Elevating Clinical Semantics: Contrastive Pre-Training Beyond Cross-Entropy in Discharge Summaries †
Abstract
1. Introduction
- We propose the Contrastive Representations Pre-Training (CRPT) model for clinical language tasks, enhancing sentence-level semantics by integrating contrastive loss and WWM;
- We demonstrate significant improvements in medical NLP tasks using the BLUE benchmark, including natural language inference and sentence similarity tasks;
- We introduce the Bio+Discharge Summary CRPT model, which combines domain-specific pre-training in biomedical and clinical corpora and outperforms existing models on various tasks;
- We discuss practical considerations for negative sampling, domain-specific masking, and model interpretability, highlighting potential avenues for improved accuracy and better clinical integration.
2. Materials and Methods
2.1. Data Collection
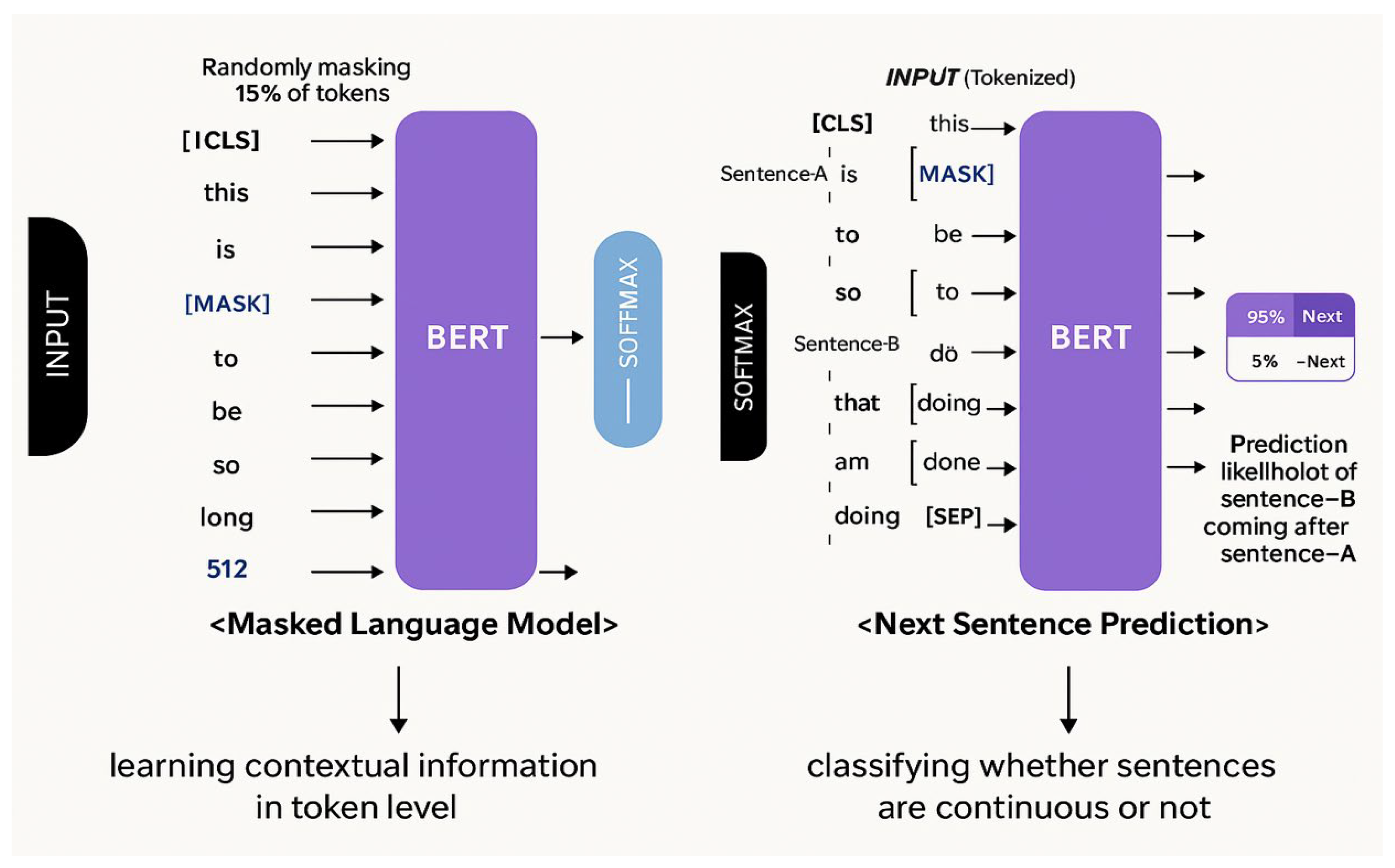
2.2. Pre-Training BERT Model
2.3. Pre-Training Discharge Summary CRPT Model
2.4. Pre-Training Bio+Discharge Summary CRPT Model
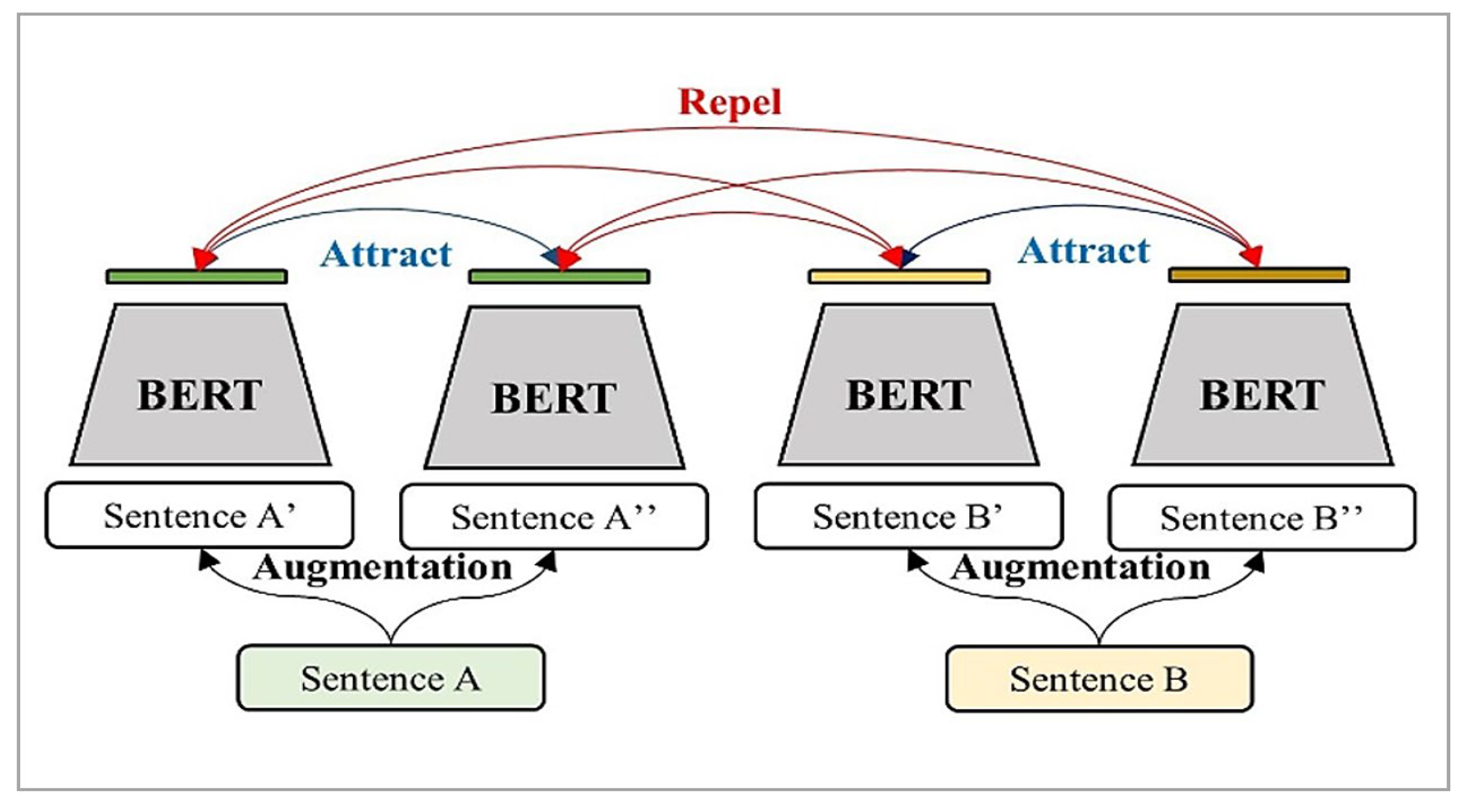
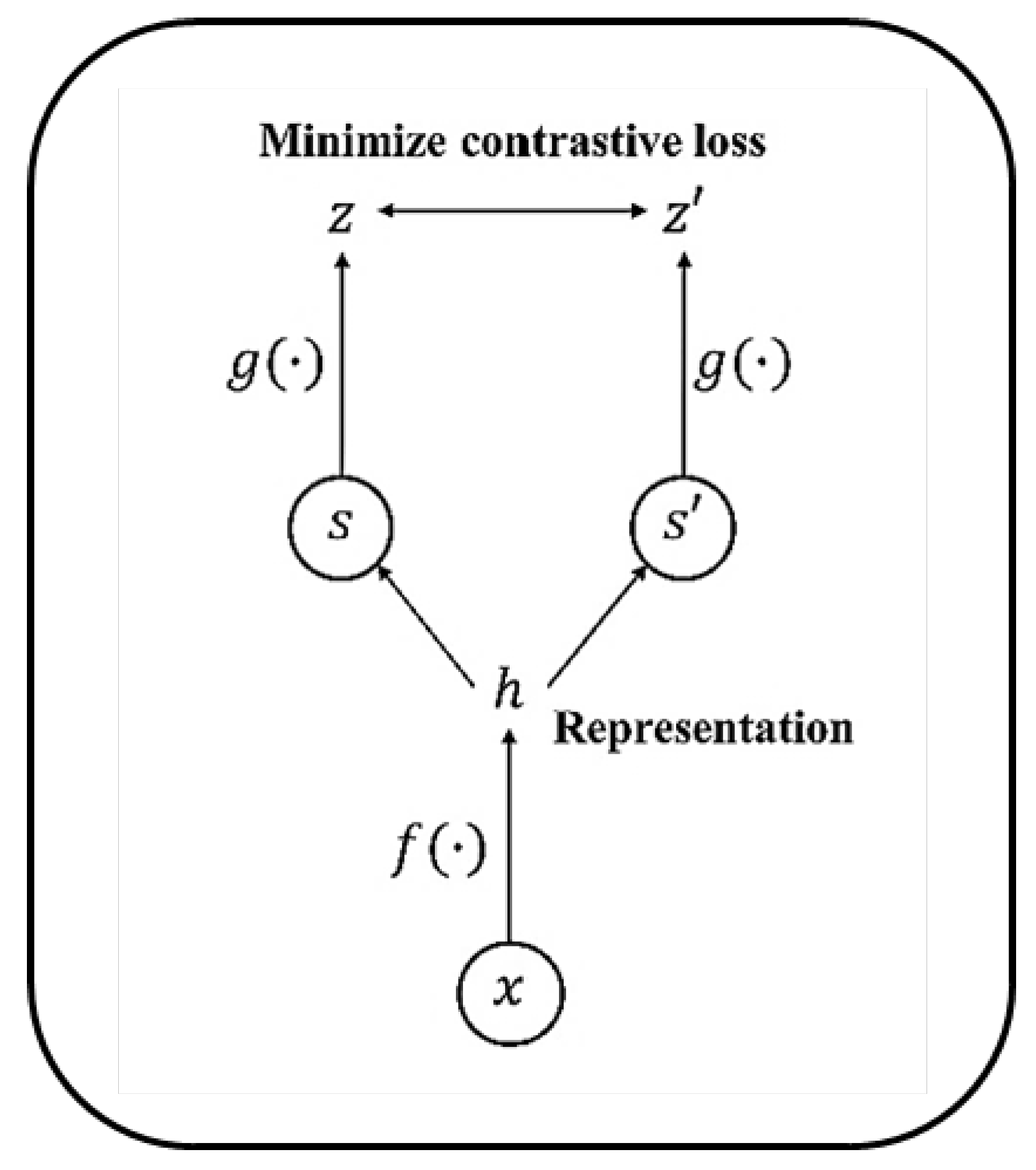
2.5. Contrastive Self-Supervised Learning
3. Proposed Approach
3.1. NSP and Its Limitations
3.2. Contrastive Representation Learning (CRL)
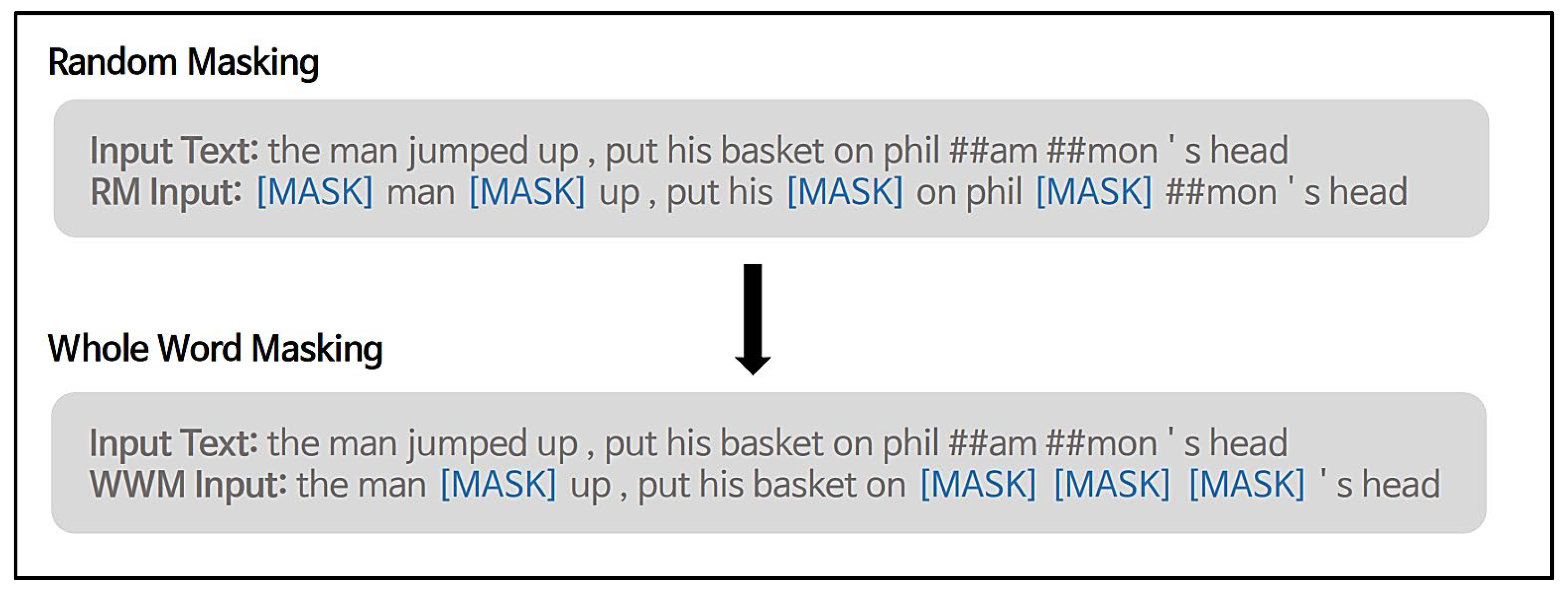
3.3. Whole-Word Masking (WWM)
4. Experimental Result
4.1. Experimental Setup
4.2. Evaluation of the CRPT Model
4.3. Quantitative Analysis
4.3.1. Sentence-Level Evaluation
4.3.2. Token-Level Evaluation
4.4. Quantitative Analysis of CRPT Models
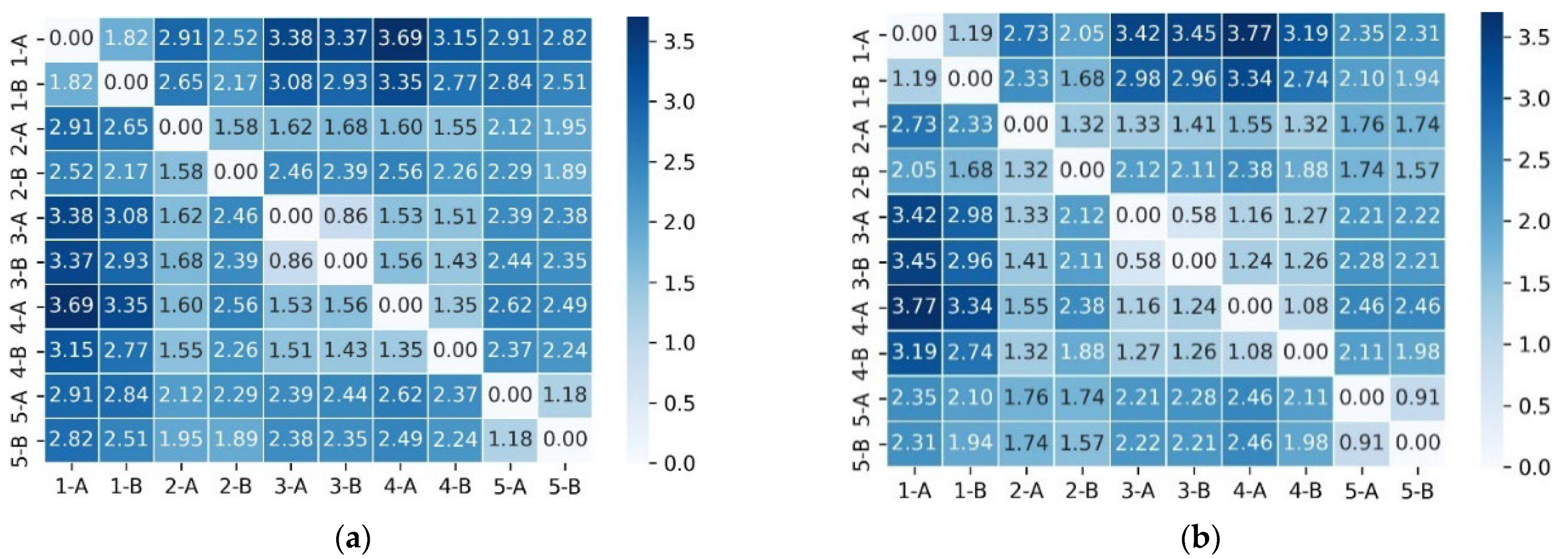
- Local coherence (main diagonal). In both models, the principal diagonal—corresponding to consecutive sentence pairs—displays the highest similarity. However, CRPT consistently yields darker cells and larger numeric values (e.g., 3.7 vs. 3.3), indicating that the contrastive objective tightens local semantic cohesion;
- Global discourse (off-diagonal intra-document blocks). Cells linking non-adjacent sentences from the same summary retain moderate similarity under CRPT, whereas BERT often collapses these to near 0. This suggests that CRPT balances sentence-level specificity with broader document context;
- True negatives (inter-document region). Rows and columns labeled ‘5-A’ and ‘5-B’ originate from different patient notes. In CRPT, these cross-document pairs cluster around 0 or even negative values (e.g., −3.7), forming a pale band that contrasts sharply with the intra-document area. The separation verifies that the mixed negative-sampling scheme effectively pushes unrelated sentences apart.
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BERT | Bidirectional encoder representations from transformers |
| NLP | Natural language processing |
| CRPT | Contrastive representations pre-training |
| CRL | Contrastive representations learning |
| WWM | Whole-word masking |
References
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Wang, A.; Singh, A.; Michael, J.; Hill, F.; Levy, O.; Bowman, S.R. GLUE: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding. arXiv 2018, arXiv:1804.07461. [Google Scholar]
- Alsentzer, E.; Murphy, J.R.; Boag, W.; Weng, W.-H.; Jin, D.; Naumann, T.; McDermott, M. Publicly Available Clinical BERT Embeddings. arXiv 2019, arXiv:1904.03323. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.-H.; Kang, J. BioBERT: A Pre-Trained Biomedical Language Representation Model for Biomedical Text Mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. arXiv 2020, arXiv:2007.15779. [Google Scholar] [CrossRef]
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: A Pretrained Language Model for Scientific Text. arXiv 2019, arXiv:1903.10676. [Google Scholar]
- Peng, Y.; Yan, S.; Lu, Z. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. arXiv 2019, arXiv:1906.05474. [Google Scholar]
- Fang, H.; Xie, P. ConSERT: Contrastive Self-Supervised Learning for Language Understanding. arXiv 2020, arXiv:2005.12766. [Google Scholar]
- Giorgi, J.M.; Nitski, O.; Bader, G.D.; Wang, B. DeCLUTR: Deep Contrastive Learning for Unsupervised Textual Representations. arXiv 2020, arXiv:2006.03659. [Google Scholar]
- Wei, J.; Zou, K. EDA: Easy Data Augmentation Techniques for Boosting Performance on Text Classification Tasks. arXiv 2019, arXiv:1901.11196. [Google Scholar]
- Sennrich, R.; Haddow, B.; Birch, A. Improving Neural Machine Translation Models with Monolingual Data. arXiv 2015, arXiv:1511.06709. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum Contrast for Unsupervised Visual Representation Learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 9729–9738. [Google Scholar]
- Chu, C.; Wang, R. A Survey of Domain Adaptation for Neural Machine Translation. arXiv 2018, arXiv:1806.00258. [Google Scholar]
- Won, D.; Lee, Y.; Choi, H.-J.; Jung, Y. Contrastive Representations Pre-Training for Enhanced Discharge Summary BERT. In Proceedings of the 2021 IEEE 9th International Conference on Healthcare Informatics (ICHI), Victoria, BC, Canada, 9–12 August 2021; pp. 507–508. [Google Scholar] [CrossRef]
- Koehn, P.; Knowles, R. Six Challenges for Neural Machine Translation. arXiv 2017, arXiv:1706.03872. [Google Scholar]
- Zhang, Z.; Han, X.; Liu, Z.; Jiang, X.; Sun, M.; Liu, Q. ERNIE: Enhanced Language Representation with Informative Entities. arXiv 2019, arXiv:1905.07129. [Google Scholar]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Li, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a Freely Accessible Critical Care Database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Wu, Y.; Schuster, M.; Chen, Z.; Le, Q.V.; Norouzi, M.; Macherey, W.; Krikun, M.; Cao, Y.; Gao, Q.; Macherey, K.; et al. Google’s Neural Machine Translation System: Bridging the Gap between Human and Machine Translation. arXiv 2016, arXiv:1609.08144. [Google Scholar]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. ALBERT: A Lite BERT for Self-Supervised Learning of Language Representations. arXiv 2019, arXiv:1909.11942. [Google Scholar]
- Kalyan, K.S.; Sangeetha, S. SecNLP: A Survey of Embeddings in Clinical Natural Language Processing. J. Biomed. Inform. 2020, 101, 103323. [Google Scholar] [CrossRef]
- Hadsell, R.; Chopra, S.; LeCun, Y. Dimensionality Reduction by Learning an Invariant Mapping. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR ’06), New York, NY, USA, 17–22 June 2006; pp. 1735–1742. [Google Scholar]
- Sogancıoğlu, G.; Ozturk, H.; Ozgür, A. BIOSSES: A Semantic Sentence Similarity Estimation System for the Biomedical Domain. Bioinformatics 2017, 33, 49–58. [Google Scholar] [CrossRef]
- Suominen, H.; Salanterä, S.; Velupillai, S.; Chapman, W.W.; Savova, G.; Elhadad, N.; Pradhan, S.; South, B.R.; Mowery, D.L.; Jones, G.J.; et al. Overview of the SHARE/CLEF eHealth Evaluation Lab 2013. In Proceedings of the International Conference on Cross-Language Evaluation Forum for European Languages, Valencia, Spain, 23–26 September 2013; pp. 212–231. [Google Scholar]
- Uzuner, O.; South, B.R.; Shen, S.; DuVall, S.L. 2010 i2b2/VA Challenge on Concepts, Assertions, and Relations in Clinical Text. J. Am. Med. Inform. Assoc. 2011, 18, 552–556. [Google Scholar] [CrossRef]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. arXiv 2016, arXiv:1603.01360. [Google Scholar]
- Settles, B. Biomedical Named Entity Recognition Using Conditional Random Fields and Rich Feature Sets. In Proceedings of the International Joint Workshop on Natural Language Processing for Biomedicine and Its Applications, Geneva, Switzerland, 28–29 August 2004; pp. 107–110. [Google Scholar]
- Habibi, M.; Weber, L.; Neves, M.; Wiegandt, D.L.; Leser, U. Deep Learning with Word Embeddings Improves Biomedical Named Entity Recognition. Bioinformatics 2017, 33, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.L. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]

| Model | MedNLI | i2b2 2006 | i2b2 2010 | i2b2 2012 | i2b2 2014 |
|---|---|---|---|---|---|
| Clinical BERT | 80.8 | 91.5 | 86.4 | 78.5 | 92.6 |
| Discharge Summary BERT | 80.6 | 91.9 | 86.4 | 78.4 | 92.8 |
| Model | Initialize From | Data and Training Steps |
|---|---|---|
| BERT-Base BioBERT-Base Discharge Summary BERT Bio+Discharge Summary BERT BlueBERT | - BERT-Base BERT-Base BioBERT-Base BERT-Base | Wikipedia + BookCorpus 1M steps PubMed 200K steps + PMC 270K steps Discharge Summary Notes 150K steps Discharge Summary Notes 150K steps PubMed 5M steps + MIMIC 200K steps |
| Discharge Summary CRPT Bio+Discharge Summary CRPT | BERT-Base BERT-Base | Discharge Summary Notes 200K steps PubMed 3M steps + Discharge Summary Notes 200K steps |
| Corpus | Task | Metrics | Train | Dev | Test |
|---|---|---|---|---|---|
| MedNLI | Inference | Accuracy | 11,232 | 1395 | 1422 |
| BIOSSES | Sentence similarity | Pearson | 64 | 16 | 20 |
| ShARe/CLEFE | NER | F1 | 4628 | 1075 | 5195 |
| i2b2-2010 | Relation extraction | F1 | 3110 | 11 | 6293 |
| Model | MedNLI (Acc) | BIOSSES (r) | ShARe/CLEFE (F1) | i2b2-2010 (F1) |
|---|---|---|---|---|
| BERT-Base | 0.757 ± 0.011 | 0.800 ± 0.089 | 0.798 ± 0.006 | 0.728 ± 0.006 |
| BioBERT | 0.819 ± 0.010 | 0.821 ± 0.086 | 0.812 ± 0.005 | 0.737 ± 0.006 |
| Discharge Summary BERT | 0.785 ± 0.011 | 0.718 ± 0.101 | 0.814 ± 0.005 | 0.736 ± 0.006 |
| Bio+Discharge Summary BERT | 0.814 ± 0.010 | 0.819 ± 0.086 | 0.819 ± 0.005 | 0.745 ± 0.005 |
| BlueBERT | 0.841 ± 0.010 | 0.839 ± 0.082 | 0.838 ± 0.005 | 0.757 ± 0.005 |
| Discharge Summary CRPT | 0.819 ± 0.010 | 0.773 ± 0.094 | 0.827 ± 0.005 | 0.754 ± 0.005 |
| Bio+Discharge Summary CRPT | 0.847 ± 0.010 | 0.848 ± 0.080 | 0.837 ± 0.005 | 0.761 ± 0.005 |
| Index | Sentences | |
|---|---|---|
| Pair 1 | A | On admission, her daughters were in disagreement over her code status, and her original, long-standing DNR/DNI status was changed to allow for intubation if needed. However, when the patient’s respiratory status continued to decline to the point of need for intubation, the patient refused intubation. |
| B | Her family was notified and agreed that their mother’s wishes should be fulfilled. She was started on IV morphine, then converted to morphine drip on HD #3 for comfort, and all other medications were discontinued. Her family was at her bedside, and their Rabbi was called. She passed away. | |
| Pair 2 | A | She was seen by renal, who felt that her increase in creatinine may have been secondary to ATN/hypotension and recommended avoiding aggressive overdiuresis. |
| B | She subsequently required aggressive diuresis given her rapid afib/chf with lasix and niseritide drips. However, her creatinine remained at baseline of 1.7–2.0 with diuresis. | |
| Pair 3 | A | She was admitted to the hospital after having a procedure to open up her trachea, and she had some difficulty breathing after the procedure. |
| B | The breathing tube was placed back in her throat, and she was admitted to the ICU for a couple of days and had the breathing tube removed. | |
| Pair 4 | A | With clearing of her mental status, patient also began expressing suicidal and homicidal ideation with auditory hallucinations. |
| B | She admitted to hearing voices telling her to kill herself by overdosing on pills, as well as voices telling her to hurt others, though no one in particular. | |
| Pair 5 | A | A section of the cell block demonstrates a lymphoid infiltrate comprised of small–medium-sized lymphocytes with small–moderate amounts of cytoplasm and round–oval nuclei with mostly vesicular chromatin. |
| B | Admixed are smaller lymphocytes with scant cytoplasm and hyperchromatic nuclei. A review of the cytology prep (alcohol fixed, pap stain) demonstrates aggregates of mildly enlarged lymphocytes. | |
| Model | Masking | Method | MedNLI | BIOSSES | ShARe/CLEFE | i2b2-2010 |
|---|---|---|---|---|---|---|
| Discharge Summary BERT | Random | NSP | 0.785 | 0.740 | 0.814 | 0.736 |
| Discharge Summary BERT | Whole Word | NSP | 0.811 | 0.761 | 0.823 | 0.750 |
| Discharge Summary CRPT | Random | CRL | 0.816 | 0.767 | 0.825 | 0.743 |
| Discharge Summary CRPT | Whole Word | CRL | 0.819 | 0.773 | 0.827 | 0.754 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Jung, Y. Elevating Clinical Semantics: Contrastive Pre-Training Beyond Cross-Entropy in Discharge Summaries. Appl. Sci. 2025, 15, 6541. https://doi.org/10.3390/app15126541
Kim S, Jung Y. Elevating Clinical Semantics: Contrastive Pre-Training Beyond Cross-Entropy in Discharge Summaries. Applied Sciences. 2025; 15(12):6541. https://doi.org/10.3390/app15126541
Chicago/Turabian StyleKim, Svetlana, and Yuchae Jung. 2025. "Elevating Clinical Semantics: Contrastive Pre-Training Beyond Cross-Entropy in Discharge Summaries" Applied Sciences 15, no. 12: 6541. https://doi.org/10.3390/app15126541
APA StyleKim, S., & Jung, Y. (2025). Elevating Clinical Semantics: Contrastive Pre-Training Beyond Cross-Entropy in Discharge Summaries. Applied Sciences, 15(12), 6541. https://doi.org/10.3390/app15126541






