PELM: A Deep Learning Model for Early Detection of Pneumonia in Chest Radiography
Abstract
1. Introduction
1.1. Background
1.2. Motivation
1.3. Objectives
- Compare the diagnostic performance of multiple CNN and Transformer-based architectures for pneumonia detection from CXR images.
- Evaluate whether ensemble learning and attention mechanisms yield tangible improvements over traditional approaches.
- Provide an empirical basis for selecting models for AI-assisted clinical decision support.
2. Literature Reviews
- Asnaoui et al. [15] conducted a performance comparison of several fine-tuned models, notably InceptionResNet_V2, ResNet50, and MobileNet_V2, and found that an ensemble of these achieved the highest F1-score (94.84%) on a COVID-era pneumonia dataset. While effective, their work focused on model-level stacking without optimized weight tuning, limiting adaptability across datasets.
- Rahman and Muhammad [16] used transfer learning with four CNNs—AlexNet, ResNet18, DenseNet201, and SqueezeNet—to distinguish between bacterial and viral pneumonia. Their comprehensive three-class evaluation (normal, bacterial, viral) demonstrated DenseNet201’s superiority, yet lacked ensemble integration or spatial attention mechanisms, which are vital for nuanced feature focus.
- Hossain et al. [17] incorporated LSTM layers with MobileNet and ResNet backbones to model temporal dependencies in X-ray sequences. Although this hybrid CNN-RNN approach achieved 90.2% accuracy, its application to static CXR data is limited and requires sequential imaging, which is not always clinically available.
- Ahmad et al. [18] proposed a temporal model using CheXNet and GRUs to track COVID-19 progression across serial CXRs. Their zone-based approach yielded a strong AUC of 0.98, showing the value of spatial granularity, though the model’s reliance on longitudinal data limits its generalizability to single-instance diagnoses.
- Irvin et al. [19], through the CheXpert dataset, demonstrated DenseNet121’s capability to achieve radiologist-level performance (AUC: 0.98). However, the study was confined to single-model evaluations and did not explore ensemble robustness or recall optimization, which are critical in minimizing missed diagnoses.
- An et al. [20] introduced multi-head attention and dynamic pooling within a DenseNet121-EfficientNetB0 hybrid, achieving 95.19% accuracy and significantly reducing false positives. Despite strong precision (98.38%), the method’s computational complexity and architectural specificity may hinder scalability in resource-constrained settings.
- Pacal et al. [21] benchmarked over 60 CNN and ViT models on cervical imaging, illustrating that ViT-B16 combined with ensemble strategies (e.g., max-voting) improved low-class performance. While not focused on pneumonia, this study underlines the value of Transformer-based architectures and voting techniques in medical imaging.
- Darici et al. [22] compared lightweight custom CNNs with ensemble strategies on pediatric CXRs. Their use of SMOTE for class balancing and separable convolutions yielded 95% accuracy in binary classification. However, their ensemble model underperformed compared to their standalone CNN, indicating potential redundancy without adaptive weighting.
- Sharma and Guleria [5] combined VGG16 feature extraction with multiple classifiers (e.g., NN, SVM, RF), achieving up to 95.4% accuracy and AUCs of 0.988. While showcasing the benefit of hybrid pipelines, the approach lacked architectural fusion, which can consolidate spatial-semantic feature richness.
- Varshni et al. [23] demonstrated that combining DenseNet169 features with classical classifiers like SVM outperformed logistic regression and basic CNNs (AUC: 0.80). However, limited dataset size (n = 2862) and absence of attention mechanisms constrained generalizability.
- Validation across a large, multi-source dataset with balanced class distributions.
- Integrating CNNs and Transformers through feature-level fusion to enhance diagnostic accuracy.
- Incorporating transfer learning, batch normalization, and dropout to mitigate overfitting.
- Tailored for real-time deployment in resource-constrained environments such as ICUs and emergency settings.
- PELM is designed to deliver a robust, interpretable, and clinically viable deep learning framework for automated pneumonia detection from chest radiographs.
3. Materials and Methods
3.1. Data Collection and Preprocessing
3.1.1. Dataset
3.1.2. Preprocessing
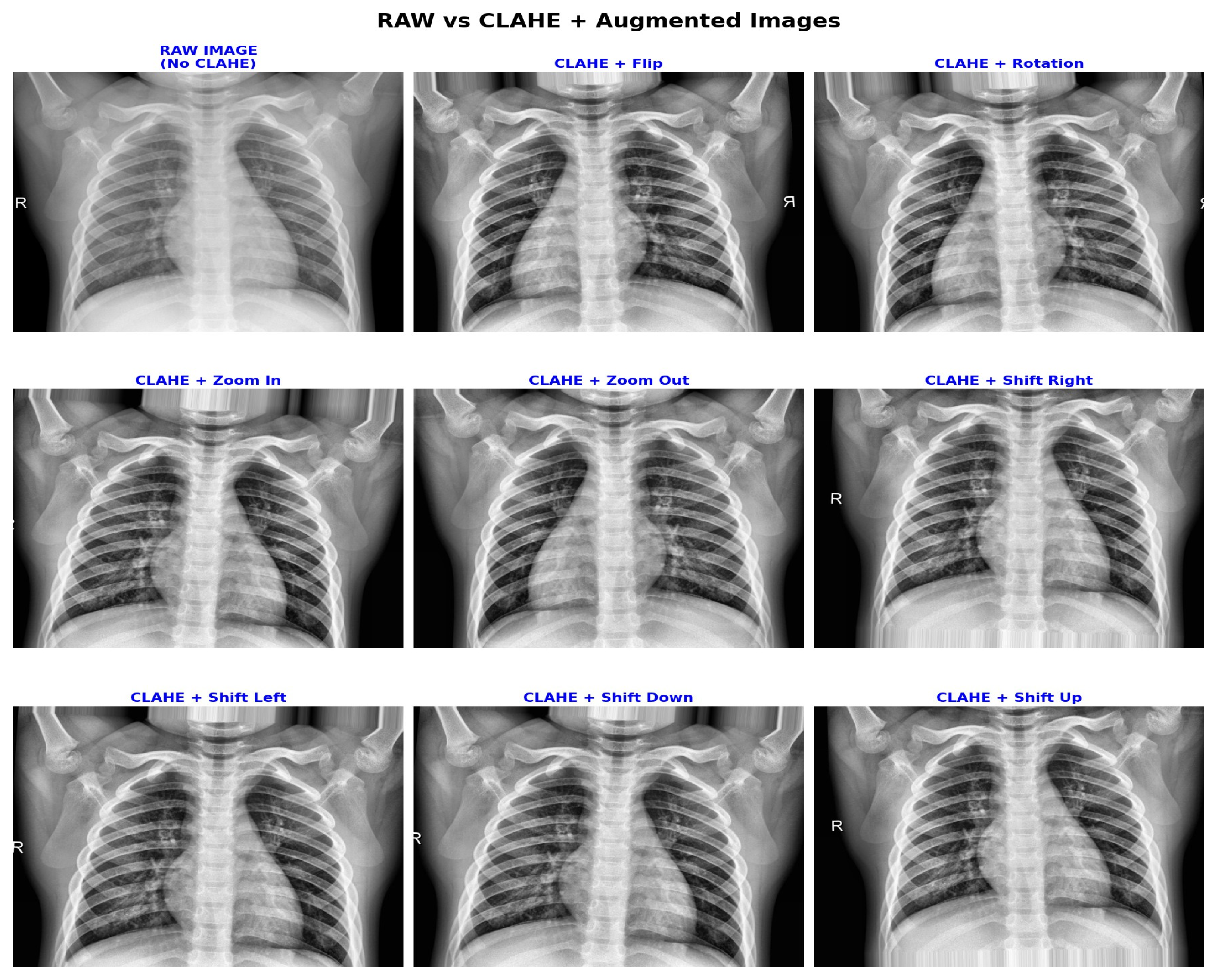
3.2. Clinically Aware Data Augmentation
3.3. Contrast Enhancement with CLAHE
3.4. Model Architectures
- VGG16 is a 16-layer deep CNN known for its uniform and straightforward architecture. It consists of stacked 3 × 3 convolutional layers followed by max-pooling layers, concluding with three fully connected layers. Despite having around 138 million parameters, VGG16 remains a popular backbone for transfer learning due to its strong performance on large-scale datasets like ImageNet. An overview of the adapted model used in our experiments is shown in Figure 6.
- ResNet50 is a 50-layer network that introduces residual connections to enable efficient training of deep networks. It uses identity and convolutional blocks with bottleneck layers for dimensionality reduction. With approximately 25.6 million parameters, ResNet50 is widely adopted for its generalization and ease of optimization. An overview of the adapted model used in this work is illustrated in Figure 7.

- InceptionV3 employs factorized and asymmetric convolutions to reduce computational cost while preserving expressiveness. It also integrates auxiliary classifiers to aid in training deeper networks. With ~23.5 million parameters, it supports larger input sizes and excels in multi-scale feature extraction. An overview of the adapted version used in this work is illustrated in Figure 8.
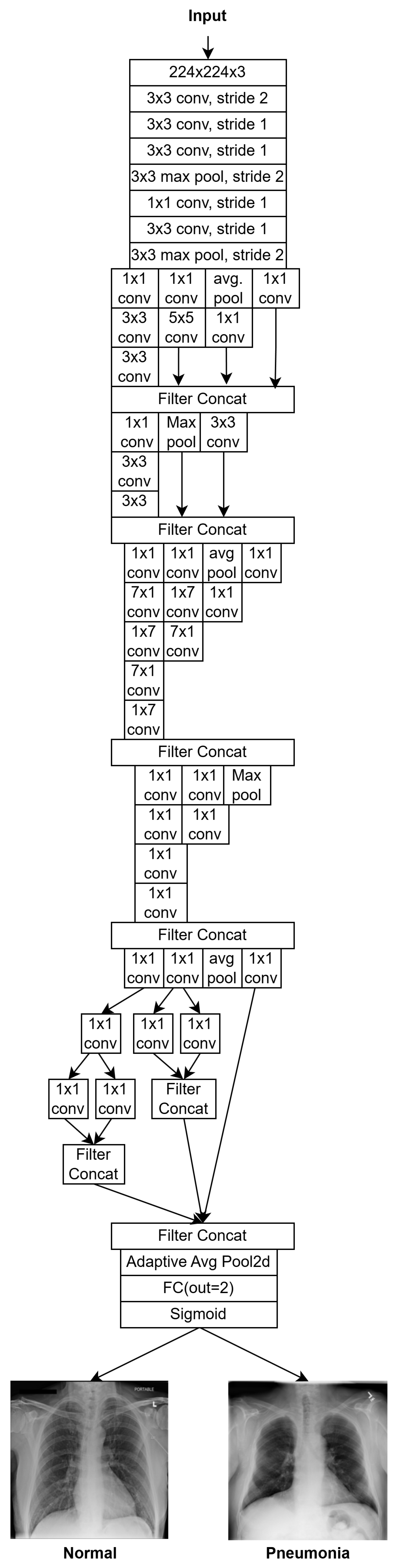
- F1: Feature vector from InceptionV3, reduced to 80 dimensions;
- F2: Feature vector from VGG16, reduced to 2048 dimensions;
- F3: Feature vector from ResNet50, reduced to 512 dimensions;
- F4: Feature vector from Vision Transformer (ViT), reduced to 768 dimensions.
- Batch Normalization layer: stabilizes feature distributions;
- Dense Layer 1;
- Dropout (rate = 0.4);
- Dense Layer 2;
- Dropout (rate = 0.3);
- Dense Layer 3;
- Output Layer: A single neuron with sigmoid activation.
otherwise (Normal/Other Findings)
3.5. Training and Validation
3.6. Hyperparameter Tuning
- Learning Rate: Tested within the range of 0.0001 to 0.01, with 0.001 yielding the most stable and accurate results when using the Adam optimizer.
- Batch Size: Values of 16, 32, and 64 were evaluated. A batch size of 32 offered the best trade-off between training stability and computational efficiency.
- Dropout Rate: Dropout values between 0.2 and 0.5 were explored in the dense layers of the classification head. The final architecture included dropout rates of 0.4 and 0.3 in successive layers, effectively reducing overfitting.
- Optimization: The Adam optimizer was used and momentum parameters were kept constant as (, ).
- Number of Dense Units: Various configurations of dense layer sizes were tested after feature concatenation. The best performance was achieved using three fully connected layers with gradually decreasing units (e.g., 1024, 512, 128).
- Activation Functions: ReLU activation was used for intermediate layers, while a sigmoid activation function was applied to the final output layer for binary classification.
3.7. Performance Metrics
4. Results
Model Performance
5. Discussion
5.1. Strengths of Deep Learning Models
5.2. Limitations and Challenges
6. Future Work
7. Conclusions
8. Innovative Contribution
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AUC | Area Under The Curve |
| CLAHE | Contrast-Limited Adaptive Histogram Equalization |
| CNN | Convolutional Neural Networks |
| CT | Computed Tomography |
| CTR | Cardiothoracic Ratio |
| DL | Deep Learning |
| DOAJ | Directory Of Open-Access Journals |
| LD | Linear Dichroism |
| LSTM | Long Short-Term Memory |
| MAC | Multiply-Add Operations |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MRI | Magnetic Resonance Imagine |
| NIH | National Institutes Of Health |
| PELM | Pneumonia Ensemble Learning Model |
| ResNet50 | Residual Network 50 |
| TLA | Three-Letter Acronym |
| ViT | Vision Transformer |
References
- Maurer, J.R. Long-term exposure to ambient air pollution and risk of hospitalization with community-acquired pneumonia in older adults. Yearb. Pulm. Dis. 2011, 2011, 150–152. [Google Scholar] [CrossRef]
- Sajed, S.; Sanati, A.; Garcia, J.E.; Rostami, H.; Keshavarz, A.; Teixeira, A. The effectiveness of deep learning vs. traditionalmethods for lung disease diagnosis using chest X-ray images: A systematic review. Appl. Soft Comput. 2023, 147, 110817. [Google Scholar] [CrossRef]
- WHO. Pneumonia “Pneumonia in Children”. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/pneumonia (accessed on 24 May 2025).
- Aljawarneh, S.A.; Al-Quraan, R. Pneumonia detection using enhanced convolutional neural network model on chest X-ray images. Big Data 2025, 13, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Guleria, K. A deep learning based model for the detection of pneumonia from chest X-ray images using VGG-16 and neural networks. Procedia Comput. Sci. 2023, 218, 357–366. [Google Scholar] [CrossRef]
- Campbell, H.; el Arifeen, S.; Hazir, T.; O’Kelly, J.; Bryce, J.; Rudan, I.; Qazi, S.A. Measuring coverage in MNCH: Challenges in monitoring the proportion of young children with pneumonia who receive antibiotic treatment. PLoS Med. 2013, 10, e1001421. [Google Scholar] [CrossRef]
- Kanwal, K.; Asif, M.; Khalid, S.G.; Liu, H.; Qurashi, A.G.; Abdullah, S. Current diagnostic techniques for pneumonia: A scoping review. Sensors 2024, 24, 4291. [Google Scholar] [CrossRef]
- Kareem, A.; Liu, H.; Sant, P. Review on pneumonia image detection: A machine learning approach. Hum.-Centric Intell. Syst. 2022, 2, 31–43. [Google Scholar] [CrossRef]
- Htun, T.P.; Sun, Y.; Chua, H.L.; Pang, J. Clinical features for diagnosis of pneumonia among adults in primary care setting: A systematic and meta-review. Sci. Rep. 2019, 9, 7600. [Google Scholar] [CrossRef]
- Ahmadova, A.; Huseynov, I.; Ibrahimov, Y. Improving pneumonia diagnosis with RadImageNet: A deep transfer learning approach. Authorea 2023, 8, 25. [Google Scholar] [CrossRef]
- Prakash, J.A.; Asswin, C.; Ravi, V.; Sowmya, V.; Soman, K. Pediatric pneumonia diagnosis using stacked ensemble learning on multi-model Deep CNN Architectures. Multimed. Tools Appl. 2022, 82, 21311–21351. [Google Scholar] [CrossRef]
- Shirwaikar, R. A machine learning application for medical image analysis using deep convolutional neural networks (cnns) and transfer learning models for pneumonia detection. J. Electr. Syst. 2024, 20, 2316–2324. [Google Scholar] [CrossRef]
- Rashed, B.M.; Popescu, N. Performance investigation for Medical Image Evaluation and diagnosis using machine-learning and deep-learning techniques. Computation 2023, 11, 63. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Romeny, B.M.T.H.; Viergever, M.A. Computer-aided diagnosis in chest radiography: A survey. IEEE Trans. Med. Imaging 2001, 20, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- El Asnaoui, K. Design ensemble deep learning model for pneumonia disease classification. Int. J. Multimed. Inf. Retr. 2021, 10, 55–68. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Hossain, S.; Rahman, R.; Ahmed, M.S.; Islam, M.S. Pneumonia detection by analyzing XRAY images using mobilenet, resnet architecture and long short term memory. In Proceedings of the 2020 30th International Conference on Computer Theory and Applications (ICCTA), Alexandria, Egypt, 12–14 December 2020; pp. 60–64. [Google Scholar] [CrossRef]
- Ahmad, J.; Saudagar, A.K.; Malik, K.M.; Ahmad, W.; Khan, M.B.; Hasanat, M.H.; AlTameem, A.; AlKhathami, M.; Sajjad, M. Disease progression detection via deep sequence learning of successive radiographic scans. Int. J. Environ. Res. Public Health 2022, 19, 480. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. Proc. AAAI Conf. Artif. Intell. 2019, 33, 590–597. [Google Scholar] [CrossRef]
- An, Q.; Chen, W.; Shao, W. A deep convolutional neural network for pneumonia detection in X-ray images with attention ensemble. Diagnostics 2024, 14, 390. [Google Scholar] [CrossRef]
- Pacal, I.; Kılıcarslan, S. Deep learning-based approaches for robust classification of cervical cancer. Neural Comput. Appl. 2023, 35, 18813–18828. [Google Scholar] [CrossRef]
- Darici, M.B.; Dokur, Z.; Olmez, T. Pneumonia detection and classification using deep learning on chest X-ray images. Int. J. Intell. Syst. Appl. Eng. 2020, 8, 177–183. [Google Scholar] [CrossRef]
- Varshni, D.; Thakral, K.; Agarwal, L.; Nijhawan, R.; Mittal, A. Pneumonia detection using CNN based feature extraction. In Proceedings of the 2019 IEEE International Conference on Electrical, Computer and Communication Technologies (ICECCT), Coimbatore, India, 20–22 February 2019; pp. 1–7. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- CXR-PadChest. Available online: https://bimcv.cipf.es/bimcv-projects/padchest (accessed on 24 May 2025).
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, P.; Meng, J.; Jiang, X. Attention-based DenseNet for pneumonia classification. Irbm 2022, 43, 479–485. [Google Scholar] [CrossRef]
- Marengo, A.; Pagano, A.; Santamato, V. An efficient cardiovascular disease prediction model through AI-driven IOT Technology. Comput. Biol. Med. 2024, 183, 109330. [Google Scholar] [CrossRef]
- Santamato, V.; Tricase, C.; Faccilongo, N.; Iacoviello, M.; Pange, J.; Marengo, A. Machine learning for evaluating hospital mobility: An Italian case study. Appl. Sci. 2024, 14, 6016. [Google Scholar] [CrossRef]
- Thakur, S.; Goplani, Y.; Arora, S.; Upadhyay, R.; Sharma, G. Chest X-ray images based automated detection of pneumonia using transfer learning and CNN. In Proceedings of International Conference on Artificial Intelligence and Applications, New Delhi, India, 6–7 February 2020; Advances in Intelligent Systems and Computing. Springer: Berlin/Heidelberg, Germany, 2020; pp. 329–335. [Google Scholar] [CrossRef]
- Jain, R.; Nagrath, P.; Kataria, G.; Sirish Kaushik, V.; Jude Hemanth, D. Pneumonia detection in chest x-ray images using convolutional neural networks and transfer learning. Measurement 2020, 165, 108046. [Google Scholar] [CrossRef]
- Chhikara, P.; Singh, P.; Gupta, P.; Bhatia, T. Deep convolutional neural network with transfer learning for detecting pneumonia on chest X-rays. In Advances in Bioinformatics, Multimedia, and Electronics Circuits and Signals; Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2019; pp. 155–168. [Google Scholar] [CrossRef]
- Liang, G.; Zheng, L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Programs Biomed. 2020, 187, 104964. [Google Scholar] [CrossRef] [PubMed]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef]
- Bhatt, H.; Shah, M. A convolutional neural network ensemble model for pneumonia detection using chest X-ray images. Healthc. Anal. 2023, 3, 100176. [Google Scholar] [CrossRef]
- Mabrouk, A.; Díaz Redondo, R.P.; Dahou, A.; Abd Elaziz, M.; Kayed, M. Pneumonia detection on chest X-ray images using ensemble of deep convolutional Neural Networks. Appl. Sci. 2022, 12, 6448. [Google Scholar] [CrossRef]
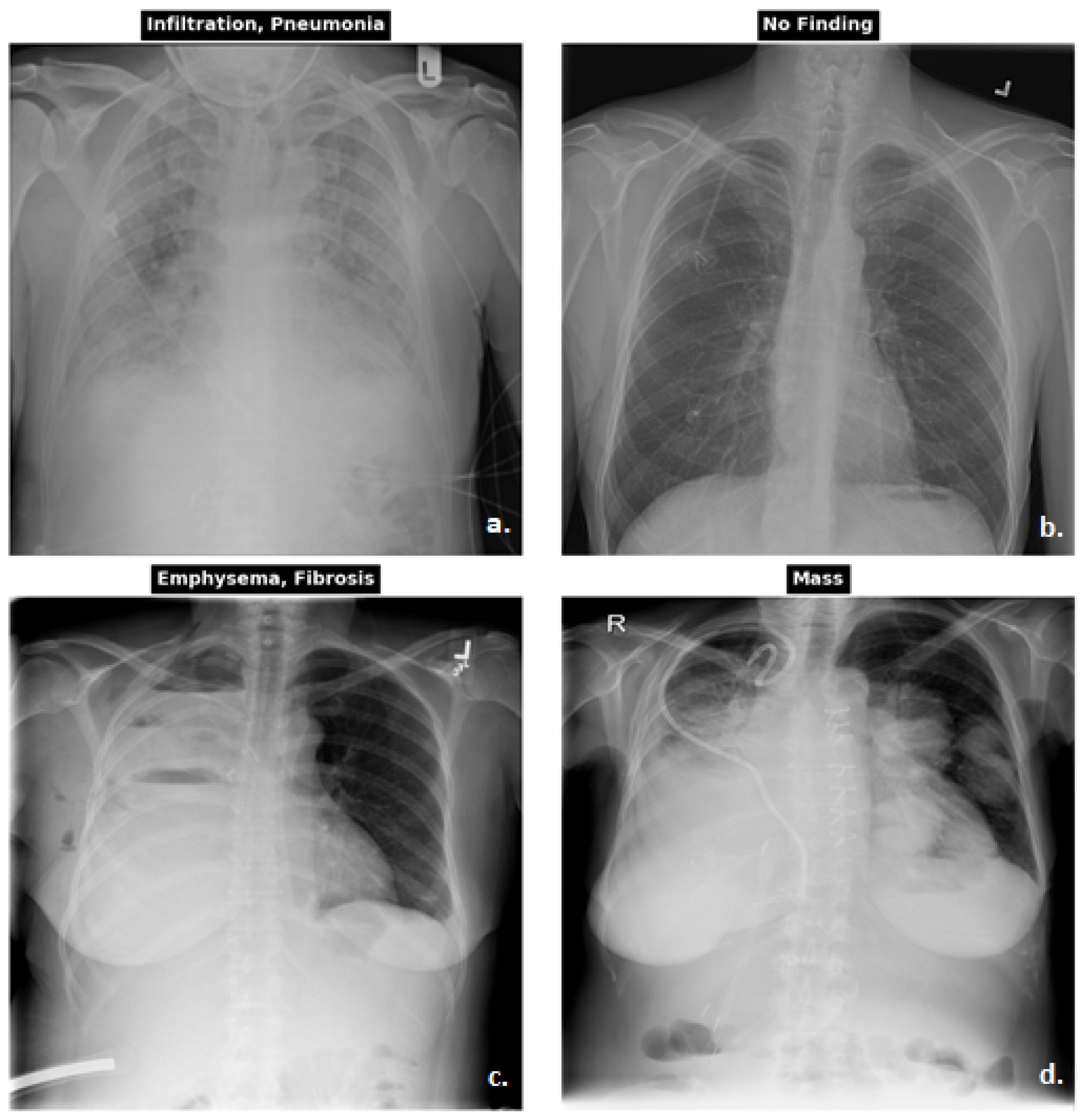

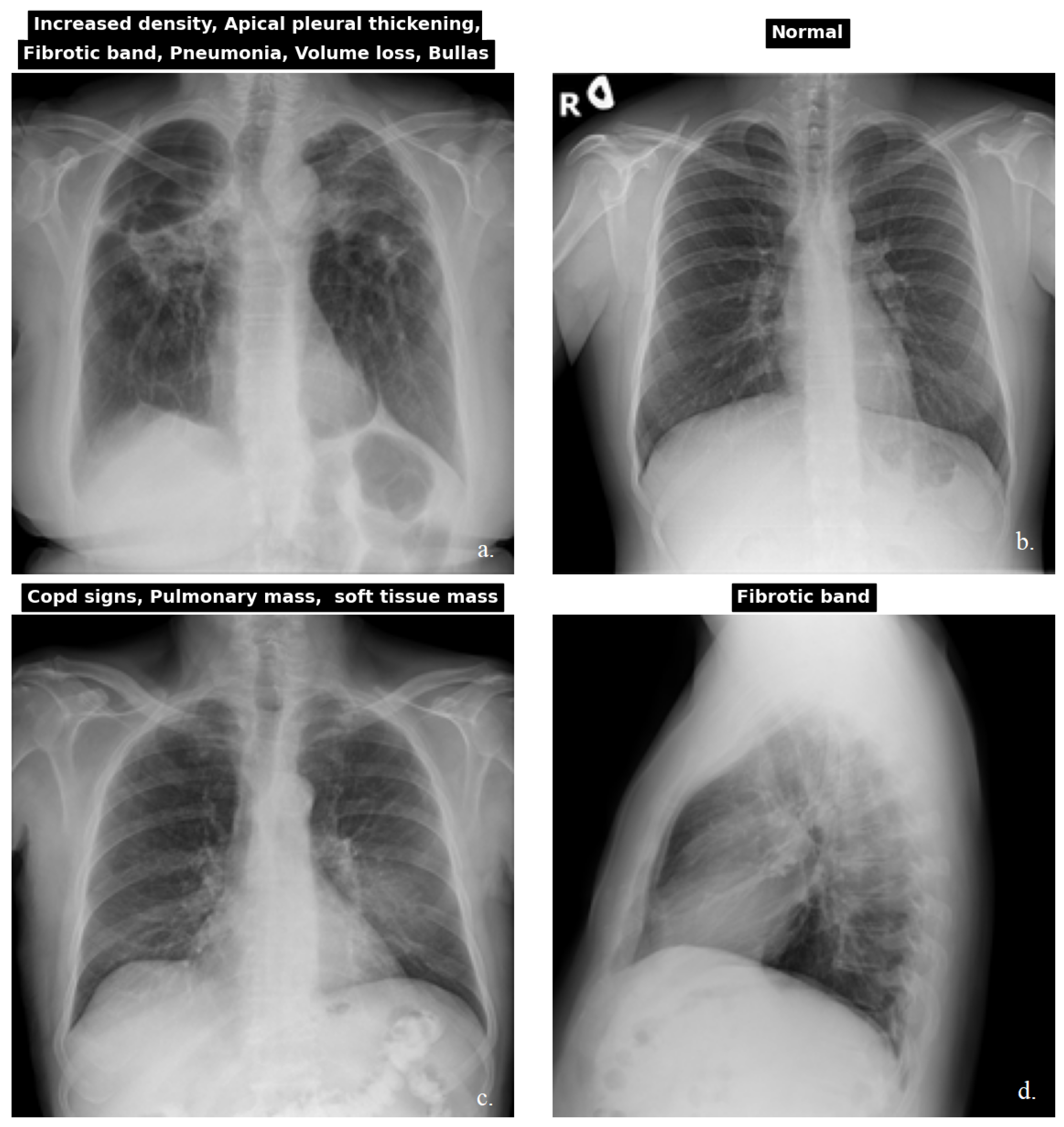




| CXR NIH | CheXpert | PadChest | Kaggle CXR Pneumonia | |
|---|---|---|---|---|
| Source | NIH Clinical Center, USA | Stanford University, Stanford ML Group | San Juan Hospital, Alicante, Spain | Guangzhou Women and Children’s Medical Center, China |
| Images | 112,120 frontal-view X-ray images from 30,805 patients | 224,316 chest radiographs from 65,240 patients | 160,000+ CXR images from 67,000 patients | 5863 PA-view pediatric chest X-rays (JPEG format) |
| Label Method | 14 thoracic pathologies mined using NLP from radiology reports | 14 observations (e.g., pneumonia, edema) with uncertainty- aware label extraction from reports | 174 radiological findings, 104 anatomical locations, 19 differential diagnoses | physician consensus with third-expert adjudication; bacterial and viral pneumonia classes included |
| Kaggle CXR Pneumonia | CXR NIH | PadChest | CheXpert | |
|---|---|---|---|---|
| Pneumonia | 4273 | 1431 | 8174 | 6047 |
| Normal | 1583 | 60,361 | 50,684 | 22,419 |
| Other Findings | - | 50,328 | 102,003 | 195,182 |
| Total | 5856 | 112,120 | 160,861 | 223,648 |
| Dataset | Pneumonia | Normal/Other Findings | Total Used | Quality Filtered | Notes |
|---|---|---|---|---|---|
| PadChest | 3000 | 3000 | 6000 | ~2000 | Downsampled for balance |
| CXR NIH | 1400 | 1400 | 2800 | ~0 | Nearly all pneumonia cases used |
| Kaggle CXR Pneumonia | 400 | 400 | 800 | ~56 | Small dataset, mostly retained |
| CheXpert | 20,200 | 20,200 | 40,400 | ~5600 | Majority source filtered and balanced |
| Total | 25,000 | 25,000 | 50,000 | ~7656 | After filtering + 1:1 balancing |
| Augmentation | Range/Setting | Clinical Purpose |
|---|---|---|
| CLAHE | clipLimit = 2.0, grid = 8 × 8 | Enhances local contrast in low-density lung regions, improving feature visibility. |
| Horizontal Flip | 50% chance | Simulates left/right orientation variable, compromising diagnostic integrity. |
| Translation | Max ±12 pixels (approximately 5%) | Models minor patient positioning variable image acquisition. |
| Rotation | Max ±5° | Preserves heart silhouette stability with slight orientation tolerance. |
| Zoom | 95–105% | Simulates small magnification changes, introducing anatomical distortion. |
| Training | Validation | Test | |
|---|---|---|---|
| Pneumonia | 20,000 | 2500 | 2500 |
| Normal | 18,000 | 2250 | 2250 |
| Other Findings | 2000 | 250 | 250 |
| Total | 40,000 | 5000 | 5000 |
| Model | Accuracy | Precision | Recall | F1-Score | Specificity | AUC |
|---|---|---|---|---|---|---|
| PELM (Proposed Model) | 0.96 | 0.99 | 0.91 | 0.95 | 0.91 | 0.91 |
| ResNet50 | 0.95 | 0.95 | 0.85 | 0.90 | 0.77 | 0.81 |
| DenseNet201 | 0.93 | 0.92 | 0.81 | 0.86 | 0.73 | 0.77 |
| MobileNetV2 | 0.93 | 0.94 | 0.83 | 0.88 | 0.76 | 0.88 |
| VGG16 | 0.93 | 0.95 | 0.85 | 0.90 | 0.77 | 0.81 |
| InceptionV3 | 0.92 | 0.97 | 0.87 | 0.92 | 0.83 | 0.85 |
| DenseNet121 | 0.92 | 0.97 | 0.88 | 0.92 | 0.82 | 0.85 |
| ViT | 0.92 | 0.97 | 0.88 | 0.92 | 0.82 | 0.85 |
| Authors | Classes | Methods | Image Number | Accuracy | Precision | Recall | F1-Score | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|---|
| (Thakur et al., 2020) [31] | Normal, Pneumonia | Pretrained VGG16 | 5856 | 0.90 | 0.98 | - | 0.93 | - | - |
| (Jain et al., 2020) [32] | Normal, Pneumonia | VGG19,ResNet50,VGG16, two customized models and Inception V3 | 5856 | 0.80 | - | - | - | - | - |
| (Chhikara et al., 2019) [33] | Normal, Pneumonia | Modified InceptionV3, Pre-processing | 5856 | 0.90 | 0.90 | 0.95 | 0.93 | - | 0.91 |
| (Liang et al., 2020) [34] | Normal, Pneumonia | CNN with residual DL architecture | 5856 | 0.90 | 0.89 | 0.96 | 0.92 | - | 0.95 |
| (Zech et al., 2018) [35] | Normal, Pneumonia | DenseNet121, ResNet50 | 158,323 | 0.95 | 0.93 | - | - | 0.71 | 0.93 |
| (Sharma et al., 2023) [5] | Normal, Pneumonia | VGG16 with CNN | 5856 | 0.92 | 0.94 | 0.93 | 0.93 | - | 0.97 |
| (Bhatt et al., 2023) [36] | Normal, Pneumonia | CNN, Ensemble | 5863 | 0.84 | 0.80 | 0.99 | 0.88 | - | 0.93 |
| (Mabrouk et al., 2023) [37] | Normal, Pneumonia | DenseNet169, Mo-bileNetV2 and Vision Trans-former | 5856 | 0.94 | 0.94 | 0.93 | 0.93 | - | - |
| (Wang et al., 2022) [28] | Normal, Pneumonia | Modified DenseNet | 5857 | 0.93 | 0.92 | 0.96 | 0.94 | - | - |
| PELM (Proposed Model) | Normal, Pneumonia | CNN, In-ceptionV3, VGG16, ResNet- 50 | 44,836 | 0.96 | 0.99 | 0.91 | 0.95 | 0.91 | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanar, E.; Hardalaç, F.; Ayturan, K. PELM: A Deep Learning Model for Early Detection of Pneumonia in Chest Radiography. Appl. Sci. 2025, 15, 6487. https://doi.org/10.3390/app15126487
Yanar E, Hardalaç F, Ayturan K. PELM: A Deep Learning Model for Early Detection of Pneumonia in Chest Radiography. Applied Sciences. 2025; 15(12):6487. https://doi.org/10.3390/app15126487
Chicago/Turabian StyleYanar, Erdem, Fırat Hardalaç, and Kubilay Ayturan. 2025. "PELM: A Deep Learning Model for Early Detection of Pneumonia in Chest Radiography" Applied Sciences 15, no. 12: 6487. https://doi.org/10.3390/app15126487
APA StyleYanar, E., Hardalaç, F., & Ayturan, K. (2025). PELM: A Deep Learning Model for Early Detection of Pneumonia in Chest Radiography. Applied Sciences, 15(12), 6487. https://doi.org/10.3390/app15126487







