Abstract
Coronaviruses (CoVs) have recently emerged as significant causes of respiratory disease outbreaks, with the novel coronavirus pneumonia of 2019, known as COVID-19, being highly infectious and triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Understanding virus–host interactions and molecular targets in host cell death signalling is crucial for inhibitor development. Among the promising targets for inhibitor development is the main protease (Mpro), which is essential for viral replication. While current research has focused mainly on covalent inhibitors, growing attention is being given to non-covalent inhibitors due to their potential for lower toxicity and improved resistance to viral mutations. This literature review provides an in-depth analysis of recent in silico approaches used to identify and optimise non-covalent inhibitors of SARS-CoV-2 Mpro. It focuses on molecular docking and robust molecular dynamics (MD) simulation technologies to discover novel scaffolds with better binding affinities. The article summarises recent studies that pre-screened several potential non-covalent inhibitors, including natural constituents like alkaloids, flavonoids, terpenoids, diarylheptanoids, and anthraquinones, using in silico methods. The in silico approach, pivotal to developing small molecules of Mpro non-covalent inhibitors, provides an efficient avenue to guide future research efforts toward developing high-performance Mpro inhibitors for SARS-CoV-2 Mpro, representing the latest advancements in drug design.
1. Introduction
1.1. The Main Protease (Mpro) Enzyme of SARS-CoV-2
The World Health Organisation (WHO) defines the coronavirus that causes severe acute respiratory syndrome (SARS-CoV-2) as COVID-19 [1]. Recently, the virus has become a global threat [2]. With a genome size of about 30 kb, it is a highly contagious positive-strand RNA virus that codes for 29 distinct proteins essential to the virus’s survival and life cycle [3]. The beta coronavirus, or SARS-CoV-2, is closely linked to the SARS-CoV virus that caused the outbreak of the disease in 2002 and 2004 [4]. The coronavirus SARS-CoV-2 is a large, enveloped virus with a single-stranded, non-segmented, positive-sense RNA genome, belonging to the Sarbecovirus subgenus of the Betacoronavirus genus, within the Coronaviridae family. It primarily infects humans but is also known to infect various mammalian species [5].
As shown in the genomic organisation of the SARS-CoV-2 virus in Figure 1, the virion contains both non-structural proteins (nsps) and structural proteins (S, E, M, N) along with several accessory proteins that are essential for its replication, assembly, and infection processes [6]. SARS-CoV-2 has four primary structural proteins: spike (S) glycoprotein, small envelope (E) glycoprotein, membrane (M) glycoprotein, and nucleocapsid (N) protein [7]. These structural proteins support the viral entry and assembly processes [3]. The spike (S) protein facilitates viral entry into host cells by binding to ACE2 receptors on the host’s cells [8]. The nucleocapsid (N) protein is involved in packaging the viral RNA genome and plays an essential role in viral replication [9]. Additionally, the membrane (M) protein interacts with other structural proteins to determine the shape of the viral envelope, while the envelope (E) protein contributes to viral production and maturation [10].
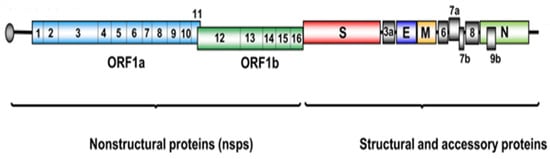
Figure 1.
Schematic representation of the SARS-CoV-2 genome highlighting non-structural proteins, structural proteins, and accessory proteins [11]. Licenced under the CC-BY licence.
However, beyond these structural components, SARS-CoV-2 also expresses several non-structural proteins (nsps) that are critical to the virus’s ability to replicate and survive within host cells [10]. Unlike structural proteins, which build the virus’s physical structure, nsps are responsible for the enzymatic and regulatory functions of the viral life cycle [12]. These proteins are initially translated as part of the large ORF1ab polyprotein, which is cleaved by two viral proteases—the main protease (Mpro or 3CLpro, 3-Chymotrypsin-like Protease or nsp5) and the papain-like protease (PLpro)—to release 16 individual nsps [13]. Each of these proteins performs specialised functions that collectively form the virus’s replication and transcription complex (RTC), essential for SARS-CoV-2’s pathogenicity [14].
Key nsps include nsp12, the RNA-dependent RNA polymerase (RdRp), which synthesises the viral RNA, and nsp13, a helicase that unwinds RNA, thus supporting efficient replication [15]. Nsp3, containing PLpro, plays a dual role in both cleaving viral polyproteins and modulating host immune responses [16]. The main protease, nsp5 (Mpro), is particularly significant, as it processes the majority of viral polyprotein cleavage sites, making it a prime target for antiviral drugs [17]. Like other coronaviruses, these proteins enclose the RNA genome [5]. The main protease (Mpro), the papain-like protease (Plpro), and the RNA-dependent RNA polymerase (RdRp) are the only three major targets for drug development against the SARS-CoV-2 virus [18].
Mpro is a cysteine protease that plays a critical role in the replication of SARS-CoV-2. Notably, it shares approximately 96% sequence and structural similarity with the Mpro of SARS-CoV, as illustrated in Figure 2A. Despite differences among coronaviruses, Mpro demonstrates intense sequence and structural conservation, distinguishing it from human proteases and making it an attractive target for drug discovery to inhibit viral replication and proliferation [19].
Functionally, Mpro operates as a homodimer composed of two protomers [9]. Structurally, Mpro consists of three domains. Domain I (residues 8–101) and domain II (residues 102–184) are responsible for the main enzymatic activity, containing the catalytic dyad composed of Cys145 and His41, located in the cleft between these two domains (P1 region) [20] as displayed in Figure 2B. Domain III (residues 201–306) supports stabilising the enzyme structure and aiding in substrate recognition.
SARS-CoV-2 and SARS-CoV share 294 amino acids out of 306 residues. The amino acid residues Phe140, Leu141, Asn142, His163, and Glu166 constitute the S1 subsite. Meanwhile, the His41–Cys145 catalytic dyad (Figure 2C), along with Ser144 and Glu143, form the S1′ subsite. The S2 subsite includes His164, Asp187, Arg188, and Met49, Tyr54, while S3 comprises Leu167, Gln189, Met165, Thr190, and Gln192 [21].
To emphasise its critical role in viral replication and pathogenesis, Mpro has become an essential target of many existing vaccines and antiviral medicines [22]. A thorough understanding of the structure–activity relationships (SAR) within its active site subpockets (S1, S1′, S2, and S3) is crucial for designing potent inhibitors (Figure 2D).
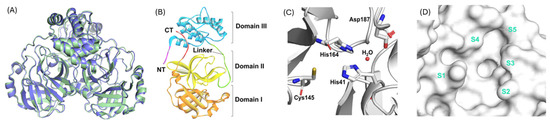
Figure 2.
(A) 3D representations of the main proteases (as a homodimer) from SARS-CoV-2 (pale green, PDB: 6XHU) and SARS-CoV (slate, PDB: 1UJ1). (B) Structural domains of SARS-CoV-2 Mpro monomer, showing domain I (orange), domain II (yellow), and domain III (blue). (C) The catalytic dyad residues in the active site of SARS-CoV-2 Mpro. (D) Surface view of the catalytic pocket and its subpockets (S1–S5) in SARS-CoV-2 Mpro [23]. Licenced under the CC-BY licence.
These subpockets provide specific sites for molecular interactions that can be targeted to enhance inhibitor binding and efficacy. Overall, the structural and functional significance of Mpro is well established, reinforcing its status as a critical target for antiviral drug development against SARS-CoV-2 and its emerging variants.
1.2. The Need for Novel Inhibitors
Existing inhibitors may experience reduced efficacy, develop resistance, or cause unfavourable side effects [24]. Therefore, developing novel inhibitors is essential for overcoming these challenges and enhancing therapeutic efficacy. Antiviral drugs such as molnupiravir, remdesivir, and PF-07321332 (nirmatrelvir), as presented in Figure 3, have demonstrated clinical effectiveness. While prolonged use may lead to the emergence of drug-resistant mutations, this highlights the ongoing need for research and development of new inhibitors [25].
Other inhibitors, such as FB2001, SIM0417, vv116, and RAY003, primarily employ a peptide-based covalent method. This method presents challenges concerning target selectivity and pharmacokinetic features [26]. Consequently, there is an increasing emphasis on developing safer coronavirus antiviral drugs that are neither covalent nor peptidomimetic, such as Shionogi’s S-217622. Compared to other inhibitors like PF-07321332, S-217622 demonstrates improved drug metabolism and pharmacokinetic properties, addressing certain limitations. While the US Food and Drug Administration (FDA)-approved antivirals such as boceprevir and telaprevir effectively suppress Mpro, some mutants have exhibited lower binding affinity to these drugs, particularly at critical residues likely to undergo positive selection [26]. Novel coronavirus strains could potentially establish endemicity, posing a significant risk of another global pandemic, necessitating the development of effective antiviral therapies [26].
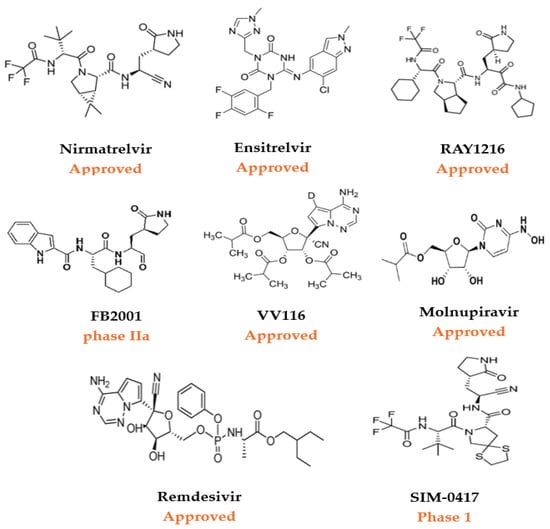
Figure 3.
A selection of approved small-molecule COVID-19 therapeutics and investigational antivirals for SARS-CoV-2 in clinical trials [27].
Despite improvements, challenges remain in advancing clinical trials and eventual approval of Mpro non-covalent inhibitors [26]. Weak inhibitory activity, the necessity for structurally diverse inhibitors, undesirable drug metabolism, pharmacokinetics (DMPK) characteristics, and toxicity concerns require the exploring of alternative approaches [28]. For instance, integrating modern computational methods with experimental practice and utilising efficient screening techniques could overcome these challenges. Therefore, drug repurposing, which leverages existing therapies, presents an attractive option for addressing the urgent need for effective antiviral solutions. The following section will delve into the role of drug repurposing in antiviral research.
1.3. The Role of Drug Repurposing
Drug repurposing is an efficient strategy for identifying new therapeutic uses for already approved drugs [13]. This approach gained prominence during the COVID-19 pandemic when speed and safety were critical. One of the key advantages of drug repurposing, particularly for identifying SARS-CoV-2 inhibitors, is its potential to accelerate the drug development process significantly. Since these drugs have already undergone extensive testing for safety and efficacy, they can bypass early-phase trials, allowing for faster progression to clinical evaluation for new indications [29].
Beyond speed, drug repurposing also minimises the financial risks associated with traditional drug discovery. It capitalises on existing pharmacological and toxicological data, which aids in predicting efficacy and side effects in new disease contexts [25]. Moreover, many of these drugs are already in production, facilitating rapid distribution during public health emergencies [30].
Recent studies indicate that the effects of SARS-CoV-2 extend beyond the respiratory system (RS) to the central nervous system (CNS), intensifying the need for rapid and effective treatments [30,31]. In this context, drug repurposing has facilitated the identification of promising Mpro inhibitors. For instance, the noncovalent inhibitor ML188, initially developed for SARS-CoV, was successfully adapted for SARS-CoV-2 [32]. Similarly, compounds such as X77, masitinib, and ML300 have emerged through repurposing pipelines as potential SARS-CoV-2 Mpro inhibitors [33]. These examples underscore the strategic value of repositioning existing drugs to accelerate antiviral drug discovery.
While several reviews have broadly explored SARS-CoV-2 Mpro inhibitors, many of these articles either conflate covalent and non-covalent classes or lack a detailed analysis of the computational strategies specifically tailored to non-covalent inhibitors. The present review addresses this gap by critically highlighting representative structure-based in silico approaches—including molecular docking, molecular dynamics (MD) simulations, and virtual screening—used in the discovery and optimisation of non-covalent Mpro inhibitors, with a particular emphasis on role of repurposed drug scaffolds, and the increasing importance of robust, high-fidelity MD simulations in elucidating binding mechanisms, and refining lead compounds. Rather than providing an exhaustive survey, this work offers a focused and critical perspective on recent computational advancements that have contributed to the development of these promising compounds, a topic that remains under-discussed in the current literature.
Importantly, the review also underscores the essential role of in vitro validation to confirm biological relevance and the value of integrated in vitro and in vivo strategies in progressing potential Mpro inhibitors from computational hits to clinically viable candidates. This reinforces the notion that while in silico techniques are powerful and widely used in early-stage drug discovery, they must be complemented by experimental studies to ensure translational impact.
2. Mpro Inhibitors for SARS-CoV-2
2.1. Overview of Mpro Inhibitors
Over the past few years, various classes of inhibitors have been explored, ranging from covalent and non-covalent small molecules to natural-product-derived inhibitors. While some have shown promise in preclinical and clinical trials, challenges related to resistance, pharmacokinetics, and selectivity persist [34]. Each class comes with distinct mechanisms of action, advantages, and challenges. This review focuses on non-covalent Mpro inhibitors due to their reversibility, reduced risk of off-target toxicity, and increasing evidence of potent antiviral efficacy.
Covalent inhibitors form a covalent bond with the target protein, often at a nucleophilic residue within the active site [35]. These inhibitors can be further categorised as reversible and irreversible inhibitors, such as nirmatrelvir, an FDA-approved drug, which forms reversible bonds with the active site of Mpro via reactive functional groups like nitriles or Michael acceptors [35].
Several Mpro covalent inhibitors have shown promising potential against SARS-CoV-2, which has been identified through both experimental assays and in silico studies (Figure 4). Among these, MG-101, a calpain inhibitor, exhibited robust inhibition of SARS-CoV-2 replication with an IC50 of 2.89 ± 0.86 μm using the FRET assay [36] and was shown to impact both Mpro activity and viral entry, with an EC50 of 0.038 µm, suggesting its potential as a dual inhibitor [37]. Lycorine HCl, a plant-derived alkaloid, demonstrated potent antiviral activity with an EC50 of 0.01 µm and a high selectivity index [38]. Additionally, the HIV protease inhibitors Atazanavir and Nelfinavir mesylate displayed significant binding affinity to Mpro, contributing to their antiviral efficacy [39]. The effectiveness of drug combination therapy was demonstrated when MG-101 was paired with other Mpro inhibitors like Lycorine HCl or Nelfinavir, achieving substantial reductions in viral titer [37]. This combination approach, targeting both Mpro and PLpro, presents a promising strategy for COVID-19 treatment [37].
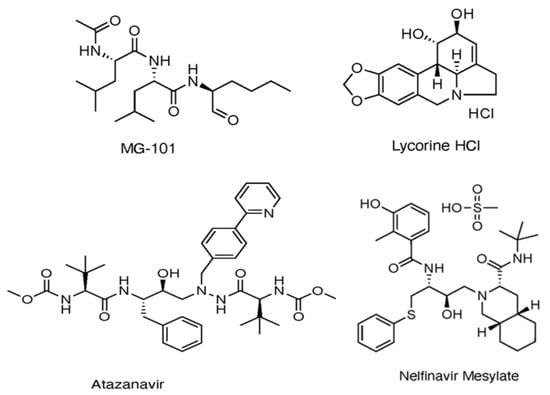
Figure 4.
Multiple Mpro covalent inhibitors targeting SARS-CoV-2 [37].
Dual inhibitors, like olgotrelvir, combine Mpro inhibition with the targeting of additional pathways, such as host proteases, to enhance antiviral potency and reduce resistance [40]. While covalent inhibitors like nirmatrelvir have demonstrated significant clinical success, non-covalent inhibitors and dual-acting compounds hold promise for addressing challenges such as resistance and toxicity [35]. Although covalent inhibitors have shown significant promise, non-covalent Mpro inhibitors are gaining attention for their potential for reversible binding [41].
2.2. Discovery Strategies for Non-Covalent Inhibitors of Mpro
Unlike covalent inhibitors, which form irreversible bonds with the catalytic dyad and which may raise concerns about off-target toxicity and resistance, non-covalent inhibitors engage the active site reversibly through hydrogen bonding, hydrophobic contacts, and van der Waals interactions. These interactions often lead to improved selectivity, reduced toxicity, better pharmacokinetic profiles, and a lower likelihood of resistance development [16]. Based on recent findings, four key guidelines have been established to design highly effective inhibitors targeting the SARS-CoV-2 Mpro enzyme. First, Rule 1 highlights that the S1 sub-pocket requires a ring of five or six members with a proton donor or acceptor (-NH or =O group) to form crucial hydrogen bonds with a particular residue. Moving to Rule 2, in the S1′ sub-pocket, which is known for hydration and design issues, it is critical to establish a water bridge between the scaffold and Thr26, as well as hydrogen/covalent bonding with the Asn142-Gly143-Ser144-Cys145 (NGSC) motif.
Furthermore, the S2 sub-pocket favours cyclic moiety interactions with His41 or hydrogen bonding with His41 and Gln189, which increases efficacy. Finally, the S3 binding site requires a significant aromatic or aliphatic component. It has been observed that while the S1 and S2 sub-pockets are significant for noncovalently bound inhibitors, the S1′ and S3 sub-pockets may influence inhibitor efficacy [42].
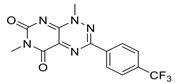
Considering these structural insights, ensitrelvir (S-217622) was approved in Japan in November 2022 under the brand name Xocova®. Its discovery involved a combination of virtual screening, biological assays, and structure-based drug design [17]. As shown in Figure 5, optimising the P1′ ligand and introducing a 6-chloro-2-methyl-2H-indazole scaffold enhanced its inhibitory activity by 90-fold, achieving an IC50 of 0.013 μm and an EC50 of 0.37 μm. The compound demonstrated broad-spectrum antiviral activity against SARS-CoV-2 variants and favourable pharmacokinetics for oral dosing, including high metabolic stability (96% in human liver microsomes) and 97% oral absorption. Clinical trials confirmed its safety and tolerability, with no significant safety concerns identified, leading to both emergency and full approval in Japan [43].
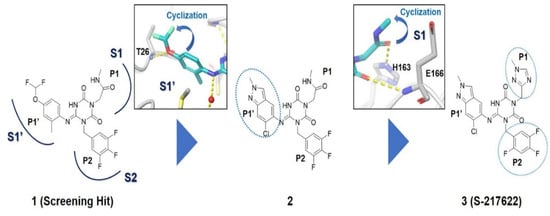
Figure 5.
Structural optimisation and representative compounds in the development of ensitrelvir [17].
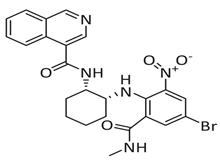
Meanwhile, mprosevir (WU-04), discovered through DNA-encoded library screening, utilises an isoquinoline-based core to inhibit SARS-CoV-2 Mpro non-covalently with an IC50 of 0.072 μm [44]. Preclinical studies showed antiviral efficacy comparable to nirmatrelvir in animal models [45]. Phase 1 clinical trials evaluated its safety, tolerability, and pharmacokinetics, whereas Phase 2 and ongoing Phase 3 trials aim to establish its efficacy in mild-to-moderate COVID-19 cases [34]. The lack of published Phase 2/3 data limits direct comparison with ensitrelvir.
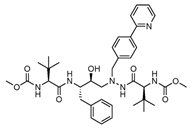
Another noteworthy Mpro non-covalent inhibitor is ML188, identified through structure-based drug design, binds the active site with an IC50 of 1.5 µm, interacts with the catalytic dyad (His41 and Cys145) via hydrogen bonds, and stabilises the S1′ subsite through hydrophobic interactions [32]. Given its promising binding characteristics, ML188 is an essential scaffold for further optimisation. Figure 6 represents the chemical optimizations of ML188, which yielded compound 23R with an IC50 of 0.20 μm, demonstrating significantly improved potency. Moreover, it exhibited antiviral activity in both Vero E6 (EC50 = 1.27 μm) and Calu-3 cells (EC50 = 3.03 μm). Structural analysis using X-ray crystallography revealed that 23R induced a novel binding pocket between the S2 and S4 subsites, contributing to its strong interaction with the enzyme [46].
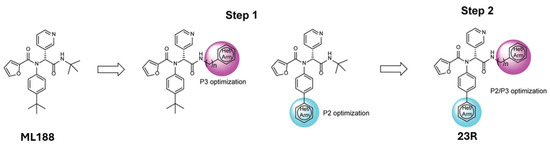
Figure 6.
Chemical optimisation of ML188 [46].
In parallel, ML300 was also subjected to optimisation, ultimately leading to the development of compound 41 (CCF981). Notably, this derivative extended into the hydrophilic S2c subsite, significantly enhancing its inhibitory potency, as reflected by an IC50 of 0.068 μm and an EC50 of 0.497 μm in Vero E6 cells [16].
Scaffold refinement emerged as a central strategy for enhancing potency, as demonstrated by the advancement of ML188 to its optimised analogue 23R (IC50 = 0.20 μm) and the progression of ML300 to CCF981 (IC50 = 0.068 μm). These improvements reflect the value of targeted modifications to key substituents, particularly those that enhance interactions within the S1′, S2, and S4 subsites. Nonetheless, these efforts remain mostly cell-line-bound, lacking validation in animal models and pharmacokinetic studies, which hinders the assessment of in vivo efficacy. For example, while 23R induced a new binding pocket, the absence of ADME or toxicity profiling remains a critical gap.
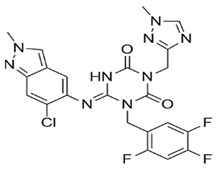
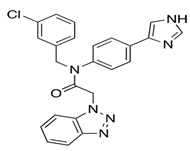
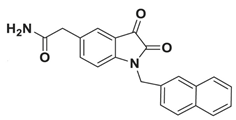
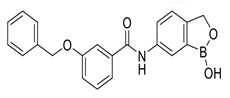
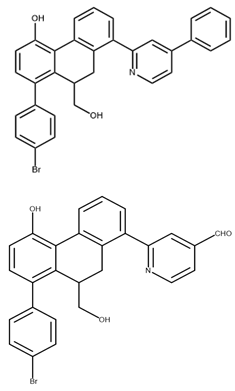
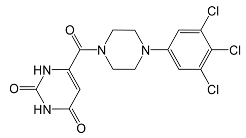
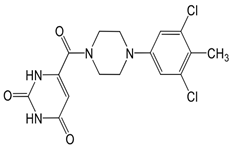
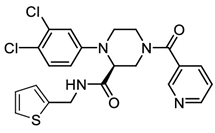
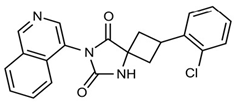
In pursuing effective SARS-CoV-2 Mpro inhibitors, Table 1 displays some representative medicinal chemistry strategies and active compounds included in this review. In the screening of structure-related compounds based on a series of N-substituted isatin derivatives, compound 26 was identified as a potent Mpro inhibitor, demonstrating the highest potency (IC50 = 0.045 μm) [43]. However, high cytotoxicity hindered further antiviral evaluation. Additionally, Benzoxaborole-based inhibitors were explored for their dual activity against SARS-CoV-2 and dengue virus proteases [47]. The resulting compound 18 exhibited an IC50 of 6.1 μm against SARS-CoV-2 Mpro with minimal cytotoxicity.

Table 1.
Chemical structures and biological activities of multiple non-covalent inhibitors using in silico and experimental methods.
Moreover, non-peptidomimetic 9,10-dihydrophenanthrene derivatives, specifically C1 and C2, demonstrated potent Mpro inhibition with IC50 values of 1.55 μm and 1.81 μm, respectively [48]. Compound C1 also exhibited excellent metabolic stability in the gastrointestinal tract, plasma, and liver microsomes. Seeking to enhance binding affinity further, researchers introduced structural modifications to Mcule-5948770040 (Figure 7), leading to the development of HL-3-68 and Mcule-CSR-494190-S1, with HL-3-68 showing the strongest binding affinity [49]. However, none of these compounds displayed measurable antiviral activity in cell-based assays. On another front, a piperazine-based inhibitor, GC-14, stood out due to its potent inhibition (IC50 = 0.40 μm) and strong antiviral activity (EC50 = 1.1 μm), along with low cytotoxicity (CC50 > 100 μm) and high selectivity for SARS-CoV-2 Mpro [50]. These findings highlight the ongoing challenges in balancing potency, cytotoxicity, and antiviral efficacy in the search for effective Mpro inhibitors.
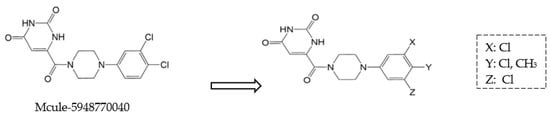
Figure 7.
Structural modifications of the hit compound (Mcule-5948770040) [41].
Compound 26 and compound 18 demonstrate that high enzymatic potency does not guarantee antiviral efficacy, particularly when cytotoxicity (as seen in compound 26) or poor cellular uptake (as likely in compound 18) limit their utility. This pattern recurs with HL-3-68, which fails in cellular assays despite its strong binding affinity—emphasising the well-known disconnect between in vitro target engagement and whole-cell antiviral efficacy.
Screening an ultra-large set of 235 million molecules led to the selection of imidazolidine-2,4-dione-based inhibitors, which were further optimised to develop compound 19 with sub-micromolar inhibitory activity and broad-spectrum anti-coronavirus properties [51]. Similarly, high-throughput screening (HTS) using FRET assays uncovered non-covalent inhibitors such as walrycin B [52] and quinazoline derivative QZ4 [53]; however, their therapeutic potential was limited due to toxicity concerns. In contrast, masitinib, identified through cell-based screening, exhibited vigorous anti-SARS-CoV-2 activity (EC50 = 3.2 μm) and was validated structurally as a Mpro inhibitor [54].
Machine learning-driven virtual screening enhanced efficiency, ultimately leading to the discovery of Z-DQMD-FMK, which exhibited an IC50 of 0.92 μm for Mpro inhibition [55]. Likewise, deep docking methods identified Z4927220858 (IC50 ~10 μm) spanning multiple Mpro subsites, further emphasising AI’s role in improving hit rates [56].
Compounds such as GC-14 and Masitinib exhibit sub-micromolar inhibitory activity and present favourable selectivity and cytotoxicity profiles, critical parameters for clinical development.
To discover new inhibitors, a set of thioester-based inhibitors was designed via virtual screening from the Tübingen Kinase Inhibitor Collection (TüKIC) and exhibited notable inhibitory activity against Mpro, supported by co-crystal structures that revealed key interactions [57]. While this study is commendable for experimentally validating hits through crystallography, it stops short of evaluating these compounds in relevant cellular or in vivo models, leaving questions about pharmacokinetics, bioavailability, and toxicity unanswered. Figure 8 summarises the synthesis and activity of these thioesters, such as compounds 3w and 3x, which showed potent inhibition in vitro.
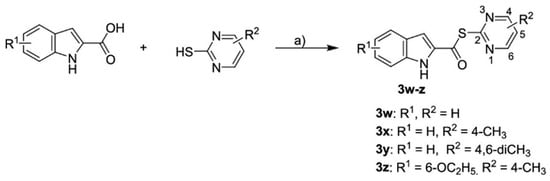
Figure 8.
Synthesis of thioesters 3w-z [57]. “a)” POCl3, pyridine, DCM, 12 h, 55–82%.
In contrast, repurposing efforts using HIV protease inhibitors like lopinavir and ritonavir initially showed promise in vitro, with lopinavir/ritonavir yielding the lowest IC50 value. However, these compounds also significantly impaired cell viability [58], suggesting a narrow therapeutic index and raising doubts about their suitability for treating COVID-19. The high concentrations required for darunavir and atazanavir, despite minimal cytotoxicity even at 200 µm, further underscore the disconnect between protease inhibition and clinical efficacy. These results suggest that the observed antiviral effects might stem from off-target interactions rather than specific Mpro inhibition, emphasising the need for mechanistic clarity and further in vivo validation. Table 2 overviews existing HIV protease inhibitors targeting SARS-CoV-2 Mpro [58].

Table 2.
Inhibition results in cell culture of HIV protease inhibitors targeting SARS-CoV-2 Mpro [58].
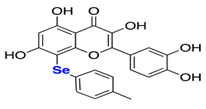
Natural products have also been extensively explored as Mpro inhibitors. While they offer chemical diversity, many of these studies suffer from an over-reliance on docking scores without a robust experimental follow-up. Table 3 outlines some natural compounds that inhibited Mpro of SARS-CoV-2. For example, shikonin emerged from high-throughput screening (HTS) as a non-covalent Mpro inhibitor with an IC50 of 0.397 μm [59]. However, its cytotoxicity limited further development [60]. Similarly, quercetin exhibited modest Mpro affinity, but structural optimisation using selenium-functionalized derivatives enhanced its antiviral activity [61]. Furthermore, selenoquercetin analogues were synthesised and screened as SARS-CoV-2 Mpro inhibitors, demonstrating improved potency over quercetin. The most effective derivative, compound 2d, exhibited an IC50 of 8 μm for antiviral activity, representing a 24-fold enhancement compared to quercetin. Consequently, in silico and experimental studies revealed non-covalent inhibition involving a selenium-mediated hydrogen bond with Gln189, stabilising the compound within the active site and blocking the His41/Cys145 catalytic dyad [61]. However, although in silico and biochemical data support its binding to the catalytic dyad, its in vivo efficacy and safety remain unproven.

Table 3.
Chemical structures and IC50 values of selected natural compounds that inhibited Mpro of SARS-CoV-2 [41].
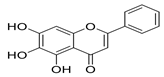
Moreover, baicalein, a polyphenolic compound found in Scutellaria baicalensis and other plants, stands out as a more balanced candidate, exhibiting strong docking (−9.8 kcal/mol), low cytotoxicity, and in vitro potency (EC50 = 1.69 µm; SI > 118) [62,63,64]. However, detailed pharmacokinetic and metabolic stability studies are needed before clinical translation.
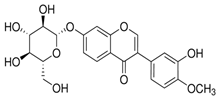
Similarly, calycosin-7-O-β-glucopyranoside, derived from natural sources, exhibits strong binding to Mpro with a docking score of −8.33 kcal/mol [65]. This compound, like Baicalein, presents an appealing option for drug development due to its structural characteristics that favour interaction with Mpro’s active site, which could lead to effective inhibition of the viral enzyme [65].
While in vitro potency remains a vital metric, its translation into clinical viability equally relies on pharmacokinetic and toxicity profiles. Shikonin, despite having strong IC50 values, faced limitations due to cytotoxicity. In contrast, baicalein and calycosin-7-O-β-glucopyranoside displayed promising drug-like behaviour with low toxicity and high selectivity.
A broader screening of 73 withanolides using Lipinski’s Rule of Five and ADME filters identified several hits with favourable drug likeness [66], but these remain in an early computational stage. While such filters are helpful, they may oversimplify complex pharmacokinetic behaviour, and reliance on them alone can result in false positives or negatives.
Rational drug development has contributed to enhanced binding affinity and specificity through methods such as virtual screening of focused libraries (e.g., TüKIC), scaffold hybridization, and the incorporation of favourable pharmacophores (e.g., piperazine rings, selenium atoms). In natural products, compounds like baicalein and selenoquercetin illustrate that modest starting activity can be significantly improved through targeted derivatization, for instance, introducing selenium to enhance hydrogen bonding with Gln189.
Together, these examples highlight the power of in silico approaches in identifying effective inhibitors of SARS-CoV-2 Mpro, utilising both natural and synthetic compounds. Computational tools play a crucial role in antiviral drug development by streamlining hit identification and optimisation. Ultimately, bridging the gap between the discovery and clinical application of non-covalent Mpro inhibitors relies on ongoing collaboration among computational modelling, rational drug design, and biological validation.
2.3. Computer-Aided Drug Design
Computer-aided drug design (CADD) has quickly become an essential tool in the battle against SARS-CoV-2, assisting researchers in identifying and optimising inhibitors aimed at the virus’s main protease [67]. By integrating structure-based (SBDD) and ligand-based (LBDD) strategies, CADD provides a rational and cost-effective framework that complements traditional discovery approaches [68]. While promising, its value lies in its ability to prioritise and optimise compounds before entering the resource-intensive experimental pipeline.
In structure-based approaches, molecular docking, molecular dynamics (MD) simulations, and free energy calculations predict how candidate molecules interact with Mpro [69]. These methods are crucial for estimating the binding affinity of potential inhibitors, providing insights into the strength and stability of drug-target interactions. High-throughput virtual screening (HTVS) accelerates the drug discovery process by enabling the evaluation of large chemical libraries, significantly reducing the time and cost typically associated with experimental testing. More advanced techniques, such as quantum mechanics/molecular mechanics (QM/MM) simulations, offer more profound insights into ligand–protein interactions, thereby enhancing the precision of drug optimisation [70].
On the other hand, ligand-based approaches, including quantitative structure–activity relationship (QSAR) modelling and pharmacophore mapping, utilise known inhibitors to predict new, similarly bioactive compounds [69]. These methods have proven effective in quickly identifying lead candidates, which can be refined into more potent inhibitors. They are crucial in guiding the rational design of novel Mpro inhibitors, ensuring that the compounds exhibit the desired biological activity.
The process of drug design, especially when enhancing existing compounds, follows key steps [22]. This structured process optimises the chemical properties of drug candidates, enhancing their efficacy, selectivity, and pharmacokinetic profiles [28]. It begins by identifying a lead compound—such as ML188—and organising these data in databases according to drug-like criteria [16]. Techniques such as X-ray crystallography, NMR, and cryo-EM are used to determine the three-dimensional structure of the target protein [58]. The compounds are then subjected to an ensemble docking approach, where conformers generated during molecular dynamics simulations are utilised to calculate binding free energy, which predicts the strength of drug–target interactions [71]. This process culminates in synthesis, validation, and preclinical trials, which are essential to ensure that the inhibitors are effective in real-world conditions.
As drug design continues to evolve, the increasing use of computer screens has amplified the significance of CADD in identifying antiviral agents. For example, the successful discovery of antiviral compounds for diseases like HIV and HCV has spurred interest in targeting Mpro for SARS-CoV-2 treatment [72]. Leveraging advanced computational techniques, researchers have identified terpene metabolites from the Red Sea coral reefs as promising Mpro inhibitors [73]. These natural compounds, including flavonoids, alkaloids, and terpenoids, exhibited strong binding affinities to Mpro, highlighting the untapped potential of marine and plant-derived metabolites in COVID-19 therapy. Notable examples include Lopinavir, Depresosterol, and Erylosides B, each offering a unique structural foundation for future drug development.
Natural product scaffolds have also inspired innovative approaches to Mpro inhibition. Compounds derived from the aloe plant, such as aloesin and aloeresin D, demonstrated inhibitory activity with IC50 values ranging from 6.8 μm to 138.0 μm (Figure 9) [74]. These findings showcase the ongoing efforts to harness the therapeutic potential of natural sources. Similarly, Table 4 reported a group of bioactive compounds from medicinal plants, such as N3, hesperidin, rutin, diosmin, and apiin, with their binding affinities exceeding the control compound Nelfinavir of −147 [75]. These discoveries highlight the promise of plant-derived molecules as a valuable resource for designing effective inhibitors. However, studies reporting IC50 values without correlating them with therapeutic indices or bioavailability parameters risk overestimating their drug-like potential.
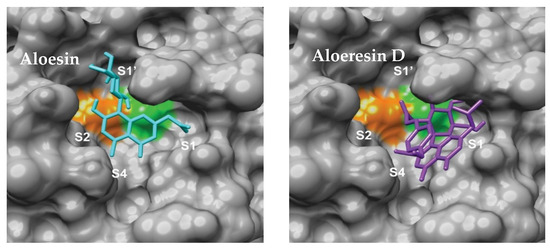
Figure 9.
Aloesin and Aloesin D binding modes [74]. The catalytic dyad of Cys 145 and His 41 are displayed in green and orange respectively.

Table 4.
Results of the docking of some phenolic compounds on the crystal structure of COVID-19 main protease, including N3, Nelfinavir, Hesperidin, Rutin, Diosmin, and Apiin [75].
Despite these promising results, the transition from computational predictions to experimental validation remains critical. A recent in silico study investigated two virtual screening methods using ultra-large chemical databases to identify Mpro inhibitors. One method utilised structure-based docking on a library of over 235 million compounds, while the other employed a fragment-based approach [51]. As represented in Figure 10, both methods identified distinct groups of inhibitors with promising antiviral effects, confirmed through crystallography and cellular models. This underscores the value of large-scale screening in identifying potent inhibitors quickly and effectively.
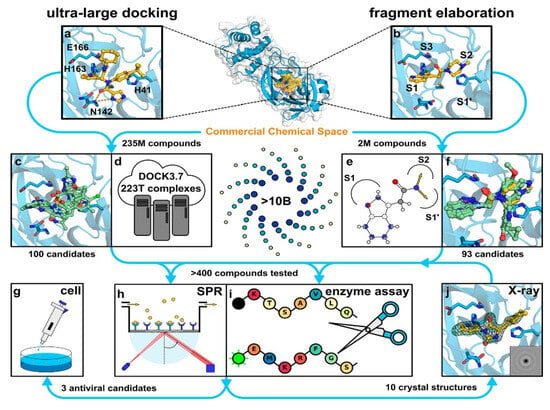
Figure 10.
Representation of the two virtual screening approaches: ultra-large docking and fragment elaboration [51]. Licenced under the CC-BY licence. (a) The crystal structure of Mpro bound to the inhibitor X77 (PDB ID: 6W63) was used for an ultralarge-scale dock-ing screen. (b) The crystal structure of Mpro in complex with a fragment molecule (compound 4, PDB ID: 5RF7) was used for docking a focused library designed for fragment elaboration. (c,d) Representative compounds identified from molecular docking of a ZINC15 library containing 235 million lead-like molecules. (e,f) Representative compounds identified from docking focused libraries derived from fragment elaboration. (g–j) Selected inhibitors were further validated through biophysical and biochemical assays, X-ray crystallography, and testing in virus-infected cell models.
Fragment-based drug discovery has also yielded valuable insights. For instance, NMR-based screening identified the uracil-containing fragment Z604, which showed time-dependent binding to Mpro [76]. By expanding Z604 through fragment-growing techniques, researchers developed a compound that occupies all four active site subpockets, offering a blueprint for dual-inhibitor drug design. Yet, while this method excels at identifying minimal binding elements, it requires careful balancing of binding potency and drug-like properties during elaboration—a challenge often overlooked in computational-only studies.
On the other hand, docking-based screenings have identified other promising inhibitors, such as pyrimidinedione derivatives, which were further optimised through medicinal chemistry and X-ray crystallography [77].
Docking provides crucial insights into how small molecules bind to the active site of target proteins, offering a foundation for lead identification and optimisation [78]. This process identified compounds such as C21, demonstrating strong binding potential through multiple hydrogen bonds and hydrophobic interactions. This discovery highlights the critical role of computational methods in predicting and optimising drug interactions.
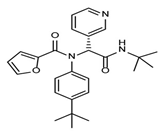
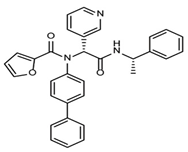
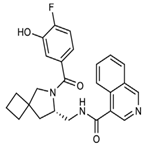
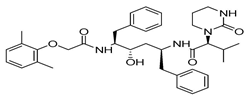
The discovery of nonpeptidic, noncovalent SARS-CoV-2 Mpro inhibitors, such as Mei derivatives, further illustrates the power of CADD. By employing advanced docking techniques and validating these compounds through re-docking studies, researchers identified essential residues involved in binding, such as Cys145 and Glu166 [79]. This information guided the optimisation of more potent derivatives, resulting in compounds like S5-27 and S5-28 (Figure 11), which exhibited improved binding affinities and inhibitory activities with IC50 values of 1.09 and 1.35 μm, respectively.
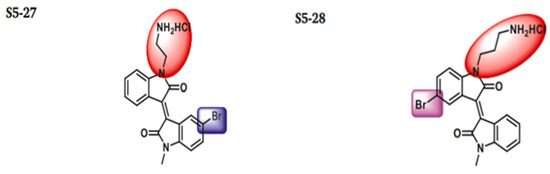
Figure 11.
The chemical structures of S5-27 and S5-28.
In contrast to the promise of in silico tools, their overuse without a proper experimental follow-up remains a critical weakness. For example, compounds like C21, S5-27, and pyrimidinedione derivatives exhibited high binding affinities and favourable interaction profiles in docking simulations; only a handful were synthesised and biologically tested.
While computational methods like docking and molecular dynamics simulations provide invaluable insights, experimental validation remains crucial. Recent studies have integrated both approaches, validating computational predictions of binding affinity and molecular interactions through synthesis and testing. For example, Kitamura et al. used molecular docking to predict the binding of a compound to the 3CLpro protease, confirming their predictions through co-crystallisation experiments and revealing a novel binding pocket [80]. Similarly, Han et al. validated their computational predictions by synthesising and testing CCF0058981, which demonstrated potent antiviral activity in cell-based assays [16].
Virtual screening has also proven effective in identifying preclinical compounds like MG-132, which exhibited sub-micromolar antiviral activity in Vero cells [81]. Integrating virtual screening with experimental assays offers a powerful strategy for rapidly identifying potential antiviral agents. In parallel, the screening of FDA-approved drugs has yielded promising compounds, such as dipyridamole and ebselen, which demonstrated antiviral activity against Mpro in vitro [60,82].
The development of triazole-based inhibitors like ensitrelvir exemplifies the successful integration of computational and experimental methods. After virtual screening and X-ray validation, ensitrelvir entered clinical trials and became the first non-covalent Mpro inhibitor approved [77]. Although compounds like GC376 show potent inhibition of SARS-CoV-2 Mpro in vitro, the in vivo efficacy remains a challenge [83,84].
While these successes are promising, challenges persist in computational drug discovery. False positives in virtual screening, inaccuracies in docking score predictions, and the need for extensive experimental validation remain significant hurdles [77]. However, by merging computational predictions with rigorous experimental testing, this integrated approach enhances the likelihood of developing effective therapies that can successfully transition from in vitro hits to in vivo candidates.
Thus, traditional drug discovery approaches are often slow and resource-intensive, while in silico docking analysis offers a quicker, more cost-effective alternative for large-scale molecular docking screening supported by molecular dynamics simulations. This method has been particularly effective in identifying potential inhibitors for SARS-CoV-2 Mpro by focusing on inhibitors previously reported for SARS-Mpro [85].
2.4. Molecular Dynamics (MD) in Drug Development
Computer-aided drug design (CADD) includes various in silico tools, with molecular dynamics (MD) simulations being one of the most essential methods, particularly in pharmaceutical development and formulation research [86]. Beyond initial hit identification, MD simulations enable researchers to assess drug behaviour at the atomic level, providing insights into solubility, conformational flexibility, and complex stability that static docking alone cannot capture. This section explores the significant contributions of MD simulations in developing inhibitors targeting the SARS-CoV-2 Mpro, illuminating how these simulations guide the design and optimisation of promising candidates.
MD simulations, by mimicking the dynamic behaviour of protein–inhibitor complexes, allow researchers to probe interactions that are key to inhibitor efficacy. These interactions—hydrogen bonds, hydrophobic contacts, and van der Waals forces—are pivotal in determining how effectively an inhibitor can bind to its target [87]. With this understanding, researchers can predict how modifications to the inhibitor structure may enhance its binding affinity, specificity, and overall therapeutic potential [86]. The true advantage of MD lies in its ability to differentiate between fundamental binding and notable stability under physiological conditions—something often overlooked in research focused solely on docking. Moreover, the progress in machine learning (ML) has increased reliance on MD simulations, streamlining the rapid development of new pharmacological agents.
One notable success in the search for effective Mpro inhibitors has been the exploration of compounds derived from withania species. Among them, Withacoagulin H (W30) emerged as a standout candidate [66]. Through 50 ns of MD simulations, it exhibited remarkable stability within the Mpro active site, demonstrating a binding energy calculated by MMGBSA of −63.463 kJ/mol, significantly outperforming other compounds (Figure 12). This impressive result suggests its potential as a valuable candidate for further in vitro evaluation, bringing it one step closer to becoming a therapeutic option.
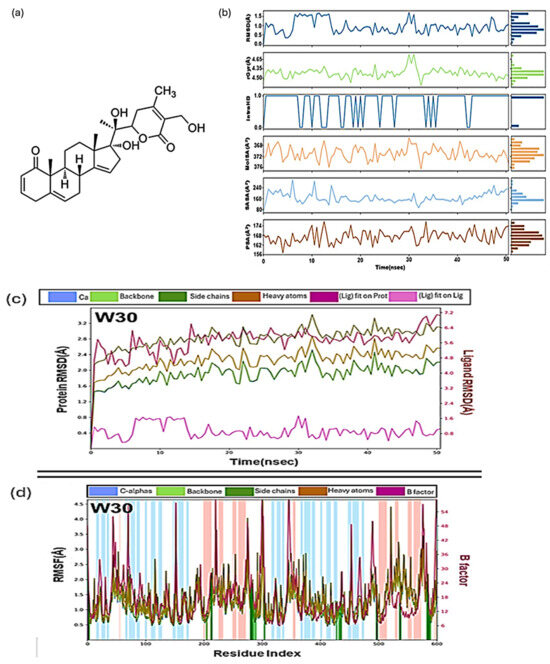
Figure 12.
(a) The structure of the most stable compound of Withacoagulin H with the lowest binding energy complex with a coagulin H (W30). (b) Values during molecular dynamics simulation of W30 with Mpro, Root mean square deviation (RMSD), radius of gyration (rGyr), intramolecular hydrogen bonds (intraHB), molecular surface area (MolSA), solvent accessible surface area (SASA), polar surface area (PSA). Colour coding: hydrogen bond (green), hydrophobic contacts (purple) and water-bridge (blue). (c) Root mean square deviation (RMSD) plot of W30 as a function of time and (d) Root mean square fluctuations (RMSF) plot for W30 [66].
While MD simulations have shown great promise, many studies have relied on relatively short simulation times (e.g., 20 ns, 50 ns) to predict the stability and binding behaviour of Mpro inhibitors. This is particularly concerning given the inherent flexibility and complexity of the Mpro active site, which may require longer simulation times to capture the dynamic nature of the protein–ligand interactions fully. For instance, the study on W30 reports a 50 ns MD simulation, which, while valuable, might not be sufficient to observe the full range of interactions and potential conformational changes in the complex. The limited duration of these simulations may overlook essential binding events or dynamic behaviours that could influence the inhibitor’s long-term stability and efficacy.
Moreover, studies combining MD simulations with ensemble docking or simulations extending to microsecond timescales (1 μs) are rare but crucial for assessing the full extent of binding dynamics. This approach has led to the identification of inhibitors such as Z1244904919 and Z1759961356, whose dynamic binding behaviours were studied in depth with simulations extending to 1 μs [88]. Binding free energy calculations using the MM/GBSA model showed that electrostatic and hydrophobic interactions are essential for stabilising these complexes, setting the stage for experimental validations. In Table 5, the positive values of ΔGpol indicate unfavourable solvation, which is a standard result in MM/GBSA calculations. These positive ΔGpol values reflect the desolvation penalty for the ligand, meaning that the system resists solvation in the binding process. Thus, polar solvation does not contribute favourably to binding but is instead a hindrance. However, while the simulation duration and methodological rigour here are notable, these studies still fall short of validating pharmacokinetic or toxicological profiles, leaving a gap between computational promise and clinical relevance.

Table 5.
The binding free energies (kcal/mol) components of Z1244904919 and Z1759961356 [88].
Exploring marine-derived natural products has also yielded promising candidates in the search for Mpro inhibitors. Screening 227 terpenes, six compounds, including erylosides B, stood out for their significant inhibitory potential [73]. MD simulations over 100 ns confirmed the stability of the erylosides B-Mpro complex, which demonstrated a binding affinity of −51.9 kcal/mol, far superior to the reference inhibitor, Lopinavir (Figure 13). This comparison adds context to the potency of natural products. Yet, despite robust MD validation over 100 ns, there remains a lack of biological characterisation—no data on cytotoxicity, metabolic stability, or viral inhibition—which limits their utility beyond theoretical models.
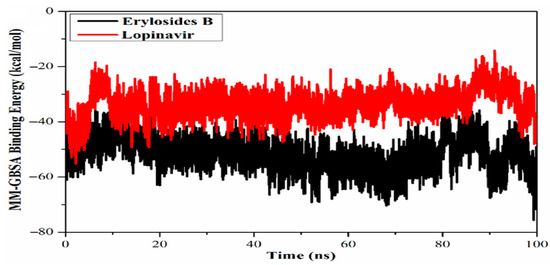
Figure 13.
Calculated MM/GBSA binding energy per frame for erylosides B (black) and lopinavir (red) with Mpro over 100 ns MD simulations [73]. Licenced under the CC-BY 4.0 licence.
Similar critiques apply to studies on natural flavonoids, which have emerged as strong contenders in Mpro inhibition. In docking studies and MD simulations, rutin, a flavonoid derivative, demonstrated the lowest predicted binding energy of −8.67 kcal/mol, aligning perfectly with the Mpro binding site (6LU7) [88]. While this robust simulation stability supports its potential as a therapeutic candidate, real-world factors such as solubility and pharmacokinetics may significantly impact its therapeutic potential. Therefore, including experimental validation, such as cell-based assays or animal models, is essential for bridging the gap between in silico predictions and therapeutic efficacy.
Most of these natural compounds suffer from issues like poor bioavailability or low stability, which complicates their transition from computational models to viable therapeutics. Other chemical scaffolds, such as benzimidazoles and benzothiazoles, have also demonstrated promising inhibitory capacities. For example, one benzimidazole-Mpro complex (Comp 1) exhibited a binding free energy of −7.3 kcal/mol [89]. Trajectories for key properties like RMSD, molecular surface area (MolSA), radius of gyration (rGyr), polar surface area (PSA), and solvent-accessible surface area (SASA) confirmed the stability and favourable dynamics of the complex over 100 ns simulations (Figure 14). These multi-parametric analyses are a strength, providing a more nuanced picture of complex stability and drug-like behaviour. While the binding free energy of –7.3 kcal/mol indicates promising interactions, it falls within a typical range for early-stage hits. It would benefit from further optimisation to enhance potency and selectivity.
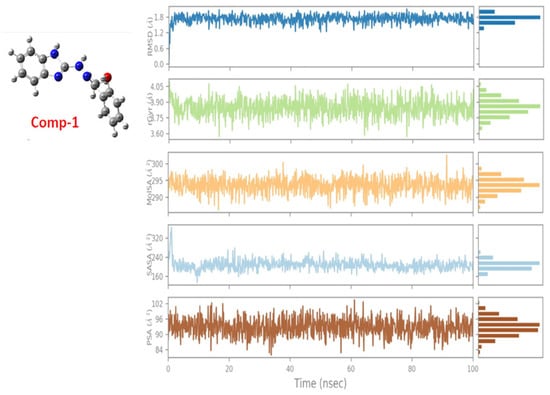
Figure 14.
Ligand property trajectories for the Comp.1-6LU7 complex [89].
An exception to these trends is the study of diselenides such as (PhSe)2, which not only underwent MD simulations but also progressed to experimental testing [90]. Experimental studies revealed that (PhSe)2 inhibited viral replication with an EC50 value of 2.4 μm, similar to other antiviral agents [91]. This compound stands out for its dual computational and biological validation.
The role of augmented intelligence in drug discovery is increasingly vital, particularly in developing quantitative structure–activity relationships (QSAR). One such innovation is the MDBinding programme module, developed by Wang and Vasilyev [92]. This tool employs MD simulations to design and pre-screen new inhibitors by modifying functional groups of FDA-approved drugs, thus enhancing their binding efficiency. By refining compounds like Adrafinil and Baicalein to target SARS-CoV-2 Mpro, MDBinding offers an efficient way to optimise FDA-approved drugs with minimal human intervention, opening new avenues for drug discovery.
However, the reliance on relatively short simulation times and limited experimental validation presents challenges that must be addressed to fully capitalise on the potential of MD simulations in drug discovery. These examples demonstrate that MD simulations, when integrated with experimental data and advanced computational tools, are a cornerstone in developing novel Mpro inhibitors, offering a path toward more effective treatments for SARS-CoV-2 and beyond [92].
2.5. Leveraging Quantum Mechanics for ADME-Optimised Drugs
Beyond the initial pre-screening of inhibitors using molecular docking, molecular dynamics (MD) simulations, and other in silico tools, these computational methods offer more profound insights into critical drug properties. They help evaluate drug-likeness, pharmacokinetics, and toxicity profiles at the molecular level—essential factors for optimising new drug candidates [91]. Many inhibitors, particularly small and flexible compounds, such as non-covalent small molecule Mpro inhibitors like ML188 and its potent derivative 23R [93], require a detailed understanding of their structural and electronic properties. Consequently, quantum mechanical studies have become indispensable in drug design.
Among these, density functional theory (DFT), a widely applied quantum mechanics (QM) method, plays a crucial role in obtaining quantitative structure–property relationships (QSPR) for inhibitors [94]. By accurately predicting electronic properties, DFT provides atomic-level insights into inhibitors’ conformation, reactivity, and interaction with their target proteins. These electronic and structural attributes are closely linked to a drug’s ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties, influencing potency and stability. However, the inherent flexibility of inhibitors introduces challenges in conformational sampling, which remains a significant hurdle in effective drug design [94].
Since most drug design algorithms operate in three-dimensional (3D) space, understanding the conformational landscape of a molecule is essential. The binding sites of proteins often reside in concave surface regions, requiring inhibitors to adopt specific 3D geometries for optimal binding [95]. However, identifying the bioactive conformations of flexible inhibitors with multiple rotatable bonds or unsaturated ring systems remains a challenge. As summarised by Habgood et al., once a molecular scaffold has been identified, three major obstacles must be addressed, namely: (1) developing methods to generate ensembles of a molecule’s bioactive conformations, (2) rationally modifying a pre-established bioactive structure, and (3) approximating real solution-phase conformational ensembles in correlation with spectroscopic data, such as IR, NMR, and UV–vis spectroscopy [96]. While crystal structures provide snapshots of single conformations, they fail to capture the complete flexibility of these molecules.
Advanced computational methods have been developed to generate conformational ensembles that more accurately represent bioactive states to bridge this gap. Robust QM-based conformational sampling techniques have become invaluable, offering insights into a wide range of conformer clusters and their associated properties, as illustrated in Figure 15 [94]. These methods enhance our understanding of an inhibitor’s structural dynamics and provide predictive spectroscopic data. For example, shifts in IR, NMR, and UV–Vis spectra derived from DFT calculations have been correlated with inhibitory potency in the case of tyrosine kinase inhibitors (TKIs), where optical spectral differences between the global minimum and crystallographic conformations aligned with activity trends [97]. However, this correlation has thus far been observed only in TKIs, and its direct applicability to structurally and mechanistically distinct systems—such as SARS-CoV-2 Mpro inhibitors—remains to be established. Therefore, while the approach holds conceptual promise, especially for flexible molecules like ML188, it should currently be regarded as exploratory in the context of Mpro-targeted drug discovery.
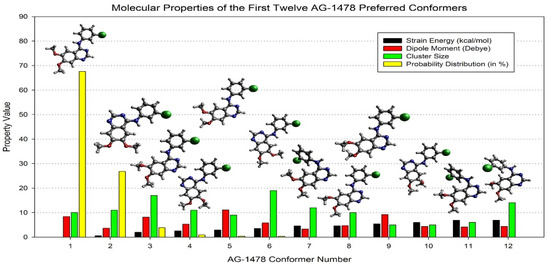
Figure 15.
Comparison of the top twelve AG-1478 preferred conformers above the strain energy cutoff (10.5 kcal/mol), dipole moment (D), cluster size, and probability distribution (in %), together with the optimised structures in the DMSO [94].
Notably, variations in optical spectra between an inhibitor’s global minimum structure and its crystal structure often correlate with its potency—where larger spectral shifts (depicted in orange in Figure 16) correspond to higher inhibitory efficacy, reflected by a more negative ln(IC50) function [97]. This trend is evident in potent tyrosine kinase inhibitors (TKIs) such as TKI-Cl and TKI-Br, which display distinct spectral behaviours compared to their less effective counterparts.
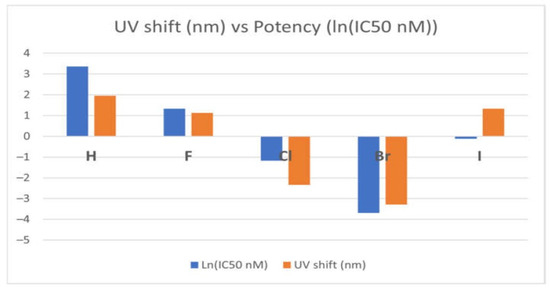
Figure 16.
Correlation between optical shift and the potency of the EGFR TKIs [97]. Licenced under the CC-BY 4.0 licence.
Among the promising drug candidates targeting SARS-CoV-2 Mpro are non-covalent small molecule inhibitors like ML188. These flexible compounds have gained attention for their potential in antiviral therapy, with many being repurposed FDA-approved drugs or novel inhibitors specifically designed against SARS-CoV-2 [92]. Recent advancements in MD-based binding prediction methods have further demonstrated the capability to enhance inhibitors such as Adrafinil and Baicalein. Expanding the application of MDBinding to compounds like ML188 presents an exciting opportunity to accelerate the development of next-generation Mpro inhibitors, paving the way for more effective antiviral therapeutics.
3. Summary and Outlook
This review provides an in-depth analysis of recent advances in computer-aided design for SARS-CoV-2 Mpro inhibitors, focusing on docking and MD simulations. Key methodologies include molecular docking, MD simulations, and DFT calculations, which have been instrumental in developing natural and synthetic inhibitors. MD simulations offer atomic-level insights into drug flexibility and binding mechanisms, while DFT and other calculations reveal complementary information about the electronic properties of the inhibitors. In silico approaches offer several advantages, such as accelerating the drug discovery process, reducing costs associated with laboratory testing, and minimising environmental impact. Molecular docking and virtual screening facilitate the identification of potential drug candidates and allow for the prediction of drug repurposing, saving valuable time. Additionally, in silico approaches offer structural and mechanistic insights that support quantitative structure and property relationship (QSPR), enhancing rational drug design in the future.
In summary, this review provides a focused critique of structure-based computational strategies employed in the discovery of non-covalent SARS-CoV-2 Mpro inhibitors, outlining both the strengths and inherent limitations of these approaches. While in silico methods are instrumental in accelerating early-stage drug discovery and prioritising promising candidates, they must be complemented by rigorous in vitro and in vivo validation to ensure translational relevance. Looking ahead, future efforts should also focus on enhancing the robustness and efficiency of MD simulations [92], which remain computationally intensive yet critical for accurately capturing protein–ligand interactions and refining lead optimisation strategies.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
I.A. acknowledges the Swinburne University of Technology Tuition Fee Scholarship (TFS).
Conflicts of Interest
The authors declare no conflicts of interest in this research.
References
- Kumar, D.; Chauhan, G.; Kalra, S.; Kumar, B.; Gill, M.S. A perspective on potential target proteins of COVID-19: Comparison with SARS-CoV for designing new small molecules. Bioorgan. Chem. 2020, 104, 104326. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, C.; Xin, L.; Ren, X.; Tian, L.; Ju, X.; Li, H.; Wang, Y.; Zhao, Q.; Liu, H. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020, 206, 112711. [Google Scholar] [CrossRef]
- Sharma, P.; Vijayan, V.; Pant, P.; Sharma, M.; Vikram, N.; Kaur, P.; Singh, T.; Sharma, S. Identification of potential drug candidates to combat COVID-19: A structural study using the main protease (mpro) of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 6649–6659. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Astuti, I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Chatterjee, S.; Agoramoorthy, G.; Lee, S.-S. Structural Landscape of nsp Coding Genomic Regions of SARS-CoV-2-ssRNA Genome: A Structural Genomics Approach Toward Identification of Druggable Genome, Ligand-Binding Pockets, and Structure-Based Druggability. Mol. Biotechnol. 2024, 66, 641–662. [Google Scholar] [CrossRef]
- Mondol, W.C. Exploring the Potential of Organic Molecules in the Treatment of COVID-19; Brac University: Dhaka, Bangladesh, 2021. [Google Scholar]
- Chan, C.C.-Y.; Guo, Q.; Chan, J.F.-W.; Tang, K.; Cai, J.-P.; Chik, K.K.-H.; Huang, Y.; Dai, M.; Qin, B.; Ong, C.P.; et al. Identification of novel small-molecule inhibitors of SARS-CoV-2 by chemical genetics. Acta Pharm. Sin. B 2024, 14, 4028–4044. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F. Structure-based design, synthesis and biological evaluation of peptidomimetic aldehydes as a novel series of antiviral drug candidates targeting the SARS-CoV-2 main protease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.-M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Geng, M.; Peng, Y.; Meng, L.; Lu, S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020, 10, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Adelusi, T.I.; Oyedele, A.-Q.K.; Monday, O.E.; Boyenle, I.D.; Idris, M.O.; Ogunlana, A.T.; Ayoola, A.M.; Fatoki, J.O.; Kolawole, O.E.; David, K.B. Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–Molecular dynamics, molecular mechanics, and density functional theory investigations. J. Mol. Struct. 2022, 1250, 131879. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Muratov, E.N.; Amaro, R.; Andrade, C.H.; Brown, N.; Ekins, S.; Fourches, D.; Isayev, O.; Kozakov, D.; Medina-Franco, J.L.; Merz, K.M. A critical overview of computational approaches employed for COVID-19 drug discovery. Chem. Soc. Rev. 2021, 50, 9121–9151. [Google Scholar] [CrossRef]
- Han, S.H.; Goins, C.M.; Arya, T.; Shin, W.-J.; Maw, J.; Hooper, A.; Sonawane, D.P.; Porter, M.R.; Bannister, B.E.; Crouch, R.D.; et al. Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro). J. Med. Chem. 2022, 65, 2880–2904. [Google Scholar] [CrossRef]
- Unoh, Y.; Uehara, S.; Nakahara, K.; Nobori, H.; Yamatsu, Y.; Yamamoto, S.; Maruyama, Y.; Taoda, Y.; Kasamatsu, K.; Suto, T.; et al. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 2022, 65, 6499–6512. [Google Scholar] [CrossRef]
- Duan, X.; Lacko, L.A.; Chen, S. Druggable targets and therapeutic development for COVID-19. Front. Chem. 2022, 10, 963701. [Google Scholar] [CrossRef]
- Liu, X.; Ren, X.; Hua, M.; Liu, F.; Ren, X.; Sui, C.; Li, Q.; Luo, F.; Jiang, Z.; Xia, Z. Progress of SARS-CoV-2 Main protease peptide-like inhibitors. Chem. Biol. Drug Des. 2024, 103, e14425. [Google Scholar] [CrossRef]
- Xu, Y.-S.; Chigan, J.-Z.; Li, J.-Q.; Ding, H.-H.; Sun, L.-Y.; Liu, L.; Hu, Z.; Yang, K.-W. Hydroxamate and thiosemicarbazone: Two highly promising scaffolds for the development of SARS-CoV-2 antivirals. Bioorgan. Chem. 2022, 124, 105799. [Google Scholar] [CrossRef]
- Tumskiy, R.S.; Tumskaia, A.V. Multistep rational molecular design and combined docking for discovery of novel classes of inhibitors of SARS-CoV-2 main protease 3CLpro. Chem. Phys. Lett. 2021, 780, 138894. [Google Scholar] [CrossRef]
- Ebenezer, O.; Shapi, M. Promising inhibitors against main protease of SARS-CoV-2 from medicinal plants: In silico identification. Acta Pharm. 2022, 72, 159–169. [Google Scholar] [CrossRef]
- Hu, Q.; Xiong, Y.; Zhu, G.-H.; Zhang, Y.-N.; Zhang, Y.-W.; Huang, P.; Ge, G.-B. The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19. MedComm 2022, 3, e151. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef]
- Cooper, M.S.; Zhang, L.; Ibrahim, M.; Zhang, K.; Sun, X.; Röske, J.; Göhl, M.; Brönstrup, M.; Cowell, J.K.; Sauerhering, L. Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease. J. Med. Chem. 2022, 65, 13328–13342. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, H.; Lin, Q.; Lyu, J.; Lu, L.; Chen, H.; Zhang, X.; Zhang, Y.; Chen, K. Progress on SARS-CoV-2 3CLpro Inhibitors: Inspiration from SARS-CoV 3CLpro Peptidomimetics and Small-Molecule Anti-Inflammatory Compounds. Drug Des. Dev. Ther. 2023, 16, 1067–1082. [Google Scholar] [CrossRef]
- Zhou, K.; Chen, D. Conventional Understanding of SARS-CoV-2 Mpro and Common Strategies for Developing Its Inhibitors. Chem.—A Eur. J. 2023, 24, e202300301. [Google Scholar] [CrossRef]
- Song, L.; Gao, S.; Ye, B.; Yang, M.; Cheng, Y.; Kang, D.; Yi, F.; Sun, J.-P.; Menéndez-Arias, L.; Neyts, J. Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors. Acta Pharm. Sin. B 2023, 14, 87–109. [Google Scholar] [CrossRef]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef]
- Alexandris, N.; Lagoumintzis, G.; Chasapis, C.T.; Leonidas, D.D.; Papadopoulos, G.E.; Tzartos, S.J.; Tsatsakis, A.; Eliopoulos, E.; Poulas, K.; Farsalinos, K. Nicotinic cholinergic system and COVID-19: In silico evaluation of nicotinic acetylcholine receptor agonists as potential therapeutic interventions. Toxicol. Rep. 2021, 8, 73–83. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms 2020, 8, 1250. [Google Scholar] [CrossRef]
- Lockbaum, G.J.; Reyes, A.C.; Lee, J.M.; Tilvawala, R.; Nalivaika, E.A.; Ali, A.; Kurt Yilmaz, N.; Thompson, P.R.; Schiffer, C.A. Crystal structure of SARS-CoV-2 main protease in complex with the non-covalent inhibitor ML188. Viruses 2021, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, W.; Zhou, X.; Zhang, J.; Li, J. Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77. Int. J. Biol. Macromol. 2024, 276, 133706. [Google Scholar] [CrossRef]
- Zagórska, A.; Czopek, A.; Fryc, M.; Jończyk, J. Inhibitors of SARS-CoV-2 Main Protease (Mpro) as Anti-Coronavirus Agents. Biomolecules 2024, 14, 797. [Google Scholar] [CrossRef]
- Nandi, A.; Asadi, M.; Zhang, A.; Chu, Z.T.; Warshel, A. Mechanistic Insights into Nitrile and Alkyne Covalent Inhibitors of the SARS-CoV-2 Main Protease. ACS Catal. 2025, 15, 1158–1169. [Google Scholar] [CrossRef]
- Zhang, R.; Yan, H.; Zhou, J.; Yan, G.; Liu, X.; Shang, C.; Chen, Y. Improved fluorescence-based assay for rapid screening and evaluation of SARS-CoV-2 main protease inhibitors. J. Med. Virol. 2024, 96, e29498. [Google Scholar] [CrossRef]
- Narayanan, A.; Narwal, M.; Majowicz, S.A.; Varricchio, C.; Toner, S.A.; Ballatore, C.; Brancale, A.; Murakami, K.S.; Jose, J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022, 5, 169. [Google Scholar] [CrossRef]
- Jayawardena, T.U.; Merindol, N.; Liyanage, N.S.; Desgagné-Penix, I. Unveiling Amaryllidaceae alkaloids: From biosynthesis to antiviral potential–a review. Nat. Prod. Rep. 2024, 41, 721–747. [Google Scholar] [CrossRef]
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A critical review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98. [Google Scholar] [CrossRef]
- Mao, L.; Shaabani, N.; Zhang, X.; Jin, C.; Xu, W.; Argent, C.; Kushnareva, Y.; Powers, C.; Stegman, K.; Liu, J. Olgotrelvir, a dual inhibitor of SARS-CoV-2 Mpro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19. Med 2024, 5, 42–61.e23. [Google Scholar] [CrossRef]
- Pang, X.; Xu, W.; Liu, Y.; Li, H.; Chen, L. The research progress of SARS-CoV-2 main protease inhibitors from 2020 to 2022. Eur. J. Med. Chem. 2023, 257, 115491. [Google Scholar] [CrossRef]
- Thakur, A.; Sharma, G.; Badavath, V.N.; Jayaprakash, V.; Merz, K.M., Jr.; Blum, G.; Acevedo, O. Primer for Designing Main Protease (Mpro) Inhibitors of SARS-CoV-2. J. Phys. Chem. Lett. 2022, 13, 5776–5786. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, H.; Sun, Q.; Liang, H.; Li, C.; Deng, X.; Liu, Y.; Lai, L. Potent inhibitors of SARS-CoV-2 3C-like protease derived from N-substituted isatin compounds. Eur. J. Med. Chem. 2020, 206, 112702. [Google Scholar] [CrossRef]
- She, Z.; Yao, Y.; Wang, C.; Li, Y.; Xiong, X.; Liu, Y. Mpro-targeted anti-SARS-CoV-2 inhibitor-based drugs. J. Chem. Res. 2023, 47, 17475198231184799. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z. Bench-to-bedside: Innovation of small molecule anti-SARS-CoV-2 drugs in China. Eur. J. Med. Chem. 2023, 257, 115503. [Google Scholar] [CrossRef]
- Kitamura, N.; Sacco, M.D.; Ma, C.; Hu, Y.; Townsend, J.A.; Meng, X.; Zhang, F.; Zhang, X.; Ba, M.; Szeto, T.; et al. Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors. J. Med. Chem. 2022, 65, 2848–2865. [Google Scholar] [CrossRef]
- Kühl, N.; Lang, J.; Leuthold, M.M.; Klein, C.D. Discovery of potent benzoxaborole inhibitors against SARS-CoV-2 main and dengue virus proteases. Eur. J. Med. Chem. 2022, 240, 114585. [Google Scholar] [CrossRef]
- Zhang, J.-W.; Xiong, Y.; Wang, F.; Zhang, F.-M.; Yang, X.; Lin, G.-Q.; Tian, P.; Ge, G.; Gao, D. Discovery of 9,10-dihydrophenanthrene derivatives as SARS-CoV-2 3CLpro inhibitors for treating COVID-19. Eur. J. Med. Chem. 2022, 228, 114030. [Google Scholar] [CrossRef]
- Kneller, D.W.; Li, H.; Galanie, S.; Phillips, G.; Labbé, A.; Weiss, K.L.; Zhang, Q.; Arnould, M.A.; Clyde, A.; Ma, H.; et al. Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease. J. Med. Chem. 2021, 64, 17366–17383. [Google Scholar] [CrossRef]
- Gao, S.; Sylvester, K.; Song, L.; Claff, T.; Jing, L.; Woodson, M.; Weiße, R.H.; Cheng, Y.; Schäkel, L.; Petry, M.; et al. Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity. J. Med. Chem. 2022, 65, 13343–13364. [Google Scholar] [CrossRef]
- Luttens, A.; Gullberg, H.; Abdurakhmanov, E.; Vo, D.D.; Akaberi, D.; Talibov, V.O.; Nekhotiaeva, N.; Vangeel, L.; De Jonghe, S.; Jochmans, D.; et al. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc. 2022, 144, 2905–2920. [Google Scholar] [CrossRef]
- Zhu, W.; Xu, M.; Chen, C.Z.; Guo, H.; Shen, M.; Hu, X.; Shinn, P.; Klumpp-Thomas, C.; Michael, S.G.; Zheng, W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol. Transl. Sci. 2020, 3, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Teoh, T.C. Cell-Based High-Throughput Screening Protocol for Discovering Antiviral Inhibitors Against SARS-CoV-2 Main Protease (3CLpro). Mol. Biotechnol. 2021, 63, 240–248. [Google Scholar] [CrossRef]
- Drayman, N.; DeMarco, J.K.; Jones, K.A.; Azizi, S.-A.; Froggatt, H.M.; Tan, K.; Maltseva, N.I.; Chen, S.; Nicolaescu, V.; Dvorkin, S. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science 2021, 373, 931–936. [Google Scholar] [CrossRef]
- Xu, T.; Xu, M.; Zhu, W.; Chen, C.Z.; Zhang, Q.; Zheng, W.; Huang, R. Efficient Identification of Anti-SARS-CoV-2 Compounds Using Chemical Structure- and Biological Activity-Based Modeling. J. Med. Chem. 2022, 65, 4590–4599. [Google Scholar] [CrossRef]
- Gentile, F.; Fernandez, M.; Ban, F.; Ton, A.-T.; Mslati, H.; Perez, C.F.; Leblanc, E.; Yaacoub, J.C.; Gleave, J.; Stern, A. Automated discovery of noncovalent inhibitors of SARS-CoV-2 main protease by consensus Deep Docking of 40 billion small molecules. Chem. Sci. 2021, 12, 15960–15974. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Flury, P.; Krüger, N.; Su, H.; Schäkel, L.; Barbosa Da Silva, E.; Eppler, O.; Kronenberger, T.; Nie, T.; Luedtke, S.; et al. Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure–Activity Relationships, Antiviral Activity, and X-ray Structure Determination. J. Med. Chem. 2022, 65, 9376–9395. [Google Scholar] [CrossRef]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virol. J. 2020, 17, 190. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhu, Q.; Zhou, X.; Li, W.; Yin, X.; Li, J. Structural Basis for the Inhibition of SARS-CoV-2 Mpro D48N Mutant by Shikonin and PF-07321332. Viruses 2024, 16, 65. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Mangiavacchi, F.; Botwina, P.; Menichetti, E.; Bagnoli, L.; Rosati, O.; Marini, F.; Fonseca, S.F.; Abenante, L.; Alves, D.; Dabrowska, A.; et al. Seleno-Functionalization of Quercetin Improves the Non-Covalent Inhibition of Mpro and Its Antiviral Activity in Cells against SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 7048. [Google Scholar]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Cui, M.; Zheng, C.; Zhang, P.; Ren, S.; Bao, J.; Gao, D.; Sun, R.; Wang, M.; Lin, J.; et al. Both Baicalein and Gallocatechin Gallate Effectively Inhibit SARS-CoV-2 Replication by Targeting Mpro and Sepsis in Mice. Inflammation 2022, 45, 1076–1088. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yao, S.; Zhao, W.; Li, M.; Liu, J.; Shang, W.; Xie, H.; Ke, C.; Gao, M.; Yu, K. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv 2020. [Google Scholar] [CrossRef]
- Duong, C.Q.; Nguyen, P.T.V. Exploration of SARS-CoV-2 Mpro Noncovalent Natural Inhibitors Using Structure-Based Approaches. ACS Omega 2023, 8, 6679–6688. [Google Scholar] [CrossRef]
- Verma, S.; Patel, C.N.; Chandra, M. Identification of novel inhibitors of SARS-CoV-2 main protease (Mpro) from Withania sp. by molecular docking and molecular dynamics simulation. J. Comput. Chem. 2021, 42, 1861–1872. [Google Scholar] [CrossRef]
- Yang, W.; Wang, Y.; Han, D.; Tang, W.; Sun, L. Recent advances in application of computer-aided drug design in anti-COVID-19 Virials Drug Discovery. Biomed. Pharmacother. 2024, 173, 116423. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Pereira, F. A Computer-Aided Drug Design Approach to Predict Marine Drug-Like Leads for SARS-CoV-2 Main Protease Inhibition. Mar. Drugs 2020, 18, 633. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wan, J.; Wang, G. A survey on computational methods in discovering protein inhibitors of SARS-CoV-2. Brief. Bioinform. 2021, 23, bbab416. [Google Scholar] [CrossRef]
- Ngo, S.T.; Nguyen, T.H.; Tung, N.T.; Vu, V.V.; Pham, M.Q.; Mai, B.K. Characterizing the ligand-binding affinity toward SARS-CoV-2 Mpro via physics-and knowledge-based approaches. Phys. Chem. Chem. Phys. 2022, 24, 29266–29278. [Google Scholar] [CrossRef]
- Sayed, A.M.; Alhadrami, H.A.; El-Gendy, A.O.; Shamikh, Y.I.; Belbahri, L.; Hassan, H.M.; Abdelmohsen, U.R.; Rateb, M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro). Microorganisms 2020, 8, 970. [Google Scholar] [CrossRef]
- Amin, S.A.; Banerjee, S.; Singh, S.; Qureshi, I.A.; Gayen, S.; Jha, T. First structure–activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: An endeavor on COVID-19 drug discovery. Mol. Divers. 2021, 25, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Mohamed, T.A.; Atia, M.A.M.; Al-Hammady, M.A.M.; Abdeljawaad, K.A.A.; Elkady, E.M.; Moustafa, M.F.; Alrumaihi, F.; Allemailem, K.S.; et al. In Silico Mining of Terpenes from Red-Sea Invertebrates for SARS-CoV-2 Main Protease (Mpro) Inhibitors. Molecules 2021, 26, 2082. [Google Scholar] [CrossRef] [PubMed]
- Hicks, E.G.; Kandel, S.E.; Lampe, J.N. Identification of Aloe-derived natural products as prospective lead scaffolds for SARS-CoV-2 main protease (Mpro) inhibitors. Bioorgan. Med. Chem. Lett. 2022, 66, 128732. [Google Scholar] [CrossRef]
- Adem, Ş.; Eyupoglu, V.; Ibrahim, I.M.; Sarfraz, I.; Rasul, A.; Ali, M.; Elfiky, A.A. Multidimensional in silico strategy for identification of natural polyphenols-based SARS-CoV-2 main protease (Mpro) inhibitors to unveil a hope against COVID-19. Comput. Biol. Med. 2022, 145, 105452. [Google Scholar] [CrossRef]
- Altincekic, N.; Jores, N.; Löhr, F.; Richter, C.; Ehrhardt, C.; Blommers, M.J.J.; Berg, H.; Öztürk, S.; Gande, S.L.; Linhard, V.; et al. Targeting the Main Protease (Mpro, nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies. ACS Chem. Biol. 2024, 19, 563–574. [Google Scholar] [CrossRef]
- Janin, Y.L. On the origins of SARS-CoV-2 main protease inhibitors. RSC Med. Chem. 2024, 15, 81–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, M.; Yuan, F.; Zhang, J.; Song, J.; Yang, L. Exploring the covalent inhibition mechanisms of inhibitors with two different warheads acting on SARS-CoV-2 Mpro by QM/MM simulations. Comput. Theor. Chem. 2024, 1242, 114979. [Google Scholar] [CrossRef]
- Gao, Q.; Liu, S.; Zhou, Y.; Fan, J.; Ke, S.; Zhou, Y.; Fan, K.; Wang, Y.; Zhou, Y.; Xia, Z.; et al. Discovery of meisoindigo derivatives as noncovalent and orally available Mpro inhibitors: Their therapeutic implications in the treatment of COVID-19. Eur. J. Med. Chem. 2024, 273, 116498. [Google Scholar] [CrossRef]
- Chen, R.; Gao, Y.; Liu, H.; Li, H.; Chen, W.; Ma, J. Advances in research on 3C-like protease (3CL pro) inhibitors against SARS-CoV-2 since 2020. RSC Med. Chem. 2023, 14, 9–21. [Google Scholar] [CrossRef]
- Kuzikov, M.; Costanzi, E.; Reinshagen, J.; Esposito, F.; Vangeel, L.; Wolf, M.; Ellinger, B.; Claussen, C.; Geisslinger, G.; Corona, A. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Liu, S.; Sun, J.; Chen, Z.; Jiang, M.; Zhang, Q.; Wei, Y.; Wang, X.; Huang, Y.-Y. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B 2020, 10, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.-C.; Ke, Y.-Y.; Huang, S.Y.; Huang, P.-N.; Kung, Y.-A.; Chang, T.-Y.; Yen, K.-J.; Peng, T.-T.; Chang, S.-E.; Huang, C.-T. Discovery of M protease inhibitors encoded by SARS-CoV-2. Antimicrob. Agents Chemother. 2020, 64, e00872-20. [Google Scholar] [CrossRef]
- Cáceres, C.J.; Cardenas-Garcia, S.; Carnaccini, S.; Seibert, B.; Rajao, D.S.; Wang, J.; Perez, D.R. Efficacy of GC-376 against SARS-CoV-2 virus infection in the K18 hACE2 transgenic mouse model. Sci. Rep. 2021, 11, 9609. [Google Scholar] [CrossRef] [PubMed]
- Motiwale, M.; Yadav, N.S.; Kumar, S.; Kushwaha, T.; Choudhir, G.; Sharma, S.; Singour, P.K. Finding potent inhibitors for COVID-19 main protease (Mpro): An in silico approach using SARS-CoV-3CL protease inhibitors for combating CORONA. J. Biomol. Struct. Dyn. 2022, 40, 1534–1545. [Google Scholar] [CrossRef]
- Salo-Ahen, O.M.H.; Alanko, I.; Bhadane, R.; Bonvin, A.M.J.J.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development. Processes 2021, 9, 71. [Google Scholar] [CrossRef]
- Sarfraz, M.; Rauf, A.; Keller, P.; Qureshi, A.M. N,N′-dialkyl-2-thiobarbituric acid based sulfonamides as potential SARS-CoV-2 main protease inhibitors. Can. J. Chem. 2021, 99, 330–345. [Google Scholar] [CrossRef]
- Yang, J.; Lin, X.; Xing, N.; Zhang, Z.; Zhang, H.; Wu, H.; Xue, W. Structure-Based Discovery of Novel Nonpeptide Inhibitors Targeting SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021, 61, 3917–3926. [Google Scholar] [CrossRef]
- Mohaptra, R.K.; Dhama, K.; El-Arabey, A.A.; Sarangi, A.K.; Tiwari, R.; Bin Emran, T.; Azam, M.; Raval, M.K.; Seidel, V.; Abdalla, M. Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and in-silico pharmacokinetic and toxicity studies. J. King Saud Univ.-Sci. 2021, 33, 101637. [Google Scholar] [CrossRef] [PubMed]
- Omage, F.B.; Madabeni, A.; Tucci, A.R.; Nogara, P.A.; Bortoli, M.; Rosa, A.d.S.; Neuza dos Santos Ferreira, V.; Teixeira Rocha, J.B.; Miranda, M.D.; Orian, L. Diphenyl Diselenide and SARS-CoV-2: In silico Exploration of the Mechanisms of Inhibition of Main Protease (Mpro) and Papain-like Protease (PLpro). J. Chem. Inf. Model. 2023, 63, 2226–2239. [Google Scholar] [CrossRef]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A potential target for peptidomimetics and small-molecule inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef]
- Wang, F.; Vasilyev, V. In Silico Tuning of Binding Selectivity for New SARS-CoV-2 Main Protease Inhibitors. 2024. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4981058 (accessed on 13 May 2025).
- Elgohary, A.M.; Elfiky, A.A.; Pereira, F.; Abd El-Aziz, T.M.; Sobeh, M.; Arafa, R.K.; El-Demerdash, A. Investigating the structure-activity relationship of marine polycyclic batzelladine alkaloids as promising inhibitors for SARS-CoV-2 main protease (Mpro). Comput. Biol. Med. 2022, 147, 105738. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Vasilyev, V. Accelerating optical reporting for conformation of tyrosine kinase inhibitors in solutions. Int. J. Quantum Chem. 2021, 121, e26765. [Google Scholar] [CrossRef]
- Ruud, K.; Helgaker, T.; Bak, K.L.; Jørgensen, P.; Olsen, J. Accurate magnetizabilities of the isoelectronic series BeH−, BH, and CH+. The MCSCF-GIAO approach. Chem. Phys. 1995, 195, 157–169. [Google Scholar] [CrossRef]
- Habgood, M.; James, T.; Heifetz, A. Conformational Searching with Quantum Mechanics. In Quantum Mechanics in Drug Discovery; Heifetz, A., Ed.; Springer: New York, NY, USA, 2020; pp. 207–229. [Google Scholar]
- Alagawani, S.; Vasilyev, V.; Wang, F. Optical spectra of EGFR inhibitor AG-1478 for benchmarking DFT functionals. Electron. Struct. 2023, 5, 024011. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).