Abstract
This research develops large language models (LLMs) to alleviate the workload of healthcare professionals by classifying medical records into their departments. The models utilize medical records as a dataset for fine-tuning and use clinical knowledge bases to enhance accuracy and efficiency in identifying appropriate departments. This study explores the integration of advanced large language models (LLMs) with quantized low-rank adaptation (QLoRA) for efficient training. The medical department classifier demonstrated impressive performance in diagnosing medical conditions, with an accuracy of 96.26. The findings suggest that LLM-based solutions could significantly improve the efficiency of clinical consultations. What is more, the trained models are hosted on GitHub and are publicly available for use, empowering the wider community to benefit from this research.
1. Introduction
The concern of medical professionals emerging from society was raised after the pandemic. A domain-specific large language model (LLM) called the medical pre-diagnosis department division model is developed in this research based on natural language processing (NLP) techniques to reduce the stress of medical professionals, which can analyze patient self-reported symptoms and identify medical departments. In this model, medical records are used as a dataset to train the model, enabling it to classify the correct department. Medical records are comprehensive documents containing patient identity, diagnosis, treatment, and services provided [1]. LLMs utilize medical records for tasks like summarization, patient education, and reasoning, enhancing their performance through fine-tuning with clinical-standard knowledge bases and diverse datasets [2]. As the feature useful for various natural language processing tasks [3], LLMs are considered in this research.
1.1. Background
The hotspot of the domain of Artificial Intelligence, GPT-3, developed by OpenAI, is a typical attempt at Artificial General Intelligence (AGI). In [4], by using 175 billion parameters, GPT-3 shocked the world with its strong performance on many NLP datasets. The use of LLMs to address real-world challenges highlights their immense potential. One promising application is disease department classification in the medical field, which leverages the vast demand for healthcare services and the abundance of medical records. By utilizing LLMs, such as LLaMA [5], researchers can develop domain-specific applications that effectively address critical issues across various industries.
1.2. Significance
The medical pre-diagnosis department division model can reduce the workload of doctors by automating routine inquiries and providing preliminary diagnoses. Additionally, it allows patients to gain initial insights into their health conditions, which can guide them in seeking appropriate medical care. The model can improve the efficiency of medical consultations by streamlining the process of gathering patient information and providing a preliminary assessment. Although the model could increase the efficiency of clinical consultations, it also fills the gap in the traditional NLP system. The clinical language model integrates GLM2 [6] with insights from GatorTron [7], which is designed for electronic health records (EHRs). This combination allows the medical pre-diagnosis department division model to effectively process and understand clinical narratives, often challenging for traditional NLP systems. Additionally, applying advanced fine-tuning methodologies like LoRA [8] and QLoRA [9] ensures the classifier’s high accuracy and responsiveness in real-world medical settings. This novel approach positions the classifier as a significant advancement in AI-driven healthcare.
2. Related Work
2.1. General Large Language Models
LLMs have exhibited remarkable proficiency in natural language processing tasks, such as language translation, text generation, and question answering [10]. Qwen, developed by Bai et al. [11], features various models with parameter counts ranging from 1.8 B to 14 B, proving itself to be competitive with other open-source models. Touvron et al. [5] introduced LLaMA, suggesting that smaller models might outperform larger ones under similar training conditions due to the effective use of more tokens. This efficiency is also demonstrated by PaLM 2, which optimizes architecture with compute investment [12]. In dialogue, mathematics, and code generation tasks, the Gemma model by the Gemma Team excels, as evidenced by MMLU and MBPP benchmarks. The GLM Team [6] introduced an autoregressive blank infilling model, enhancing ChatGLM’s performance among conversational models. However, LLMs may reflect social biases and toxicity, raising ethical concerns [13]. Furthermore, further model architecture and objective improvements are essential as datasets and model parameters grow.
2.2. Medical Large Language Models
Generic LLMs tend to generate factually inconsistent summaries and make overly convincing or uncertain statements when answering medical questions, potentially causing misinformation-related harm [14]. Additionally, general LLMs’ focus on broad, preference-aligned settings limits their ability to seek information proactively, hindering the development of reliable clinical LLMs for specific medical contexts [15]. Researchers have created medical LLMs to address these issues, leveraging specialized knowledge, clinical datasets, and fine-tuned architectures. Xi Yang et al. [7] introduced GatorTron, a large clinical LLM with billions of parameters, marking a significant increase in model size. The accuracy of GatorTron-large rose to 0.9020, which is a significant increase compared with other biomedical and clinical transformer models. In clinical language modeling, the accuracy of ClinicalBERT in predicting held-out tokens is around 0.857 and 0.994 in the following sentence prediction task. ClinicalBERT, developed by Kexin Huang et al. [16], expands its dataset beyond lab values and medications to capture more intricate medical relationships. Yunxiang Li et al. [17] proposed ChatDoctor, which has a precision of around 0.8444, incorporating a self-directed information retrieval mechanism that enhances beyond traditional fine-tuning. Innovations in training include Qilin-Med, introduced by Qichen Ye et al. [18], which employs a multi-stage training process combining Direct Preference Optimization (DPO) and Domain-specific Continued Pre-Training (DCPT) to mitigate overconfident predictions. The best-performing model, Qilin-Med-7B-SFT, has an accuracy of 53.56, much higher than that of other LLMs, such as LLaMA-CMExam [18]. Despite advancements in medical LLMs, their ability to process longer, complex texts requires further improvement.
3. Methodology
3.1. Dataset
Building upon a dataset named Huatuo26M-Lite (a large-scale medical QA dataset; available at https://huggingface.co/datasets/FreedomIntelligence/Huatuo26M-Lite, accessed on 10 November 2024) [19], the processed version, referred to as zeng981/nlpdataset (processed version of Huatuo26M-Lite; available at https://huggingface.co/datasets/zeng981/nlpdataset, accessed on 12 November 2024), is hosted on Hugging Face [20]. This dataset, consisting of approximately 178,000 entries, is primarily in Chinese and focuses on the medical records domain. It provides comprehensive descriptions of various diseases, with columns named id, answer, score, label, question, and related diseases, facilitating the development of models with robust capabilities for medical classification tasks. The dataset has 16 department classes, and the label distribution diagram is shown in Figure 1.
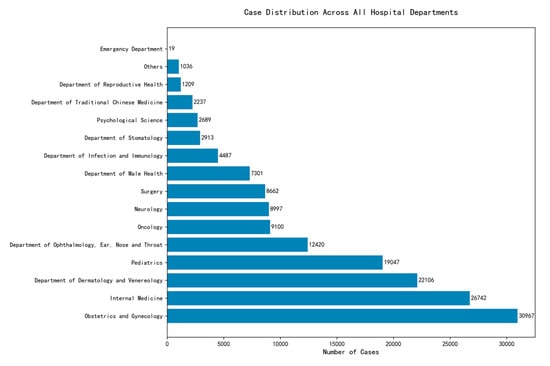
Figure 1.
Label distribution chart for department labels.
3.2. Data Processing
Columns named id, answer, score, and related_diseases are deleted. One instruction column is added to guide the model and fit the dataset into the model—the column name changes from “question” to “input” and from “label” to “output”. The dataset is divided into two groups. Eighty percent is used for training, and 20% for testing. A sample from the dataset is shown in Figure 2.

Figure 2.
Dataset sample with corresponding English translation.
3.3. Model Development and Training
3.3.1. Loss Function
The loss function measures the gap between the model’s prediction and the actual label [12]. Cross-entropy loss aimed at classifying the medical department, which can simplify training and improve test performance in multiclass classification tasks [13], is implemented. Optimization algorithms update the model’s parameters to minimize the loss function.
3.3.2. Optimization Algorithms
Parameters must be learned from the optimized algorithms to realize the objective: reduce the loss produced within the learning process. The output generated by the model will be compared with the desired output. According to the difference between the generated and desired outputs, the model will minimize the gap between the desired and generated outputs through the optimization algorithms [21].
This study adopts low-rank adaptation (LoRA) and its enhanced variant, QLoRA, for efficient fine-tuning of large language models. LoRA preserves the original model’s parameters while reducing the number of trainable components [8]. The number of trainable parameters of the downstream tasks is significantly reduced by distributing the trainable rank decomposition matrices to every layer of the Transformer architecture. QLoRA further improves upon LoRA by combining low-rank adaptation with quantization techniques, reducing memory and computation costs through lower-precision representations (e.g., 8-bit or 4-bit) while maintaining model performance. These techniques enable scalable and resource-efficient fine-tuning, making them well suited for the medical classification tasks in this research. The software environment used to integrate LoRA and QLoRA during the fine-tuning process is summarized in Table 1.

Table 1.
Software environment used for model training.
In this research, the models were fine-tuned using QLoRA to minimize training time and memory usage. All model training operations were conducted locally using an NVIDIA V100 GPU. The AdamW optimizer was employed across all model training processes.
To provide a clearer understanding of the implementation pipeline described above, the model development process is visualized in Figure 3.

Figure 3.
An architectural overview diagram illustrating the classifier model development process.
Gemma-2-9B (fine-tuned model for medical classifying tasks; available at https://huggingface.co/zeng981/nlpgemma29B, accessed on 27 May 2025) was fine-tuned with a learning rate of 5 × 10−5. Training was conducted with a batch size of 4, and the evaluation used a batch size of 8. The model was trained for 3 epochs.
GLM-4-9B (fine-tuned model for medical classifying tasks; available at https://huggingface.co/zeng981/NLPGLM-4-9B, accessed on 27 May 2025) was fine-tuned with a learning rate of 5 × 10−5. The training and evaluation batch sizes were both set to 8. The model was trained for 1 epoch.
Qwen2-7B (fine-tuned model for medical classifying tasks; available at https://huggingface.co/zeng981/NLPQwen2-7b, accessed on 27 May 2025) was fine-tuned with a learning rate of 6 × 10−5. The training batch size was set to be 16, and the evaluation batch size was set to be 8. Three epochs were used for training the model.
Qwen-1.8B (fine-tuned model for medical classifying tasks; available at https://huggingface.co/zeng981/NLPqwen-1.8b, accessed on 27 May 2025) was fine-tuned with a learning rate of 5 × 10−5. The training and evaluation batch sizes were both set to 8. Training was performed for a single epoch.
Llama-3.2-3B-Instruct (fine-tuned model for medical classifying tasks; available at https://huggingface.co/zeng981/NLPLlama3.2-3b, accessed on 27 May 2025) was fine-tuned with a learning rate of 5 × 10−5. The training and evaluation batch sizes were both set to 8. The model was trained for 1 epoch.
To mitigate overfitting, a dropout rate of 0.5 was applied uniformly across all models. Most models in this study were trained with a limited number of epochs, typically only 1. However, Qwen-1.8B and Gemma-2-9B were trained for 3 epochs, which enabled them to achieve better performance by improving generalization.
3.3.3. System Architecture of the Medical Department Classification Models
The chatbot is implemented as a single-stage end-to-end generation system. All user inputs are directly processed by a fine-tuned LLM without explicit intent or entity extraction. This design allows the model to learn domain-specific conversational patterns holistically. The model is fine-tuned on medically relevant QA data to compensate for the lack of explicit knowledge integration.
3.3.4. Pretrained LLMs
The Transformer architecture of Gemma2 [22] applies new technical modifications, such as group-query attention [23] and local–global attention [24]. Gemma2 provides an available application programming interface (API), which helps developers integrate and develop quickly. Researchers could scale the architecture of Gemma2 to adapt to varying tasks.
LLaMA achieves comparable or even better performance than larger models with fewer parameters. By training with accessible and common datasets [5], LLaMA becomes more transparent and convenient for researchers. Compared with other models, the computational needs of LLaMA are lower, which improves its efficiency.
With different models and distinct parameter counts, Qwen is developed as an all-around language model series, such as Qwen, Qwen-Chat, Code-Qwen, Code-Qwen-Chat, and Math-Qwen-Chat [11]. The model performed well in the mathematical reasoning and programming tasks.
Compared with Qwen, the application scenarios are expanded. Using state-of-the-art quantization techniques, such as Int4 and Int8 [25], Qwen2 could significantly minimize the computational resources.
3.3.5. Methodological Details
All models were evaluated using a standardized set of natural language prompts in Chinese, simulating typical user queries in a medical chatbot scenario. Each prompt consists of two parts: an instruction and an input derived from the dataset. Prompts were kept consistent across all models to ensure fair comparison. Both zero-shot and few-shot prompting scenarios were considered, depending on the model’s context window size.
The ground truth labels were derived from a publicly available medical consultation dataset, where each user query is annotated with the correct target department.
4. Evaluation
4.1. Evaluation Methods
ROUGE and BLEU are used to evaluate the model’s performance in this research. These two methods provide a comprehensive evaluation by considering word and length matching to assess sentence similarity, which helps instruct the model’s training process.
Recall-Oriented Understudy for Gisting Evaluation (ROUGE) calculates the number of overlapping units, such as n-grams, word sequences, and word pairs, between the computer-generated summary and the reference summaries created by humans. ROUGE-N (4) and ROUGE-L (1)–(3) are adopted to evaluate performance in this research.
The recall-related measure (4) uses the parameter n to represent the n-gram length, comparing the total number of n-grams and the number of matching n-grams [26].
Except for the n-gram matches, the similarity between sentences also depends heavily on the order of the words. (3) Emphasizes the sequence order and the length of the subsequence, which can work together with ROUGE-N to evaluate the model’s performance.
The main difference between Bilingual Evaluation Understudy (BLEU) and ROUGE is that BLEU (5) incorporates a brevity penalty. The brevity penalty considers sentence length and encourages the generated sentence to have a length similar to the reference sentence, which can avoid generating short sentences [27].
4.2. Evaluation Against Baselines
The fine-tuned LLMs (Qwen-1.8B, Gemma-2-9B, etc.) were compared against traditional classification baselines, including SVM and a BERT-based classifier trained on the same dataset.
Table 2 shows that, except for ClinicalBERT, other baseline models perform worse than the fine-tuned model Gemma-2-9B. The baseline models, such as SVM or un-fine-tuned BERT relying on fixed or shallow features, differ from the large pre-trained models, which have learned rich knowledge through large-scale corpora. Hence, the baseline models may be less capable of understanding complex contextual information.

Table 2.
Performance comparison between best-performing fine-tuned LLM and baseline models.
5. Results
5.1. Accuracy of Diagnosis
The classifier trained with Gemma-2-9B achieved the best accuracy of 96.26 in diagnosing potential diseases based on user-inputted symptoms.
The recall and F1 scores were also high, indicating the model’s ability to correctly identify relevant medical conditions from various symptoms, as shown in Table 3.

Table 3.
Performance comparison of different pretrained models.
By comparing Figure 4a and Figure 4b, a pattern of similar performance across different models, evaluated by ROUGE and BLEU, is observed, as indicated by the identical ranking of ROUGE and BLEU. This result may be due to the same dataset the models were trained on and the same test the models were asked to perform, demonstrating the classifiers’ stable performance.
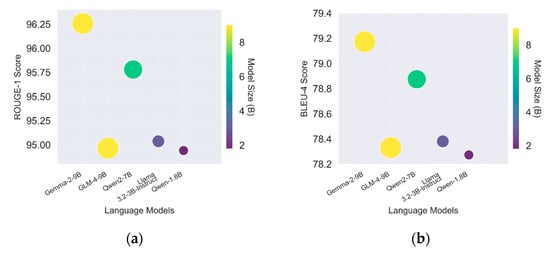
Figure 4.
(a) ROUGE score for Gemma 2 (9B), GLM 4 (9B), Qwen2 (7B), Llama 3.2 (3B), and Qwen 1 (1.8B); (b) BLUE score for Gemma 2 (9B), GLM 4 (9B), Qwen2 (7B), Llama 3.2 (3B), and Qwen 1 (1.8B).
Additionally, it can be observed that the BLEU score is lower than the ROUGE score. This is due to the different criteria used in the evaluation strategies. BLEU is more suitable for estimating machine translation performance based on its precision and brevity penalty mechanism [27]. In contrast, ROUGE estimates question-answering performance based on its accuracy mechanism [26]. According to the different mechanisms of these two evaluation methods, the lower BLEU score of the medical department classifier may be explained by the penalty applied to the length of the generated text, which does not affect the accuracy of the generated answers, as shown by the ROUGE score.
5.2. Resource Efficiency
The impressive performance of Qwen2 should be noted, which may be attributed to using ROUGE, which evaluates scores based on character matching rates [26].
5.3. Multilingual Support
The classifier successfully handled inquiries in multiple languages, demonstrating its potential for broad applicability in diverse linguistic settings.
Table 4 shows that Gemma-2 performs better than other models in ROUGE and BLUE scores, which the different characteristics of languages can explain. According to [28], over 70% of simplified Chinese’s most frequently used words are two-character words, which may differ from English expression habits.

Table 4.
Evaluation metrics for Gemma 2 (9B), GLM 4 (9B), Qwen2 (7B), Llama 3.2 (3B), and Qwen 1 (1.8B).
The relationship between speed (in steps/s, samples/s) and model accuracy is often governed by a trade-off. Larger models typically achieve higher accuracy but tend to be slower, while smaller models run faster but are less accurate.
Based on Figure 5 and Figure 6, Qwen 1.8B exhibits a shorter runtime. Its faster performance is primarily due to its smaller parameter size, but this results in lower accuracy due to underfitting. This study finds that Gemma-2, despite having fewer steps and samples per second, achieves higher accuracy.
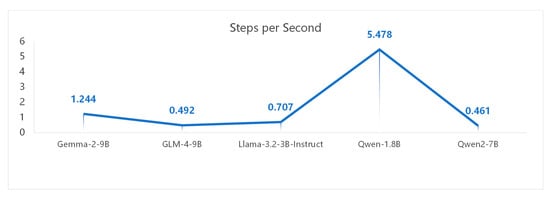
Figure 5.
Performance comparison of steps per second for various models.
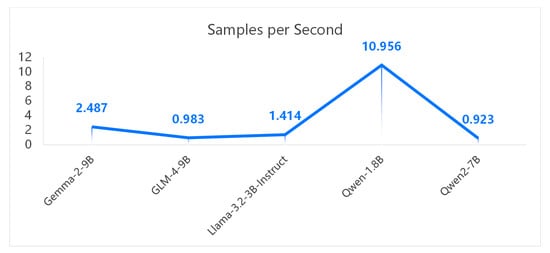
Figure 6.
Performance comparison of samples per second for multiple models.
6. Discussion
The results of this study indicate that integrating Natural Language Processing (NLP) techniques and Large Language Models (LLMs) in developing medical department classifiers is both feasible and effective. The high accuracy in diagnosing various medical conditions suggests that the model has learned to interpret and analyze symptoms consistent with standard medical practices. This demonstrates the potential of LLMs to assist in preliminary diagnosis and patient education, offering users an accessible platform for reliable medical information. However, this study also revealed several challenges and areas for improvement that must be addressed for broader application and reliability.
6.1. Limitations in Complex Cases
While the classifier demonstrated impressive performance in handling common medical scenarios, its accuracy was notably lower when diagnosing complex or rare conditions. This limitation reflects the model’s dependency on the training dataset, which may lack sufficient representation of less common diseases. Enhancing the model’s capabilities in these areas will require further training using more specialized datasets, such as those focusing on rare diseases or complex comorbidities, and potentially incorporating expert systems for more nuanced decision making [29]. Research has shown that hybrid models, which combine LLMs with rule-based expert systems, can improve performance in handling intricate medical cases [30]. Future development should explore this integration to enhance diagnostic accuracy and reliability.
Figure 7 presents the top 10 error output types of the classifier. Based on this figure, the project identifies four medical departments—Internal Medicine, Obstetrics and Gynecology, Pediatrics, and Dermatology and Venereology—where the classifier is more likely to produce incorrect outputs. This finding suggests potential future research directions aimed at improving classification accuracy among these four departments.
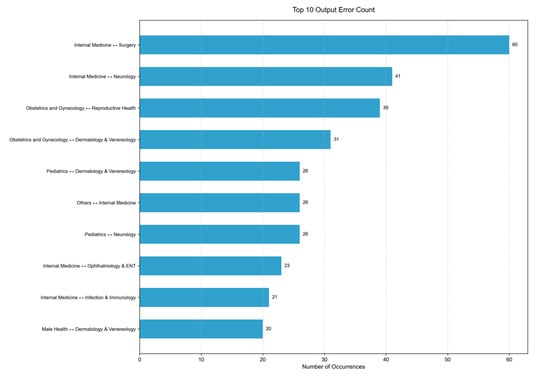
Figure 7.
Top 10 output error type the classifier made.
Table 5 presents two examples of error cases from the classifier, in both Chinese and English.

Table 5.
An example of model misclassification with the corresponding English translation.
The example at index 4528 shows a top-ranked error, where the classifier incorrectly assigns the Surgery department to Internal Medicine. This error suggests that the classifier may be affected by an unbalanced training data distribution in the database. Additionally, it may indicate the model’s limited ability to extract relevant features. This error highlights promising future research directions, such as constructing a more balanced dataset and refining the model architecture.
The example index at 75 shows an error output of the Gemma-2-9B model regarding the Department of Dermatology and Venereology. The correct output should be “Department of Dermatology and Venereology”. This error reveals the model’s limitation in distinguishing between departments with overlapping clinical functions, which may stem from relying solely on LLMs. While LLMs can capture linguistic patterns effectively, they may struggle to differentiate between departments with similar functional scopes.
To address this issue, future work could incorporate hybrid models that combine LLMs with expert systems, enabling more precise reasoning in ambiguous cases. For example, an expert system could encode clinical logic such as “widespread rashes, regardless of patient age, are typically treated by dermatologists”, thereby guiding the model toward more accurate predictions in such scenarios.
6.2. Demographic and Clinical Bias Considerations
The current evaluation does not investigate whether the model performs differently across patient subgroups defined by age, sex, ethnicity, or symptomatology. Future studies should include a thorough analysis of potential demographic and clinical biases to ensure fairness and reliability.
6.3. Ethical and Privacy Concerns
One of the most critical aspects of deploying medical classifiers is ensuring compliance with data privacy regulations, such as the Health Insurance Portability and Accountability Act (HIPAA) in the United States. This study successfully implemented data anonymization techniques to protect sensitive patient information, a necessary step in any medical application. However, the fast-evolving nature of privacy concerns in digital health requires continuous monitoring and periodic updates to the system’s privacy protocols to address new challenges [31]. Regular audits and adherence to global data protection standards, such as the General Data Protection Regulation (GDPR) in Europe, are essential to maintain user trust and prevent data breaches. Additionally, ethical considerations, including the potential for bias in classifier responses, must be carefully managed to avoid discrepancies in care [32].
6.4. Scalability and Adaptation
For the classifier to be successful over the long term, its ability to scale and adapt to new medical knowledge and evolving user needs is essential. Implementing a feedback loop where the model can learn from real-world interactions could significantly enhance its accuracy and relevance. Continuous learning systems have been shown to improve the performance of AI models by allowing them to adapt to new inputs and scenarios over time [33]. This approach could also help the classifier stay updated with the latest medical research and treatment protocols, ensuring that it provides the most current and accurate information. Developing robust mechanisms for validating and incorporating user feedback without compromising data privacy will be key to achieving this scalability.
6.5. Expert Feedback
To evaluate the clinical applicability of our proposed system, this project has consulted medical experts regarding the clinical value of the medical department classifier.
The responses from the experts are positive. It is stated that this classifier could be applied at the information desk in the outpatient department of the hospital to reduce the workload of general practitioners and nurses. Moreover, it is also considered promising as a clinical decision support system (CDSS) for doctors in medical diagnosis, which could improve the accuracy and efficiency of diagnoses made by medical professionals.
For future work, real-world experiments are needed to quantitatively assess potential improvements in clinical efficiency.
7. Conclusions
This study successfully developed a medical department classifier using advanced NLP techniques and LLMs. The classifier demonstrated high accuracy in diagnosing diseases and providing medical advice, with significant potential for real-world application. Integrating resource-efficient training methods like LoRA further enhances the classifier’s developability across various platforms.
Despite these promising results, the deployment of LLMs in healthcare raises critical concerns that must be addressed. While privacy regulations such as HIPAA and GDPR are acknowledged, this study does not provide concrete strategies for ensuring data security and protecting patient confidentiality in real-world scenarios. Moreover, the potential for algorithmic bias and the lack of discussion on data representativeness highlight the need for more rigorous evaluation to ensure fairness across diverse populations. The absence of clinical oversight in the model’s deployment process also presents significant risks, particularly in high-stakes medical decision-making contexts.
Future research should therefore prioritize improving model performance in complex clinical scenarios, implementing robust privacy-preserving mechanisms, and ensuring ethical compliance throughout development and deployment. Establishing interdisciplinary collaboration with clinical experts will be essential to validate the model’s reliability and safety. Given the model’s effectiveness in processing Chinese-language data, future work should also explore its extension to multilingual settings to enhance generalizability across diverse linguistic and cultural contexts. With these improvements, the classifier could become a valuable tool in global healthcare, offering accessible and equitable medical guidance to users worldwide.
Author Contributions
Conceptualization, B.I. and X.Z.; methodology, B.I.; software, X.Z.; validation, B.I., H.Y., and X.Z.; formal analysis, H.Y. and F.F.; investigation, X.Z.; resources, B.I.; data curation, H.Y.; writing—original draft preparation, H.Y.; writing—review and editing, B.I., A.R.S., and H.Y.; visualization, H.Y. and F.F.; supervision, B.I. and A.R.S.; project administration, B.I.; funding acquisition, B.I. All authors have read and agreed to the published version of the manuscript.
Funding
This project was made possible through funding from Wenzhou-Kean University under the grant number IRSPK2023005. Our sincere appreciation extends to the university for its financial support, which was instrumental in advancing our research. We also gratefully acknowledge the support provided by the WKU Institute of Advanced Natural Language Processing (IANLP), whose resources and collaborative environment significantly contributed to the successful execution of this work.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anisa, D.; Farhansyah, F.; Wulandari, S. Sosialisasi Pentingnya Rekam Medis Di Fasilitas Layanan Kesehatan Pada Siswa Sman 1 Teluk Sebong. Prima Portal Ris. DAN Inov. Pengabdi. Masy. 2023, 2, 253–256. [Google Scholar] [CrossRef]
- Liu, F.; Li, Z.; Zhou, H.; Yin, Q.; Yang, J.; Tang, X.; Luo, C.; Zeng, M.; Jiang, H.; Gao, Y.; et al. Large Language Models in the Clinic: A Comprehensive Benchmark. arXiv 2024, arXiv:2405.00716. [Google Scholar]
- Yang, J.; Jin, H.; Tang, R.; Han, X.; Feng, Q.; Jiang, H.; Zhong, S.; Yin, B.; Hu, X. Harnessing the Power of LLMs in Practice: A Survey on ChatGPT and Beyond. arXiv 2023, arXiv:2304.13712. [Google Scholar] [CrossRef]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. arXiv 2020, arXiv:2005.14165. [Google Scholar]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and Efficient Foundation Language Models. arXiv 2023, arXiv:2302.13971. [Google Scholar]
- Zeng, A.; Xu, B.; Wang, B.; Zhang, C.; Yin, D.; Zhang, D.; Rojas, D.; Feng, G.; Zhao, H.; Lai, H.; et al. ChatGLM: A Family of Large Language Models from GLM-130B to GLM-4 All Tools. arXiv 2024, arXiv:2406.12793. [Google Scholar]
- Yang, X. A large language model for electronic health records. npj Digit. Med. 2022, 5, 194. [Google Scholar] [CrossRef]
- Hu, E.J.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W. LoRA: Low-Rank Adaptation of Large Language Models. arXiv 2021, arXiv:2106.09685. [Google Scholar]
- Dettmers, T.; Pagnoni, A.; Holtzman, A.; Zettlemoyer, L. QLORA: Efficient Finetuning of Quantized LLMs. Adv. Neural Inf. Process. Syst. 2023, 36, 10088–10115. [Google Scholar]
- Raiaan, M.A.K.; Mukta, S.H.; Fatema, K.; Fahad, N.M.; Sakib, S.; Mim, M.M.J.; Ahmad, J.; Ali, M.E.; Azam, S. A Review on Large Language Models: Architectures, Applications, Taxonomies, Open Issues and Challenges. IEEE Access 2024, 12, 26839–26874. [Google Scholar] [CrossRef]
- Bai, J.; Bai, S.; Chu, Y.; Cui, Z.; Dang, K.; Deng, X.; Fan, Y.; Ge, W.; Han, Y.; Huang, F.; et al. Qwen Technical Report. arXiv 2023, arXiv:2309.16609. [Google Scholar]
- Zhang, G.; Piccardi, M.; Borzeshi, E.Z. Sequential Labeling with Structural SVM Under Nondecomposable Losses. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 4177–4188. [Google Scholar] [CrossRef] [PubMed]
- Demirkaya, A.; Chen, J.; Oymak, S. Exploring the Role of Loss Functions in Multiclass Classification. In Proceedings of the 2020 54th Annual Conference on Information Sciences and Systems (CISS), Princeton, NJ, USA, 18–20 March 2020; pp. 1–5. [Google Scholar]
- Tang, L. Evaluating large language models on medical evidence summarization. npj Digit. Med. 2023, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Balachandran, V.; Feng, S.; Ilgen, J.; Pierson, E.; Koh, P.W.; Tsvetkov, Y. MEDIQ: Question-Asking LLMs for Adaptive and Reliable Clinical Reasoning. arXiv 2024, arXiv:2406.00922. [Google Scholar]
- Huang, K.; Altosaar, J.; Ranganath, R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv 2020, arXiv:1904.05342. [Google Scholar]
- Li, Y.; Li, Z.; Zhang, K.; Dan, R.; Jiang, S.; Zhang, Y. ChatDoctor: A Medical Chat Model Fine-Tuned on a Large Language Model Meta-AI (LLaMA) Using Medical Domain Knowledge. Cureus 2023, 15, e40895. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, J.; Chong, D.; Zhou, P.; Hua, Y.; Liu, F.; Cao, M.; Wang, Z.; Cheng, X.; Lei, Z.; et al. Qilin-Med: Multi-stage Knowledge Injection Advanced Medical Large Language Model. arXiv 2024, arXiv:2310.09089. [Google Scholar]
- FreedomIntelligence. Huatuo26M-Lite. [Dataset]. Available online: https://huggingface.co/datasets/FreedomIntelligence/Huatuo26M-Lite (accessed on 10 November 2024).
- Zeng, X. Zeng981/Nlpdataset. [Dataset]. Available online: https://huggingface.co/datasets/zeng981/nlpdataset (accessed on 12 November 2024).
- Zaheer, R.; Shaziya, H.; College, N. A Study of the Optimization Algorithms in Deep Learning. In Proceedings of the 2019 Third International Conference on Inventive Systems and Control (ICISC), Coimbatore, India, 10–11 January 2019. [Google Scholar] [CrossRef]
- Gemma Team; Riviere, M.; Pathak, S.; Sessa, P.G.; Hardin, C.; Bhupatiraju, S.; Hussenot, L.; Mesnard, T.; Shahriari, B.; Ramé, A.; et al. Gemma 2: Improving Open Language Models at a Practical Size. arXiv 2024, arXiv:2408.00118. [Google Scholar]
- Ainslie, J.; Lee-Thorp, J.; Jong, M.; Zemlyanskiy, Y.; Lebrón, F.; Sanghai, S. GQA: Training Generalized Multi-Query Transformer Models from Multi-Head Checkpoints. arXiv 2023, arXiv:2305.13245. [Google Scholar]
- Beltagy, I.; Peters, M.E.; Cohan, A. Longformer: The Long-Document Transformer. arXiv 2020, arXiv:2004.05150. [Google Scholar]
- Yang, A.; Yang, B.; Hui, B.; Zheng, B.; Yu, B.; Zhou, C.; Li, C.; Li, C.; Liu, D.; Huang, F.; et al. Qwen2 Technical Report. arXiv 2024, arXiv:2407.10671. [Google Scholar]
- Lin, C.-Y. ROUGE: A Package for Automatic Evaluation of Summaries. Available online: https://aclanthology.org/W04-1013/ (accessed on 27 May 2025).
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. BLEU: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting on Association for Computational Linguistics—ACL ’02. Association for Computational Linguistics, Philadelphia, PA, USA, 6–12 July 2002; p. 311.
- Song, D.; Li, D. Psycholinguistic Norms for 3,783 Two-Character Words in Simplified Chinese. Sage Open 2021, 11, 21582440211054495. [Google Scholar] [CrossRef]
- Topol, E. Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again; Hachette: London, UK, 2019. [Google Scholar]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Rieke, N. The future of digital health with federated learning. npj Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, N. A Survey on Bias and Fairness in Machine Learning. ACM Comput. Surv. 2021, 54, 1–35. [Google Scholar] [CrossRef]
- Lu, J. Towards continual learning: A survey. Artif. Intell. Rev. 2022, 57, 1–29. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).