MSA K-BERT: A Method for Medical Text Intent Classification
Abstract
1. Introduction
- A knowledge-supported Language Representation (LR) model compatible with BERT is proposed in this paper, namely MSA K-BERT, which can combine knowledge in the medical field and alleviate the problems of HES and KN.
- Through a fine-grained injection of a Knowledge Graph (KG), the performance of MSA K-BERT in the medical text intent classification task is superior to that of mainstream models such as BERT.
- Using the multi-scale attention mechanism to enhance the feature layers at different stages and selectively assign different weights to the text content, MSA K-BERT can enhance the text content with the attention mechanism, which not only makes the results more accurate but also makes them more interpretable.
2. Background
2.1. Knowledge Graph
2.2. Knowledge Noise
2.3. Heterogeneity of Embedding Spaces
3. Related Work
3.1. Text Classification
3.2. Pretrained Language Models
3.3. Knowledge Noise and Heterogeneity of Embedding Spaces
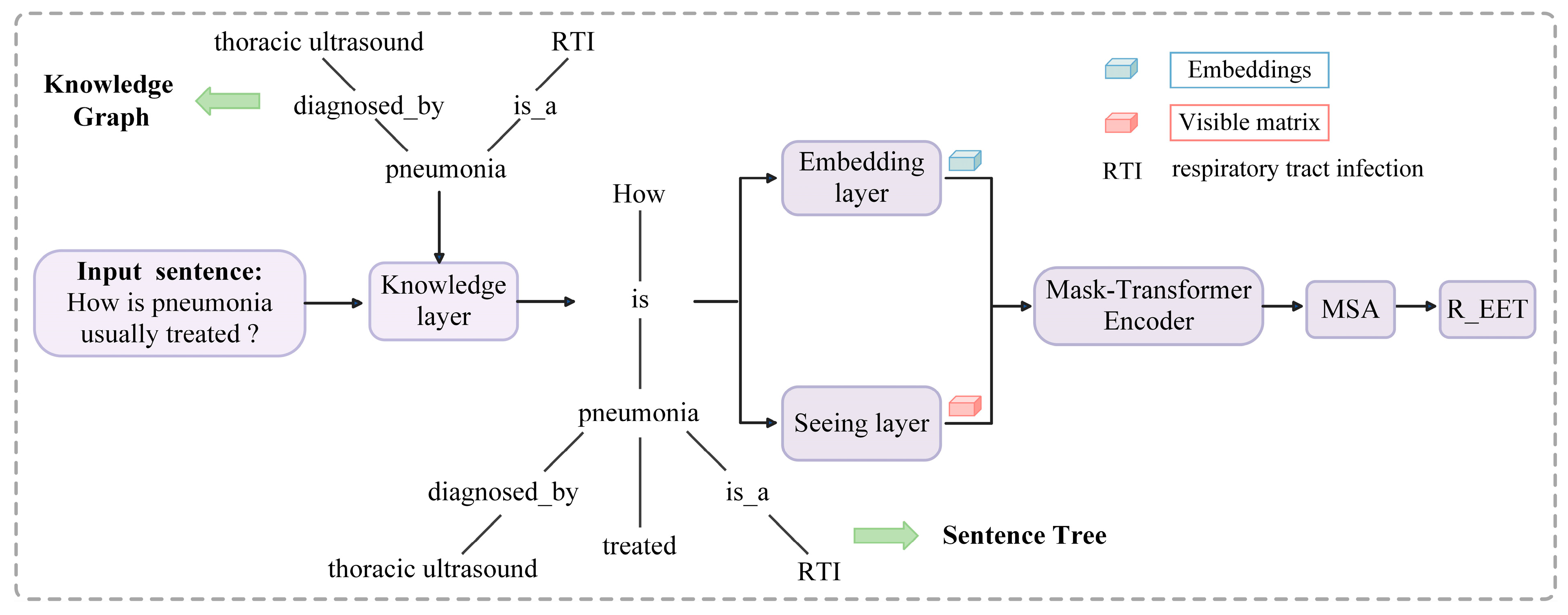
4. Methods
4.1. Model Architecture
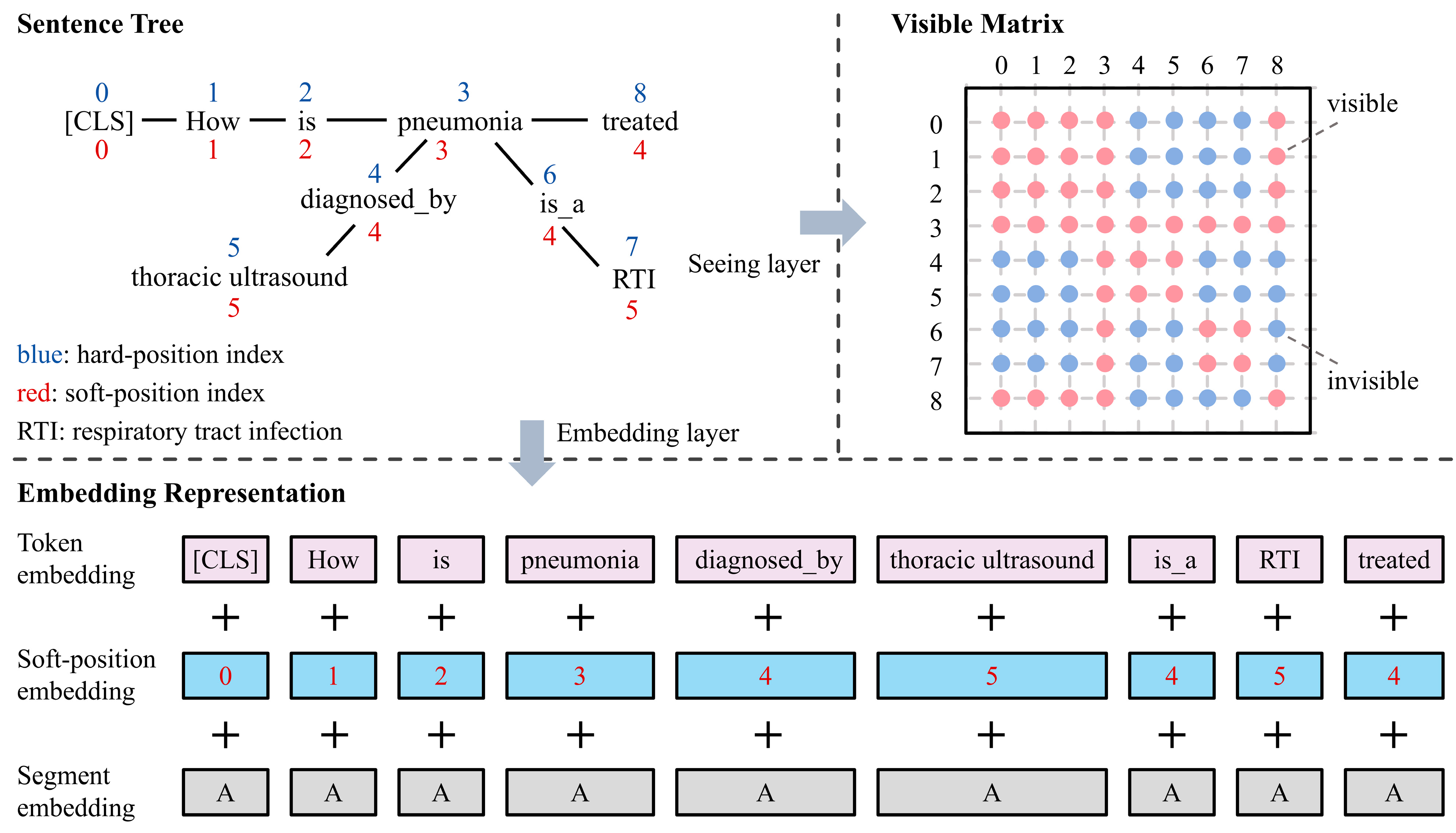
4.2. Knowledge Layer
4.3. Embedding Layer
4.4. Seeing Layer
4.5. Mask-Transformer
4.6. Multi-Scale Attention Layer
4.7. Loss Function
5. Results
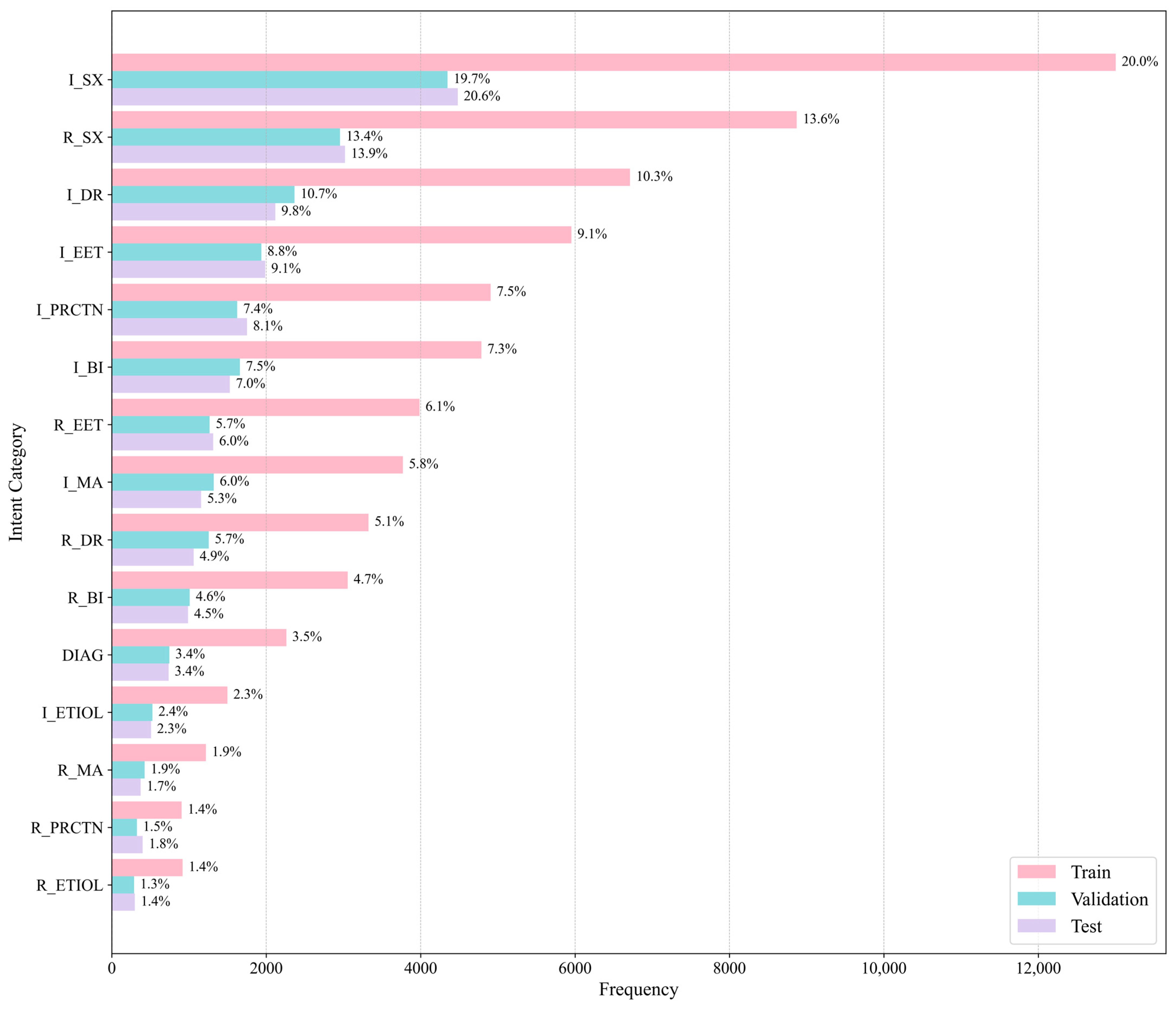
5.1. Dataset
Class Distribution
5.2. Experimental Setup
5.3. Comparative Experiment
5.4. Ablation Study
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, N.; Su, X.; Liu, T.; Hao, Q.; Wei, M. A Benchmark Dataset and Case Study for Chinese Medical Question Intent Classification. BMC Med. Inf. Decis. Mak. 2020, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Yahia, H.S.; Abdulazeez, A.M. Medical Text Classification Based on Convolutional Neural Network: A Review. Int. J. Sci. Bus. 2021, 5, 27–41. [Google Scholar]
- Yao, L.; Mao, C.; Luo, Y. Clinical Text Classification with Rule-Based Features and Knowledge-Guided Convolutional Neural Networks. BMC Med. Inf. Decis. Mak. 2019, 19, 71. [Google Scholar] [CrossRef]
- Mollaei, N.; Cepeda, C.; Rodrigues, J.; Gamboa, H. Biomedical Text Mining: Applicability of Machine Learning-Based Natural Language Processing in Medical Database. In Proceedings of the Biosignals, Caparica, Portugal, 5 March 2022; pp. 159–166. [Google Scholar]
- Lenivtceva, I.; Slasten, E.; Kashina, M.; Kopanitsa, G. Applicability of Machine Learning Methods to Multi-Label Medical Text Classification. In Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 509–522. ISBN 978-3-030-50422-9. [Google Scholar]
- Hassan, S.U.; Ahamed, J.; Ahmad, K. Analytics of Machine Learning-Based Algorithms for Text Classification. Sustain. Oper. Comput. 2022, 3, 238–248. [Google Scholar] [CrossRef]
- Minaee, S.; Kalchbrenner, N.; Cambria, E.; Nikzad, N.; Chenaghlu, M.; Gao, J. Deep Learning—Based Text Classification: A Comprehensive Review. ACM Comput. Surv. 2022, 54, 1–40. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, S. A Novel Deep-Learning-Based Model for Medical Text Classification. In Proceedings of the 2020 9th International Conference on Computing and Pattern Recognition, Xiamen, China, 30 October 2020; pp. 267–273. [Google Scholar]
- Shiri, F.M.; Perumal, T.; Mustapha, N.; Mohamed, R. A Comprehensive Overview and Comparative Analysis on Deep Learning Models: CNN, RNN, LSTM, GRU. JAI 2024, 6, 301–360. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MI, USA, 1 June 2019; Volume 1, pp. 4171–4186. [Google Scholar]
- Luan, Y.; Lin, S. Research on Text Classification Based on CNN and LSTM. In Proceedings of the 2019 IEEE International Conference on Artificial Intelligence and Computer Applications (ICAICA), Dalian, China, 29–31 March 2019; pp. 352–355. [Google Scholar]
- Kim, Y. Convolutional Neural Networks for Sentence Classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1746–1751. [Google Scholar]
- He, T.; Huang, W.; Qiao, Y.; Yao, J. Text-Attentional Convolutional Neural Network for Scene Text Detection. IEEE Trans. Image Process 2016, 25, 2529–2541. [Google Scholar] [CrossRef] [PubMed]
- Kalchbrenner, N.; Grefenstette, E.; Blunsom, P. A Convolutional Neural Network for Modelling Sentences. arXiv 2014, arXiv:1404.2188. [Google Scholar]
- Zhang, J.; Li, Y.; Tian, J.; Li, T. LSTM-CNN Hybrid Model for Text Classification. In Proceedings of the 2018 IEEE 3rd Advanced Information Technology, Electronic and Automation Control Conference (IAEAC), Chongqing, China, 12–14 October 2018; pp. 1675–1680. [Google Scholar]
- Guo, M.-H.; Xu, T.-X.; Liu, J.-J.; Liu, Z.-N.; Jiang, P.-T.; Mu, T.-J.; Zhang, S.-H.; Martin, R.R.; Cheng, M.-M.; Hu, S.-M. Attention Mechanisms in Computer Vision: A Survey. Comp. Visual. Med. 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Liang, W. CNN-RNN Based Intelligent Recommendation for Online Medical Pre-Diagnosis Support. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 912–921. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, H.; Lu, C.; Lyu, S. Multichannel CNN with Attention for Text Classification. arXiv 2020, arXiv:2006.16174. [Google Scholar]
- Dernoncourt, F.; Lee, J.Y. PubMed 200k RCT: A Dataset for Sequential Sentence Classification in Medical Abstracts. arXiv 2017, arXiv:1710.06071. [Google Scholar]
- Tsatsaronis, G.; Balikas, G.; Malakasiotis, P.; Partalas, I.; Zschunke, M.; Alvers, M.R.; Weissenborn, D.; Krithara, A.; Petridis, S.; Polychronopoulos, D.; et al. An Overview of the BIOASQ Large-Scale Biomedical Semantic Indexing and Question Answering Competition. BMC Bioinform. 2015, 16, 138. [Google Scholar] [CrossRef] [PubMed]
- Qasim, R.; Bangyal, W.H.; Alqarni, M.A.; Ali Almazroi, A. A Fine-Tuned BERT-Based Transfer Learning Approach for Text Classification. J. Healthc. Eng. 2022, 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Tu, X.; Huang, J.X. Utilizing BERT for Biomedical and Clinical Text Mining. In Data Analytics in Biomedical Engineering and Healthcare; Elsevier: Amsterdam, The Netherlands, 2021; pp. 73–103. [Google Scholar]
- Gardazi, N.M.; Daud, A.; Malik, M.K.; Bukhari, A.; Alsahfi, T.; Alshemaimri, B. BERT Applications in Natural Language Processing: A Review. Artif. Intell. Rev. 2025, 58, 166. [Google Scholar] [CrossRef]
- Talaat, A.S. Sentiment Analysis Classification System Using Hybrid BERT Models. J. Big Data 2023, 10, 110. [Google Scholar] [CrossRef]
- Kaur, K.; Kaur, P. BERT-CNN: Improving BERT for Requirements Classification Using CNN. Procedia Comput. Sci. 2023, 218, 2604–2611. [Google Scholar] [CrossRef]
- Li, W.; Gao, S.; Zhou, H.; Huang, Z.; Zhang, K.; Li, W. The Automatic Text Classification Method Based on Bert and Feature Union. In Proceedings of the 2019 IEEE 25th International Conference on Parallel and Distributed Systems (ICPADS), Tianjin, China, 4–6 December 2019; pp. 774–777. [Google Scholar]
- Wang, X.; Bo, D.; Shi, C.; Fan, S.; Ye, Y.; Yu, P.S. A Survey on Heterogeneous Graph Embedding: Methods, Techniques, Applications and Sources. IEEE Trans. Big Data 2022, 9, 415–436. [Google Scholar] [CrossRef]
- Naseem, U.; Thapa, S.; Zhang, Q.; Hu, L.; Masood, A.; Nasim, M. Reducing Knowledge Noise for Improved Semantic Analysis in Biomedical Natural Language Processing Applications. In Proceedings of the 5th Clinical Natural Language Processing Workshop, Toronto, ON, Canada, 14 July 2023; pp. 272–277. [Google Scholar]
- Thirunavukarasu, A.J.; Ting, D.S.J.; Elangovan, K.; Gutierrez, L.; Tan, T.F.; Ting, D.S.W. Large Language Models in Medicine. Nat. Med. 2023, 29, 1930–1940. [Google Scholar] [CrossRef]
- OpenAI; Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; et al. GPT-4 Technical Report. arXiv 2024, arXiv:2303.08774. [Google Scholar]
- Mullick, A.; Gupta, M.; Goyal, P. Intent Detection and Entity Extraction from BioMedical Literature. arXiv 2024, arXiv:2404.03598. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Cheng, Y.; Zhao, H.; Zhou, X.; Zhao, J.; Cao, Y.; Yang, C.; Cai, X. A Large Language Model for Advanced Power Dispatch. Sci. Rep. 2025, 15, 8925. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, S.; Zuo, S.; Chen, Y.; Sun, W.; Luo, J.; Duan, J.; Wang, H.; Wang, D. U-Net Combined with Multi-Scale Attention Mechanism for Liver Segmentation in CT Images. BMC Med. Inf. Decis. Mak. 2021, 21, 283. [Google Scholar] [CrossRef] [PubMed]
- Hogan, A.; Blomqvist, E.; Cochez, M.; D’amato, C.; Melo, G.D.; Gutierrez, C.; Kirrane, S.; Gayo, J.E.L.; Navigli, R.; Neumaier, S.; et al. Knowledge Graphs. ACM Comput. Surv. 2022, 54, 1–37. [Google Scholar] [CrossRef]
- Sengupta, S.; Krone, J.; Mansour, S. On the Robustness of Intent Classification and Slot Labeling in Goal-Oriented Dialog Systems to Real-World Noise. arXiv 2021, arXiv:2104.07149. [Google Scholar]
- Kalinowski, A.; An, Y. A Survey of Embedding Space Alignment Methods for Language and Knowledge Graphs. arXiv 2020, arXiv:2010.13688. [Google Scholar]
- Khalid, B.; Dai, S.; Taghavi, T.; Lee, S. Label Supervised Contrastive Learning for Imbalanced Text Classification in Euclidean and Hyperbolic Embedding Spaces. In Proceedings of the Ninth Workshop on Noisy and User-generated Text (W-NUT 2024), San Ġiljan, Malta, 15 November 2024; pp. 58–67. [Google Scholar]
- Mitra, V.; Wang, C.-J.; Banerjee, S. Text Classification: A Least Square Support Vector Machine Approach. Appl. Soft Comput. 2007, 7, 908–914. [Google Scholar] [CrossRef]
- Isa, D.; Lee, L.H.; Kallimani, V.P.; Rajkumar, R. Text Document Preprocessing with the Bayes Formula for Classification Using the Support Vector Machine. IEEE Trans. Knowl. Data Eng. 2008, 20, 1264–1272. [Google Scholar] [CrossRef]
- Wu, Q.; Ye, Y.; Zhang, H.; Ng, M.K.; Ho, S.-S. ForesTexter: An Efficient Random Forest Algorithm for Imbalanced Text Categorization. Knowl.-Based Syst. 2014, 67, 105–116. [Google Scholar] [CrossRef]
- Dou, B.; Zhu, Z.; Merkurjev, E.; Ke, L.; Chen, L.; Jiang, J.; Zhu, Y.; Liu, J.; Zhang, B.; Wei, G.-W. Machine Learning Methods for Small Data Challenges in Molecular Science. Chem. Rev. 2023, 123, 8736–8780. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. Sn Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Guo, J. Bidirectional LSTM with Attention Mechanism and Convolutional Layer for Text Classification. Neurocomputing 2019, 337, 325–338. [Google Scholar] [CrossRef]
- Wang, J.; Yu, L.-C.; Lai, K.R.; Zhang, X. Tree-Structured Regional CNN-LSTM Model for Dimensional Sentiment Analysis. IEEE/ACM Trans. Audio Speech Lang. Process 2019, 28, 581–591. [Google Scholar] [CrossRef]
- Han, Y.; Guo, J.; Yang, H.; Guan, R.; Zhang, T. SSMA-YOLO: A Lightweight YOLO Model with Enhanced Feature Extraction and Fusion Capabilities for Drone-Aerial Ship Image Detection. Drones 2024, 8, 145. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Han, Y.; Wang, Y.; Yuan, J.; Song, J.; Zheng, B.; Zhang, L.; Huang, S.; Chen, H. Global Compression Commander: Plug-and-Play Inference Acceleration for High-Resolution Large Vision-Language Models. arXiv 2025, arXiv:2501.05179. [Google Scholar]
- Han, Y.; Duan, B.; Guan, R.; Yang, G.; Zhen, Z. LUFFD-YOLO: A Lightweight Model for UAV Remote Sensing Forest Fire Detection Based on Attention Mechanism and Multi-Level Feature Fusion. Remote Sens. 2024, 16, 2177. [Google Scholar] [CrossRef]
- Liu, X.; Liu, T.; Huang, S.; Xin, Y.; Hu, Y.; Qin, L.; Wang, D.; Wu, Y.; Chen, H. M2IST: Multi-Modal Interactive Side-Tuning for Efficient Referring Expression Comprehension. IEEE Transactions on Circuits and Systems for Video Technology. arXiv 2025, arXiv:2407.01131v3. [Google Scholar]
- Sun, Y.; Wang, S.; Li, Y.; Feng, S.; Chen, X.; Zhang, H.; Tian, X.; Zhu, D.; Tian, H.; Wu, H. ERNIE: Enhanced Representation through Knowledge Integration. arXiv 2019, arXiv:1904.09223. [Google Scholar]
- Clark, K.; Luong, M.-T.; Le, Q.V.; Manning, C.D. ELECTRA: Pre-Training Text Encoders as Discriminators Rather Than Generators. arXiv 2020, arXiv:2003.10555. [Google Scholar]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. ALBERT: A Lite BERT for Self-Supervised Learning of Language Representations. arXiv 2020, arXiv:1909.11942. [Google Scholar]
- Rajpurkar, P.; Zhang, J.; Lopyrev, K.; Liang, P. SQuAD: 100,000+ Questions for Machine Comprehension of Text. arXiv 2016, arXiv:1606.05250. [Google Scholar]
- Wang, A.; Singh, A.; Michael, J.; Hill, F.; Levy, O.; Bowman, S.R. GLUE: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding. arXiv 2019, arXiv:1804.07461. [Google Scholar]
- Moradi, M.; Blagec, K.; Samwald, M. Deep Learning Models Are Not Robust against Noise in Clinical Text. arXiv 2021, arXiv:2108.12242. [Google Scholar]
- Si, Y.; Wang, J.; Xu, H.; Roberts, K. Enhancing Clinical Concept Extraction with Contextual Embeddings. J. Am. Med. Inform. Assoc. 2019, 26, 1297–1304. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, A.X.; Abdalla, M.; McDermott, M.; Ghassemi, M. Hurtful Words: Quantifying Biases in Clinical Contextual Word Embeddings. In Proceedings of the ACM Conference on Health, Inference, and Learning, Toronto, ON, Canada, 2 April 2020; pp. 110–120. [Google Scholar]
- Liu, W.; Zhou, P.; Zhao, Z.; Wang, Z.; Ju, Q.; Deng, H.; Wang, P. K-Bert: Enabling Language Representation with Knowledge Graph. AAAI Conf. Artif. Intell. 2020, 34, 2901–2908. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Chen, W.; Shi, K. Multi-Scale Attention Convolutional Neural Network for Time Series Classification. Neural Netw. 2021, 136, 126–140. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Fang, H.; Yao, Q.; Zhong, C.; Hao, J.; Zhang, Q.; Huang, X.; Peng, J.; Wei, Z. A Benchmark for Automatic Medical Consultation System: Frameworks, Tasks and Datasets. Bioinformatics 2023, 39, btac817. [Google Scholar] [CrossRef]
- Tian, S.; Yang, W.; Le Grange, J.M.; Wang, P.; Huang, W.; Ye, Z. Smart Healthcare: Making Medical Care More Intelligent. Glob. Health J. 2019, 3, 62–65. [Google Scholar] [CrossRef]
- Bright, T.J.; Wong, A.; Dhurjati, R.; Bristow, E.; Bastian, L.; Coeytaux, R.R.; Samsa, G.; Hasselblad, V.; Williams, J.W.; Musty, M.D.; et al. Effect of Clinical Decision-Support Systems: A Systematic Review. Ann. Intern Med. 2012, 157, 29–43. [Google Scholar] [CrossRef] [PubMed]
| Intent Category | Abbreviation | Example |
|---|---|---|
| Request Symptom | R_SX | Patient: Why do I keep coughing? |
| Inform Symptom | I_SX | Patient: I have a slight fever and sore throat. |
| Request Etiology | R_ETIOL | Patient: Is the fever due to catching a cold? |
| Inform Etiology | I_ETIOL | Doctor: It is likely caused by a viral infection. |
| Request Basic Information | R_BI | Doctor: What is your age? |
| Inform Basic Information | I_BI | Patient: I am 19 years old. |
| Request Existing Exam and Treatment | R_EET | Doctor: Did you handle the wound before coming to the hospital? |
| Inform Existing Exam and Treatment | I_EET | Patient: I did some basic bandaging. |
| Request Drug Recommendation | R_DR | Patient: What medicine should I take? |
| Inform Drug Recommendation | I_DR | Doctor: Just take some Lianhua Qingwen capsules. |
| Request Medical Advice | R_MA | Patient: Do I need to go to the hospital for tests? |
| Inform Medical Advice | I_MA | Doctor: If the fever lasts more than three days, get a blood test. |
| Request Precautions | R_PRCTN | Patient: What else should I be aware of? |
| Inform Precautions | I_PRCTN | Doctor: Rest adequately and avoid catching a chill. |
| Diagnose | DIAG | Doctor: You have gastritis. |
| Other | OTR | Patient: Thank you, doctor. |
| Intent Category | Training Set | Validation Set | Test Set |
|---|---|---|---|
| Request Symptom | 2957 | 8871 | 3020 |
| Inform Symptom | 4348 | 13,001 | 4481 |
| Request Etiology | 289 | 917 | 298 |
| Inform Etiology | 527 | 1498 | 508 |
| Request Basic Information | 1010 | 3056 | 988 |
| Inform Basic Information | 1659 | 4787 | 1528 |
| Request Existing Exam and Treatment | 1268 | 3986 | 1313 |
| Inform Existing Exam and Treatment | 1938 | 5953 | 1987 |
| Request Drug Recommendation | 1256 | 3325 | 1061 |
| Inform Drug Recommendation | 2366 | 6712 | 2118 |
| Request Medical Advice | 425 | 1219 | 373 |
| Inform Medical Advice | 1320 | 3770 | 1157 |
| Request Precautions | 326 | 904 | 400 |
| Inform Precautions | 1624 | 4906 | 1752 |
| Diagnose | 746 | 2261 | 737 |
| Other | 11,208 | 33,363 | 11,214 |
| Total | 33,267 | 98,529 | 32,935 |
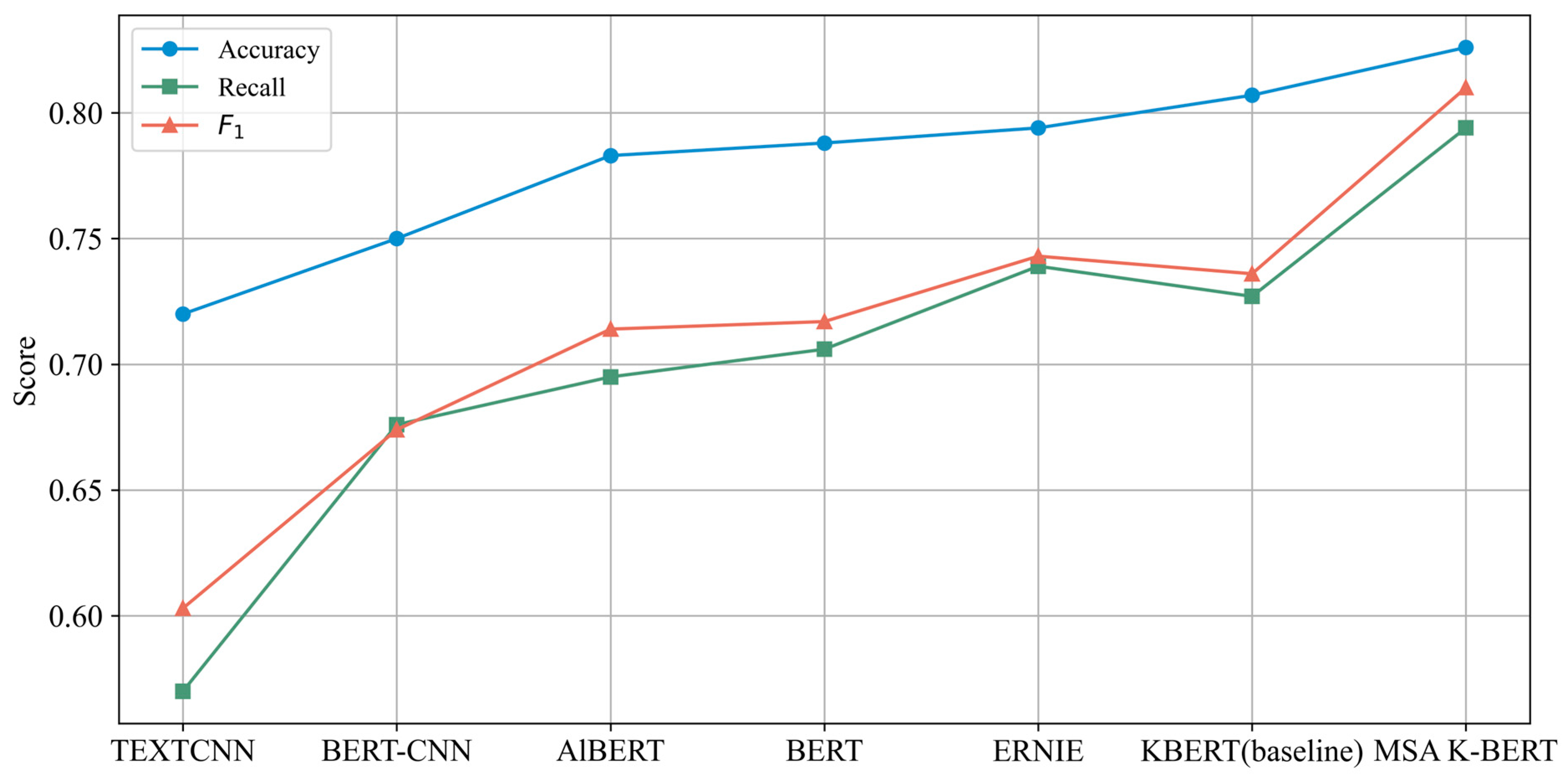
| Model | Parameters | Accuracy | Recall | F1 |
|---|---|---|---|---|
| TEXTCNN | 2 M | 0.720 | 0.570 | 0.603 |
| BERT-CNN | 110 M | 0.750 | 0.676 | 0.674 |
| AlBERT | 11 M | 0.783 | 0.695 | 0.714 |
| BERT | 110 M | 0.788 | 0.706 | 0.717 |
| ERNIE | 193.7 M | 0.794 | 0.739 | 0.743 |
| K-BERT (baseline) | 340 M | 0.807 ± 0.004 | 0.727 ± 0.006 | 0.736 ± 0.005 |
| MSA K-BERT | 310 M | 0.826 ± 0.003 | 0.794 ± 0.004 | 0.810 ± 0.003 |
| Split Ratio | Accuracy | Recall | F1 |
|---|---|---|---|
| 7:2:1 | 0.819 | 0.788 | 0.803 |
| 8:1:1 | 0.814 | 0.781 | 0.797 |
| 5:3:2 | 0.808 | 0.775 | 0.791 |
| Model | Parameters | Average Inference Time per Sample (Seconds) | Throughput (Samples/Second) |
|---|---|---|---|
| K-BERT (baseline) | 340 M | 1.42 | 0.70 |
| MSA K-BERT | 310 M | 1.18 | 0.85 |
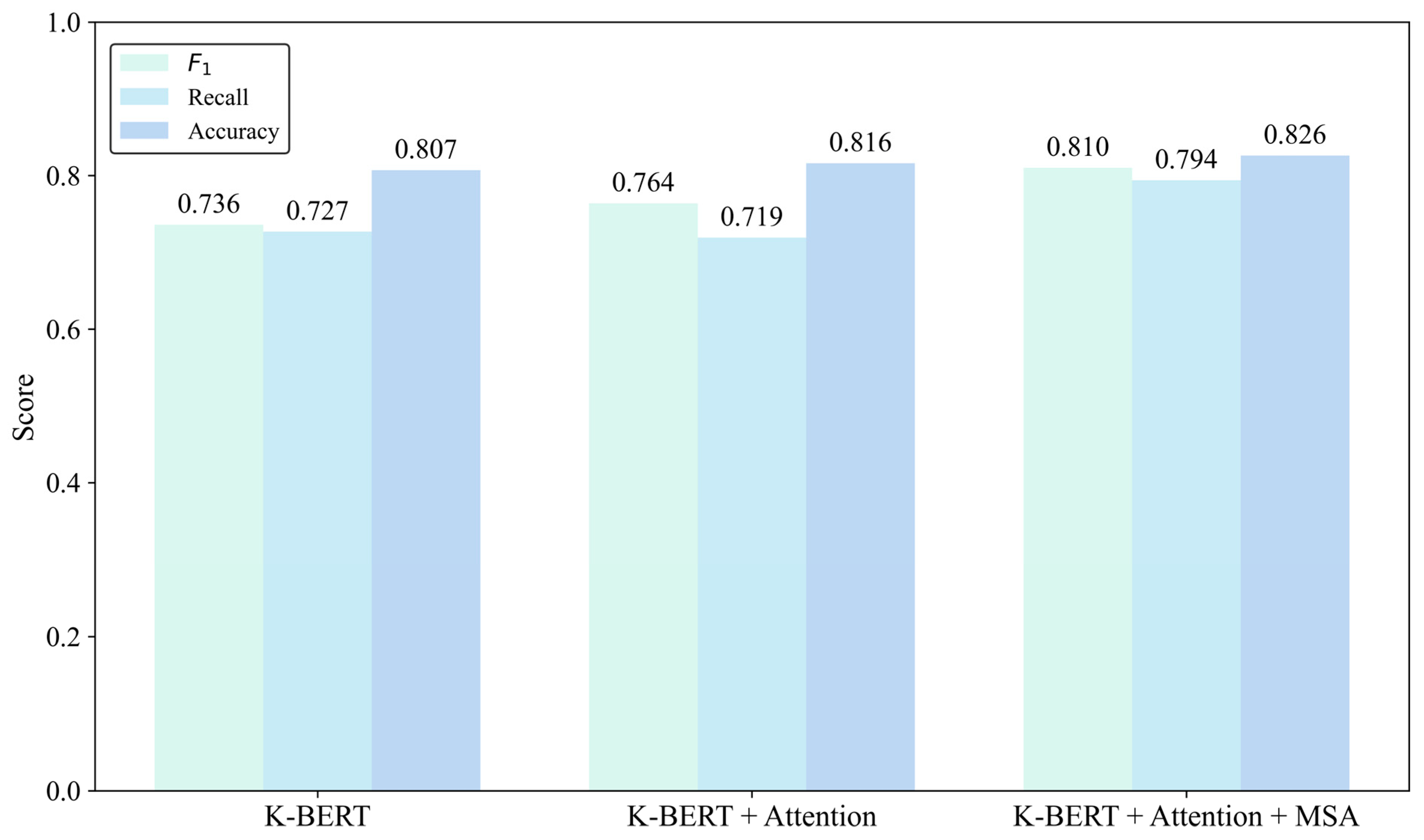
| Baseline (KBERT) | Attention | MSA | Accuracy | Recall | F1 |
|---|---|---|---|---|---|
| ★ 1 | 0.807 | 0.727 | 0.736 | ||
| ★ | ★ | 0.816 | 0.719 | 0.764 | |
| ★ | ★ | ★ | 0.826 | 0.794 | 0.810 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Xi, G. MSA K-BERT: A Method for Medical Text Intent Classification. Appl. Sci. 2025, 15, 6834. https://doi.org/10.3390/app15126834
Yuan Y, Xi G. MSA K-BERT: A Method for Medical Text Intent Classification. Applied Sciences. 2025; 15(12):6834. https://doi.org/10.3390/app15126834
Chicago/Turabian StyleYuan, Yujia, and Guan Xi. 2025. "MSA K-BERT: A Method for Medical Text Intent Classification" Applied Sciences 15, no. 12: 6834. https://doi.org/10.3390/app15126834
APA StyleYuan, Y., & Xi, G. (2025). MSA K-BERT: A Method for Medical Text Intent Classification. Applied Sciences, 15(12), 6834. https://doi.org/10.3390/app15126834





