M.; Hajianfar, G.; Hosseini, Z.; Amini, M.; Mohebi, M.; Ghaedian, T.; Madadi, S.; Rastgou, F.; Oveisi, M.; Rajabi, A.B.; et al. Myocardial Perfusion SPECT Imaging Radiomic Features and Machine Learning Algorithms for Cardiac Contractile Pattern Recognition. J. Digit. Imaging 2023, 36, 497–509. [Google Scholar] [CrossRef]
- Mohebi, M.; Amini, M.; Alemzadeh-Ansari, M.J.; Alizadehasl, A.; Rajabi, A.B.; Shiri, I.; Zaidi, H.; Orooji, M. Post-revascularization Ejection Fraction Prediction for Patients Undergoing Percutaneous Coronary Intervention Based on Myocardial Perfusion SPECT Imaging Radiomics: A Preliminary Machine Learning Study. J. Digit. Imaging 2023, 36, 1348–1363. [Google Scholar] [CrossRef]
- Hai, P.N.; Thanh, N.C.; Trung, N.T.; Kien, T.T. Transfer Learning for Disease Diagnosis from Myocardial Perfusion SPECT Imaging. CMC—Comput. Mater. Contin. 2022, 73, 5925–5941. [Google Scholar] [CrossRef]
- Kusumoto, D.; Akiyama, T.; Hashimoto, M.; Iwabuchi, Y.; Katsuki, T.; Kimura, M.; Akiba, Y.; Sawada, H.; Inohara, T.; Yuasa, S.; et al. A deep learning-based automated diagnosis system for SPECT myocardial perfusion imaging. Sci. Rep. 2024, 14, 13583. [Google Scholar] [CrossRef]
- Chen, J.J.; Su, T.Y.; Huang, C.C.; Yang, T.H.; Chang, Y.H.; Lu, H.H.S. Classification of coronary artery disease severity based on SPECT MPI polarmap images and deep learning: A study on multi-vessel disease prediction. Digit. Health 2024, 10, 20552076241288430. [Google Scholar] [CrossRef]
- Spielvogel, C.P.; Haberl, D.; Mascherbauer, K.; Ning, J.; Kluge, K.; Traub-Weidinger, T.; Davies, R.H.; Pierce, I.; Patel, K.; Nakuz, T.; et al. Diagnosis and prognosis of abnormal cardiac scintigraphy uptake suggestive of cardiac amyloidosis using artificial intelligence: A retrospective, international, multicentre, cross-tracer development and validation study. Lancet Digit. Health 2024, 6, e251–e260. [Google Scholar] [CrossRef]
- Kiso, K.; Nakajima, K.; Nimura, Y.; Nishimura, T. A novel algorithm developed using machine learning and a J-ACCESS database can estimate defect scores from myocardial perfusion single-photon emission tomography images. Ann. Nucl. Med. 2024, 38, 980–988. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.K.; Zhang, Q.N.; Khan, A.; Zhao, L.; Yang, Q.; Adeel, M.; Khan, R.; Ullah, I. 3D convolutional neural networks-based multiclass classification of Alzheimer’s and Parkinson’s diseases using PET and SPECT neuroimaging modalities. Brain Inform. 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Saini, B.S.; Gupta, S. A deep 1-D CNN learning approach with data augmentation for classification of Parkinson’s disease and scans without evidence of dopamine deficit (SWEDD). Biomed. Signal Process. Control. 2024, 91, 106008. [Google Scholar] [CrossRef]
- Magesh, P.R.; Myloth, R.D.; Tom, R.J. An Explainable Machine Learning Model for Early Detection of Parkinson’s Disease using LIME on DaTSCAN Imagery. Comput. Biol. Med. 2020, 126, 104041. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Kluge, A.; Apostolova, I.; Klutmann, S.; Kimiaei, S.; Schroeder, M.; Buchert, R. Data-driven identification of diagnostically useful extrastriatal signal in dopamine transporter SPECT using explainable AI. Sci. Rep. 2021, 11, 22932. [Google Scholar] [CrossRef]
- Ding, J.E.; Chu, C.H.; Huang, M.N.L.; Hsu, C.C. Dopamine Transporter SPECT Image Classification for Neurodegenerative Parkinsonism via Diffusion Maps and Machine Learning Classifiers. In Proceedings of the 25th Conference on Medical Image Understanding and Analysis (MIUA 2021), Oxford, UK, 12–14 July 2021; Volume 12722, pp. 377–393. [Google Scholar] [CrossRef]
- Pahuja, G.; Nagabhushan, T.N.; Prasad, B. Early Detection of Parkinson’s Disease by Using SPECT Imaging and Biomarkers. J. Intell. Syst. 2020, 29, 1329–1344. [Google Scholar] [CrossRef]
- Wang, W.; Lee, J.; Harrou, F.; Sun, Y. Early Detection of Parkinson’s Disease Using Deep Learning and Machine Learning. IEEE Access 2020, 8, 147635–147646. [Google Scholar] [CrossRef]
- Khachnaoui, H.; Chikhaoui, B.; Khlifa, N.; Mabrouk, R. Enhanced Parkinson’s Disease Diagnosis Through Convolutional Neural Network Models Applied to SPECT DaTSCAN Images. IEEE Access 2023, 11, 91157–91172. [Google Scholar] [CrossRef]
- Shiiba, T.; Arimura, Y.; Nagano, M.; Takahashi, T.; Takaki, A. Improvement of classification performance of Parkinson’s disease using shape features for machine learning on dopamine transporter single photon emission computed tomography. PLoS ONE 2020, 15, e0228289. [Google Scholar] [CrossRef]
- Khachnaoui, H.; Khlifa, N.; Mabrouk, R. Machine Learning for Early Parkinson’s Disease Identification within SWEDD Group Using Clinical and DaTSCAN SPECT Imaging Features. J. Imaging 2022, 8, 97. [Google Scholar] [CrossRef]
- Huang, G.H.; Lin, C.H.; Cai, Y.R.; Chen, T.B.; Hsu, S.Y.; Lu, N.H.; Chen, H.Y.; Wu, Y.C. Multiclass machine learning classification of functional brain images for Parkinson’s disease stage prediction. Stat. Anal. Data Min. 2020, 13, 508–523. [Google Scholar] [CrossRef]
- Pianpanit, T.; Lolak, S.; Sawangjai, P.; Sudhawiyangkul, T.; Wilaiprasitporn, T. Parkinson’s Disease Recognition Using SPECT Image and Interpretable AI: A Tutorial. IEEE Sensors J. 2021, 21, 22304–22316. [Google Scholar] [CrossRef]
- Paranjothi, K.; Ghouse, F.; Vaithiyanathan, R. Detection of Parkinson’s Disease on DaTSCAN Image Using Multi-kernel Support Vector Machine. Int. J. Intell. Eng. Syst. 2024, 17, 45–55. [Google Scholar] [CrossRef]
- Aggarwal, N.; Saini, B.S.; Gupta, S. Feature engineering-based analysis of DaTSCAN-SPECT imaging-derived features in the detection of SWEDD and Parkinson’s disease. Comput. Electr. Eng. 2024, 117, 109241. [Google Scholar] [CrossRef]
- Gorji, A.; Jouzdani, A.F. Machine learning for predicting cognitive decline within five years in Parkinson’s disease: Comparing cognitive assessment scales with DAT SPECT and clinical biomarkers. PLoS ONE 2024, 19, e0304355. [Google Scholar] [CrossRef]
- Lin, Q.; Man, Z.X.; Cao, Y.C.; Wang, H.J. Automated Classification of Whole-Body SPECT Bone Scan Images with VGG-Based Deep Networks. Int. Arab J. Inf. Technol. 2023, 20, 1–8. [Google Scholar] [CrossRef]
- Gao, R.; Lin, Q.; Man, Z.; Cao, Y. Automatic Lesion Segmentation of Metastases in SPECT Images Using U-Net-Based Model. In Proceedings of the 2nd International Conference on Signal Image Processing and Communication (ICSIPC 2022), Qingdao, China, 20–22 May 2022. [Google Scholar] [CrossRef]
- Man, Z.; Lin, Q.; Cao, Y. CNN-Based Automated Classification of SPECT Bone Scan Images. In Proceedings of the International Conference on Neural Networks, Information, and Communication Engineering (NNICE 2022), Qingdao, China, 25–27 March 2022. [Google Scholar] [CrossRef]
- Lin, Q.; Li, T.T.; Cao, C.G.; Cao, Y.C.; Man, Z.X.; Wang, H.J. Deep learning based automated diagnosis of bone metastases with SPECT thoracic bone images. Sci. Rep. 2021, 11, 4223. [Google Scholar] [CrossRef]
- Lin, Q.; Luo, M.Y.; Gao, R.T.; Li, T.T.; Man, Z.X.; Cao, Y.C.; Wang, H.J. Deep learning based automatic segmentation of metastasis hotspots in thorax bone SPECT images. PLoS ONE 2020, 15, e0243253. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, L.; Chen, X.; Man, Z.; Lin, Q.; Zeng, X.; Huang, X. Segmentation of lung cancer-caused metastatic lesions in bone scan images using self-defined model with deep supervision. Biomed. Signal Process. Control. 2023, 79, 104068. [Google Scholar] [CrossRef]
- Magdy, O.; Elaziz, M.A.; Dahou, A.; Ewees, A.A.; Elgarayhi, A.; Sallah, M. Bone scintigraphy based on deep learning model and modified growth optimizer. Sci. Rep. 2024, 14, 25627. [Google Scholar] [CrossRef]
- Ji, X.; Zhu, G.; Gou, J.; Chen, S.; Zhao, W.; Sun, Z.; Fu, H.; Wang, H. A fully automatic deep learning-based method for segmenting regions of interest and predicting renal function in pediatric dynamic renal scintigraphy. Ann. Nucl. Med. 2024, 38, 382–390. [Google Scholar] [CrossRef]
- Ryden, T.; Essen, M.V.; Marin, I.; Svensson, J.; Bernhardt, P. Simultaneous segmentation and classification of 99mTc-DMSA renal scintigraphic images with a deep learning approach. J. Nucl. Med. 2021, 62, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chang, Y.C.; Chiu, H.Y.; Cheng, C.H.; Huang, H.M. Differentiation between normal and abnormal kidneys using 99mTc-DMSA SPECT with deep learning in paediatric patients. Clin. Radiol. 2023, 78, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Phu, M.L.; Pham, T.V.; Duc, T.P.; Thanh, T.N.; Quoc, L.T.; Minh, D.C.; Ngoc, H.L.; Hong, S.M.; Thi, P.N.; Thi, N.N.; et al. RR-HCL-SVM: A two-stage framework for assessing remaining thyroid tissue post-thyroidectomy in SPECT images. Int. J. Imaging Syst. Technol. 2024, 34, e23066. [Google Scholar] [CrossRef]
- Masud, M.A.; Ngali, M.Z.; Othman, S.A.; Taib, I.; Osman, K.; Salleh, S.M.; Khudzari, A.Z.M.; Ali, N.S. Variation Segmentation Layer in Deep Learning Network for SPECT Images Lesion Segmentation. J. Adv. Res. Appl. Sci. Eng. Technol. 2024, 36, 83–92. [Google Scholar] [CrossRef]
- Kavitha, M.; Lee, C.H.; Shibudas, K.; Kurita, T.; Ahn, B.C. Deep learning enables automated localization of the metastatic lymph node for thyroid cancer on 131I post-ablation whole-body planar scans. Sci. Rep. 2020, 10, 7738. [Google Scholar] [CrossRef]
- Nadimi-Shahraki, M.H.; Fatahi, A.; Zamani, H.; Mirjalili, S. Binary Approaches of Quantum-Based Avian Navigation Optimizer to Select Effective Features from High-Dimensional Medical Data. Mathematics 2022, 10, 2770. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, B.; Xie, H.; Guo, X.; Liu, Q.; Sinusas, A.J.; Liu, C. Cross-Domain Iterative Network for Simultaneous Denoising, Limited-Angle Reconstruction, and Attenuation Correction of Cardiac SPECT. In Proceedings of the 14th International Workshop, MLMI 2023, Vancouver, BC, Canada, 8 October 2023; Volume 14348, pp. 12–22. [Google Scholar] [CrossRef]
- Anbarasu, P.N.; Suruli, T.M. Deep Ensemble Learning with GAN-based Semi-Supervised Training Algorithm for Medical Decision Support System in Healthcare Applications. Int. J. Intell. Eng. Syst. 2022, 15, 1–12. [Google Scholar] [CrossRef]
- Huxohl, T.; Patel, G.; Zabel, R.; Burchert, W. Deep learning approximation of attenuation maps for myocardial perfusion SPECT with an IQ · SPECT collimator. EJNMMI Phys. 2023, 10, 49. [Google Scholar] [CrossRef]
- Mostafapour, S.; Gholamiankhah, F.; Maroufpour, S.; Momennezhad, M.; Asadinezhad, M.; Zakavi, S.R.; Arabi, H.; Zaidi, H. Deep learning-guided attenuation correction in the image domain for myocardial perfusion SPECT imaging. J. Comput. Des. Eng. 2022, 9, 434–447. [Google Scholar] [CrossRef]
- Xie, H.; Zhou, B.; Chen, X.; Guo, X.; Thorn, S.; Liu, Y.H.; Wang, G.; Sinusas, A.; Liu, C. Transformer-Based Dual-Domain Network for Few-View Dedicated Cardiac SPECT Image Reconstructions. In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023), Vancouver, BC, Canada, 8–12 October 2023; Volume 14229, pp. 163–172. [Google Scholar] [CrossRef]
- Werner, R.A.; Higuchi, T.; Nose, N.; Toriumi, F.; Matsusaka, Y.; Kuji, I.; Kazuhiro, K. Generative adversarial network-created brain SPECTs of cerebral ischemia are indistinguishable to scans from real patients. Sci. Rep. 2022, 12, 18787. [Google Scholar] [CrossRef]
- Zhou, Y.; Tagare, H.D. Self-Normalized Classification of Parkinson’s Disease DaTscan Images. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2021), Virtual, 9–12 December 2021. [Google Scholar] [CrossRef]
- Chrysostomou, C.; Koutsantonis, L.; Lemesios, C.; Papanicolas, C.N. SPECT Angle Interpolation Based on Deep Learning Methodologies. In Proceedings of the 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC 2020), Virtual, 31 October–7 November 2020. [Google Scholar] [CrossRef]
- Ichikawa, S.; Sugimori, H.; Ichijiri, K.; Yoshimura, T.; Nagaki, A. Acquisition time reduction in pediatric 99mTc-DMSA planar imaging using deep learning. J. Appl. Clin. Med. Phys. 2023, 24, e13978. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.; Oh, D.; Kim, J.H.; Yoo, J.; Lee, W.W. Deep-learning-based attenuation map generation in kidney single photon emission computed tomography. EJNMMI Phys. 2024, 11, 84. [Google Scholar] [CrossRef]
- Lin, C.; Chang, Y.C.; Chiu, H.Y.; Cheng, C.H.; Huang, H.M. Reducing scan time of paediatric 99mTc-DMSA SPECT via deep learning. Clin. Radiol. 2021, 76, 315.e13–315.e20. [Google Scholar] [CrossRef] [PubMed]
- Leube, J.; Gustafsson, J.; Lassmann, M.; Salas-Ramirez, M.; Tran-Gia, J. Analysis of a deep learning-based method for generation of SPECT projections based on a large Monte Carlo simulated dataset. EJNMMI Phys. 2022, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Marek, K.; Jennings, D.; Lasch, S.; Siderowf, A.; Tanner, C.; Simuni, T.; Coffey, C.; Kieburtz, K.; Flagg, E.; Chowdhury, S. Parkinson Progression Marker Initiative. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 2011, 95, 629–635. [Google Scholar] [CrossRef]
- Cios, K.; Kurgan, L.; Goodenday, L. SPECT Heart. UCI Machine Learning Repository. 2001. Available online: https://archive.ics.uci.edu/dataset/95/spect+heart (accessed on 23 February 2025).
- Medical Imaging Informatics Lab. SPECTMPISeg: SPECT for Left Ventricular Segmentation. 2024. Available online: https://github.com/MIILab-MTU/SPECTMPISeg (accessed on 23 February 2025).
- Kaplan, S. SPECT MPI Dataset. 2025. Available online: https://www.kaggle.com/datasets/selcankaplan/spect-mpi (accessed on 9 February 2025).
- Aggarwal, N.; Saini, B.S.; Gupta, S. Role of Artificial Intelligence Techniques and Neuroimaging Modalities in Detection of Parkinson’s Disease: A Systematic Review. Cogn. Comput. 2023, 16, 2078–2115. [Google Scholar] [CrossRef]
- Khachnaoui, H.; Mabrouk, R.; Khlifa, N. Machine learning and deep learning for clinical data and PET/SPECT imaging in Parkinson’s disease: A review. IET Image Process. 2020, 14, 4013–4026. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, H.; Caobelli, F.; Huang, Y.; Li, Y.; Zhang, J.; Shi, K.; Yu, F. The role of deep learning in myocardial perfusion imaging for diagnosis and prognosis: A systematic review. iScience 2024, 27, 111374. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Yan, P.; Li, G.; Jiang, Y.; Luo, H.; Yin, S. Deep Learning Attention Mechanism in Medical Image Analysis: Basics and Beyonds. Int. J. Netw. Dyn. Intell. 2023, 2, 93–116. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Zhou, C.; Pen, H.; Zheng, Z.; Chen, J.; Ding, W. A review of cancer data fusion methods based on deep learning. Inf. Fusion 2024, 108, 102361. [Google Scholar] [CrossRef]
- Li, X.; Li, L.; Jiang, Y.; Wang, H.; Qiao, X.; Feng, T.; Luo, H.; Zhao, Y. Vision-Language Models in medical image analysis: From simple fusion to general large models. Inf. Fusion 2025, 118, 102995. [Google Scholar] [CrossRef]
- Topol, E. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Morley, J.; Machado, C.C.V.; Burr, C.; Cowls, J.; Joshi, I.; Taddeo, M.; Floridi, L. The ethics of AI in health care: A mapping review. Soc. Sci. Med. 2020, 260, 113172. [Google Scholar] [CrossRef]
- Mennella, C.; Maniscalco, U.; Pietro, G.D.; Esposito, M. Ethical and regulatory challenges of AI technologies in healthcare: A narrative review. Heliyon 2024, 10, e26297. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. npj Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef]
- Mongan, J.; Chen, M.Z.; Halabi, S.; Kalpathy-Cramer, J.; Langlotz, M.P.; Langlotz, C.P.; Kohli, M.D.; Erickson, B.J.; Kim, W.; Auffermann, W.; et al. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Tejani, A.S.; Klontzas, M.E.; Gatti, A.A.; Mongan, J.T.; Moy, L.; Park, S.H.; Kahn, C.E., Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): 2024 Update. Radiol. Artif. Intell. 2024, 6, e240300. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.M.; Dhiman, P.; Riley, R.D.; Beam, A.L.; Calster, B.V.; Ghassemi, M.; Liu, X.; Reitsma, J.B.; van Smeden, M.; et al. TRIPOD+AI statement: Updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024, 385, e078378. [Google Scholar] [CrossRef]
- Sadeghi, Z.; Alizadehsani, R.; CIFCI, M.A.; Kausar, S.; Rehman, R.; Mahanta, P.; Bora, P.K.; Almasri, A.; Alkhawaldeh, R.S.; Hussain, S.; et al. A review of Explainable Artificial Intelligence in healthcare. Comput. Electr. Eng. 2024, 118, 109370. [Google Scholar] [CrossRef]
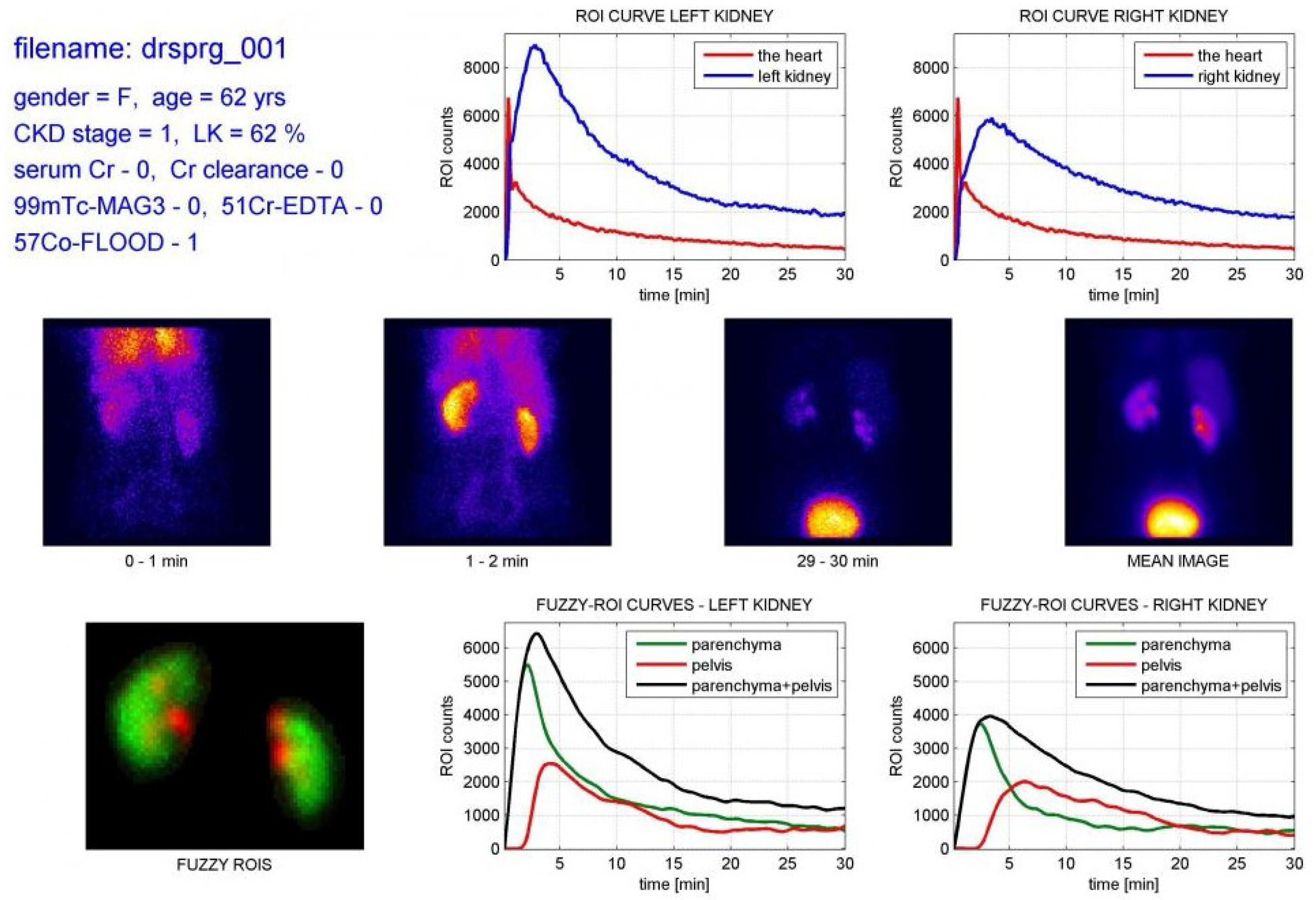
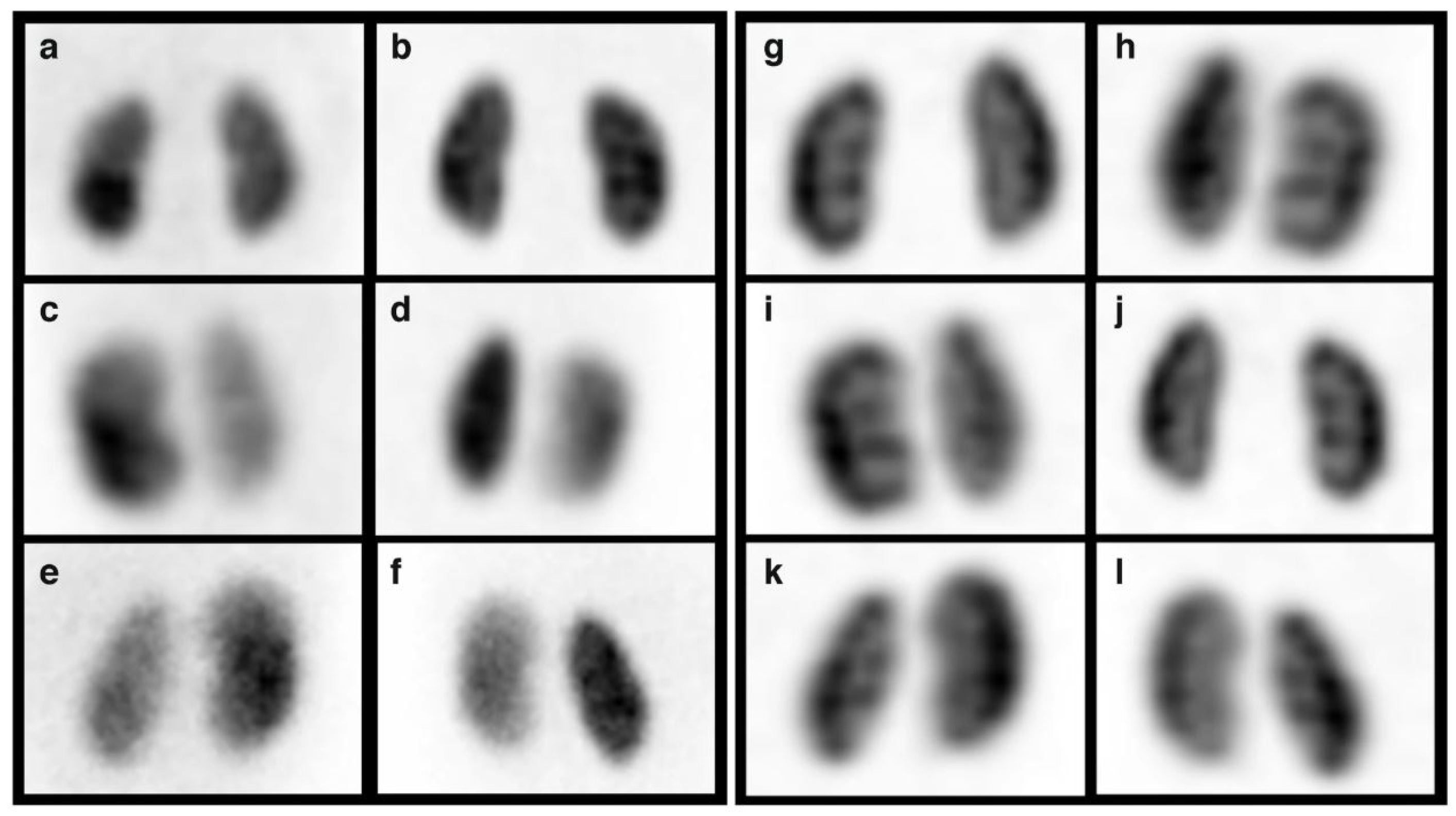

| Method | Organ | N | Publications |
|---|---|---|---|
| Diagnostic methods (N = 45) | Heart | 17 | [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36] |
| Brain | 15 | [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51] | |
| Bones | 7 | [52,53,54,55,56,57,58] | |
| Kidney | 3 | [59,60,61] | |
| Thyroid glands | 2 | [62,63] | |
| Lymph nodes | 1 | [64] | |
| General imaging methods (N = 13) | Heart | 6 | [65,66,67,68,69,70] |
| Brain | 3 | [71,72,73] | |
| Kidney | 3 | [74,75,76] | |
| N/A * | 1 | [77] |
| Data | Name | Country | N |
|---|---|---|---|
| Open dataset (N = 18) | PPMI | Multicenter | 14 |
| UCI SPECT Heart | USA | 2 | |
| SPECTMPISeg | China | 1 | |
| SPECT MPI | Turkey | 1 | |
| In house (N = 38) | China | 9 | |
| Japan | 5 | ||
| Taiwan | 5 | ||
| Greece | 3 | ||
| Iran | 3 | ||
| South Korea | 2 | ||
| Vietnam | 2 | ||
| Algeria | 1 | ||
| Egypt | 1 | ||
| Germany | 1 | ||
| USA | 1 | ||
| Multicenter | 1 | ||
| Not reported | 4 | ||
| Software phantoms | 1 | ||
| None | 1 | ||
| Classification | N = 34 | ||
|---|---|---|---|
| Planar | DenseNet21 | [35] | |
| MobileViT + GO | [58] | ||
| SPECT | CNNs | ||
| Custom CNN | [22,26,33,37,38,40,61], | ||
| Custom CNN, VGG16, DenseNet, MobileNet, Inception | [24] | ||
| Custom CNN, EfficientNet-B0, MobileNet-V2 | [44] | ||
| VGG16 | [39,52] | ||
| VGG16 from scratch and with transfer lerning | [54] | ||
| Custom feature-fusion VGG19 | [28] | ||
| ResNet50V2 | [27] | ||
| EfficientNet V2 | [34] | ||
| VGG16, VGG19, DenseNet, AlexNet, GoogleNet, NASNet-Large, ResNet, | [25] | ||
| VGG16, VGG19, DensNet, ResNet and custom VGG7, VGG21 and VGG24 | [55] | ||
| VGG, Xception, MobileNet, EfficientNet, Inception, DenseNet, ResNet | [32] | ||
| VGG16, LDA, SVN, DT, MLP, RF, AB | [47] | ||
| VGG16, AlexNet + Multi-kernel SVM | [49] | ||
| PD Net (previously published model) | [48] | ||
| DETR (previously published model) | [62] | ||
| Other Deep Learning | |||
| AE + AB, SVM, KNN, RF, GB, BC, MLP, DT, LR | [51] | ||
| Stacked AE | [42] | ||
| FNN, DT, RF, LR, KNN, SVM, LDA | [43] | ||
| Other Machine Learning | |||
| DM + LDA | [41] | ||
| Density-Based Spatial, K-means and Hierarchical Clustering | [46] | ||
| SVM | [45] | ||
| SVM, KNN, DT, BT, RF | [31] | ||
| SVM, DT, RF, LR, MLP, GB, XGB | [30] | ||
| SVM, KNN, DT, RF, LR, NB, AB, GB, SGD | [50] | ||
| SVM, KNN, DT, RF, LR, MLP, NB, GB, XGB | [29] | ||
| Segmentation | N = 10 | ||
| Planar | FNN | [64] | |
| Custom Model (Swin-Unet + DeepLab) | [59] | ||
| SPECT | Custom CNN | [57] | |
| U-net | [20,23,36,53,63] | ||
| V-net + dynamic programming | [21] | ||
| U-net, Mask R-CNN | [56] | ||
| Classification and Segmentation | N = 1 | ||
| Planar | Mask R-CNN | [60] | |
| SPECT | - | ||
| Synthetic data | ||
| Planar | - | |
| SPECT | U-net | [77] |
| GAN | [67,71] | |
| Reconstruction | ||
| Planar | DnCNN, Win5RB, ResUnet | [74] |
| SPECT | Custom Transformer-based Dual-domain Network | [70] |
| Custom Residual Network | [76] | |
| Attenuation generation or enhancement | ||
| Planar | - | |
| SPECT | cGAN (U-net + PatchGAN) | [68] |
| ResNet, U-net | [69] | |
| U-net | [75] | |
| Normalization | ||
| Planar | - | |
| SPECT | Self-normalization via a projection of voxels | [72] |
| U-net | [73] | |
| Feature selection | ||
| Planar | - | |
| SPECT | Quantum-Based Avian Navigation Optimizer | [65] |
| Mixed | ||
| Planar | - | |
| SPECT | Custom Cross-domain Iterative Network (U-net) | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vrbaški, D.; Vesin, B.; Mangaroska, K. Machine Learning for Chronic Kidney Disease Detection from Planar and SPECT Scintigraphy: A Scoping Review. Appl. Sci. 2025, 15, 6841. https://doi.org/10.3390/app15126841
Vrbaški D, Vesin B, Mangaroska K. Machine Learning for Chronic Kidney Disease Detection from Planar and SPECT Scintigraphy: A Scoping Review. Applied Sciences. 2025; 15(12):6841. https://doi.org/10.3390/app15126841
Chicago/Turabian StyleVrbaški, Dunja, Boban Vesin, and Katerina Mangaroska. 2025. "Machine Learning for Chronic Kidney Disease Detection from Planar and SPECT Scintigraphy: A Scoping Review" Applied Sciences 15, no. 12: 6841. https://doi.org/10.3390/app15126841
APA StyleVrbaški, D., Vesin, B., & Mangaroska, K. (2025). Machine Learning for Chronic Kidney Disease Detection from Planar and SPECT Scintigraphy: A Scoping Review. Applied Sciences, 15(12), 6841. https://doi.org/10.3390/app15126841








