Abstract
Proteins regulate various cellular processes and are of great biological interest. The protein search engine is a crucial tool in proteomics research, used to analyze high-throughput tandem mass spectrometry data and to identify protein sequence information. A core step in protein search engines is constructing sequence tag indexes and performing the rapid retrieval of protein databases. However, as the scale of protein sequence data continues to grow, traditional protein search engines face the dual challenges of the high storage cost of sequence tag indexes and low retrieval efficiency. To address these issues, we propose a sequence tag index scheme named STIP, which is based on an inverted index and compression techniques. Based on STIP, we design a peptide retrieval algorithm named STIP-Search. This algorithm utilizes the sequence tag index constructed by STIP for peptide sequence retrieval. STIP uses the greedy algorithm to partition the tag index into blocks; in this way, STIP can generate tag indexes for very large protein databases, such as NCBI-nr. Compared to the current four mainstream tag index generation algorithms used in Open-pFind, MODplus, TIIP and PIPI2, STIP has the lowest storage and time consumption. It utilizes delta encoding, index reduction, and dynamic bit width encoding to compress the tag index, reducing the storage cost by 76.2%. Compared to TIIP, which is currently the algorithm with the lowest time complexity, the time cost of the peptide sequence retrieval of STIP-Search is reduced by 8.94% to 23.31%.
1. Introduction
The sequence tag concept was introduced by Mann in 1994 [1]. It refers to the partial sequence of amino acids derived from a series of continuous fragment ions. Sequence tag searching combines the advantages of database search [2] and de novo sequencing [3] in mass spectrometry analysis. In protein identification, protein databases are frequently searched but updated infrequently. The construction of an index can significantly enhance retrieval speed [4]. The sequence tag index, abbreviated as tag index, is an indexing technique that reduces the search time complexity by establishing a mapping between sequence tags and corresponding sites in the protein database. In recent years, the tag index, as an important acceleration technique, has been adopted by many modern protein search engines.
The FM-index is an index scheme based on the Burrows–Wheeler transform [5]. To facilitate the implementation of the string matching algorithm, TagGraph uses the FM-index to construct an index for protein databases [6], and the index is referred to as the FM-indexed Protein. TagGraph performs sequence-splitting searches on de novo sequencing results obtained from PEAKS [7]. Then, it uses the FM-indexed Protein to search for sequence tags in the protein database. In 2024, PIPI2 continued to use this method, and it constructed tag indexes based on the FM-index to achieve the rapid retrieval of protein databases [8].
A tag index based on an inverted index is essentially a hash table where the substrings in protein sequences serve as keys and values contain information such as the position of the substring in the original protein sequence. The inverted index solves the memory expansion issue that occurs during the construction of an FM-index, making it more practical for applications. In a tag index, each sequence tag corresponds to an inverted list that consists of protein IDs and the starting positions of the sequence tag within the protein. Compared to traditional pattern-matching algorithms, this method significantly improves efficiency and optimizing time complexity from to , where N represents the length of the protein sequence and M represents the length of the sequence tag.
To enhance the efficiency of protein identification, an ion index design scheme based on an inverted index was proposed [9], which accelerates the protein search by constructing an ion mass index. In 2010, a protein sequence database organization algorithm based on the longest common prefix (ABLCP) was proposed [10]. This method eliminates redundant candidate peptide segments in the database, reducing the number of peptide spectrum matches. Many protein search engines have continued to use inverted indexes to construct ion mass indexes, such as Interrogator [11], pFind-alioth [12], and MSFragger [13]. In order to meet the demand for the rapid retrieval of sequence tags in protein sequence databases, Open-pFind [14] designs a tag index architecture using an inverted index model. Sequence tags are stored in the inverted index in a lexicographic order. The correspondence between tags and hash values is as follows:
where T represents a sequence tag, represents the i-th amino acid in the tag, is the function for calculating ASCII, and is the function that calculates the length of each tag.
In order to reduce space consumption during the protein identification process, MODplus [15] improves on MODa [16] by using a global site encoding strategy to construct tag indexes, concatenating protein sequences for storage, and reducing the index space by 25.79% to 38.79% on the original structure through a single site mapping. In 2022, the tag index TIIP was proposed, which supports the rapid recall of candidate peptides [17]. TIIP adds specific enzyme cleavage sites to the tag index and optimizes the recall time complexity of candidate peptides from to a constant level , where N represents the length of the protein sequence. Most protein search engines use sequence tag indexes to reduce the search space and lower the computational load [18,19,20].
Constructing indexes for protein databases often requires a large amount of memory. In order to solve the bottleneck problem of insufficient memory during the index generation process, Acquaye and co-authors proposed Tide [21], which detects the size of free memory in real time during the process of constructing peptide sequence indexes. Tide utilizes limited memory resources to construct peptide sequence indexes for large protein databases, removing redundant peptide sequences from the indexes and improving the efficiency of protein identification [22].
As the scale of protein data increases, tag index construction algorithms face dual technical challenges: on the one hand, the existing tag index structures have low storage efficiency; on the other hand, existing protein database retrieval algorithms require multiple traversals of protein sequences, resulting in long retrieval time [23,24]. In order to reduce the storage space of biological sequences, many sequence encoding methods have been proposed, such as BioCompress [25] and PatternHunter [26] for DNA sequences. DNA sequences have the characteristic of approximate repetition; Burrows–Wheeler transform (BWT) [27,28,29] and FM-index [30,31,32,33] are commonly used to reduce space consumption. In the field of proteomics, the existing methods for constructing the protein database index lack specialized algorithms for compressing the index, which is one of the urgent problems that need to be solved.
The currently proposed peptide sequence retrieval algorithms still suffer from the problems of high storage cost and time cost. To address the issue of high tag index storage costs, this paper improves the tag index structure and innovatively applies a hybrid compression strategy combining the delta encoding, data dimensionality reduction, and dynamic bit-width encoding into the tag index construction algorithm, thus reducing the storage cost of the tag index. To solve the problem of long retrieval time, this paper designs and implements the STIP-based protein database retrieval algorithm STIP-Search, providing a new technical path for protein database retrieval and aiding large-scale protein database searching.
We designed a protein index construction method to calculate specific enzyme cleavage site and residue mass, then store them in the protein index to avoid redundant calculation of relevant information during the retrieval stage. To solve the bottleneck problem of high memory requirement in the traditional tag index construction process, a tag index blocking algorithm is designed. Tags are grouped before tag index construction based on greedy algorithm, and the tag index is constructed in batches based on the grouping results. In order to reduce storage cost, we designed a tag index construction and compression algorithm. The sequence tags are located on the protein sequence using the tag index constructed by STIP.
2. Materials and Methods
Protein search engines face challenges with high storage costs and low retrieval efficiency of sequence tag indexes. To address these, we propose STIP, a sequence tag indexing scheme based on inverted indexing and compression algorithm, and design the STIP Search algorithm, which uses an index created by STIP for peptide sequence retrieval, significantly improving retrieval efficiency.
2.1. Data Sets
To quantify the compression efficiency of STIP on different species, this paper compares the storage cost occupied by the tag index across ten species protein databases. The ten protein databases were all downloaded from the UniProt official website on 9 November 2024, covering major evolutionary branches and containing complete annotation information. The protein database format is the standard FASTA. The species information is provided in Table 1. Figure 1 shows the distribution of different amino acids in the protein database, which was calculated by determining the frequency of each amino acid across all protein sequences in the dataset. This distribution provides insight into the overall composition of the dataset and highlights the prevalence of certain amino acids in the proteins. To verify the efficiency of protein identification, this paper uses the synthetic peptide data PXD009449, published by Kuster et al. in 2018 [24]. This dataset contains 25 MGF files and one protein database. Extract 5-mer sequence tags from the spectral files for tag localization and recall of peptide time cost testing.

Table 1.
The size of FASTA files, as well as the number of proteins, amino acids, and tags of protein database of 10 common species.
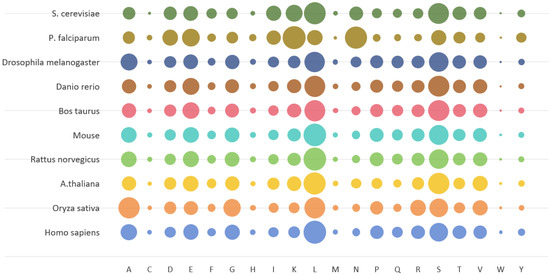
Figure 1.
Statistics on the proportion of different amino acids in a database of 10 common species. The 10 bubble colors in the figure correspond to different species categories, and the bubble size represents the frequency of occurrence of amino acids in different databases.
The NCBI-nr database is a comprehensive collection of non-redundant protein sequences curated by NCBI. It integrates data from multiple sources, including GenPept, SwissProt, and other sequence databases, to provide a unified resource for protein sequence analysis. There are 222,957,080 proteins in this protein database, and the database file size is 78.4 GB. We use this protein database to validate the performance of STIP in constructing a tag index for large-scale protein databases.
2.2. STIP
Mainstream protein identification algorithms utilize indexing to accelerate the process of sequence and protein matching, but there are still challenges. We propose a protein index and tag index construction algorithm, STIP. In this algorithm, an index partitioning algorithm is used to address the issue of high memory consumption during tag index construction, and a compressed tag indexing algorithm is proposed to address the issue of high storage index space resource consumption.
2.2.1. Overall Workflow of STIP
The overall workflow of STIP (sequence tag index for protein) is shown in Figure 2. First, the protein database is preprocessed to generate a protein index, and the greedy algorithm is used to partition the tag indexes. The tag index is partitioned using the greedy algorithm. This step is aimed at supporting the construction of tag indexes for large-scale protein databases and adapting to machines with varying memory capacities. STIP is a memory-adaptive tag index architecture. In the second step, the Rabin–Karp algorithm is used to construct a tag index of user-specified length. In the index entries of STIP, it records the protein ID and starting position of the tag in the protein sequence. In the third step, a three-level compression strategy is applied to compress the tag index, reducing its storage cost while ensuring that the search time complexity remains . Finally, the tag index is stored on disk, supporting reuse during searches.
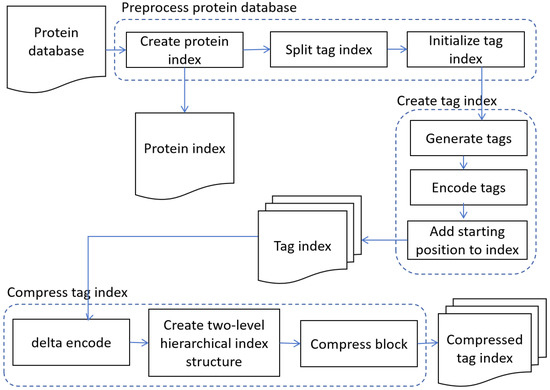
Figure 2.
The overall workflow of STIP algorithm. STIP first preprocesses the protein database and then constructs the tag index for the protein database, and finally compresses the tag index.
STIP designs an efficient tag index architecture using an inverted index, partitions the tag indexes with a greedy algorithm, optimizes the tag index construction process, and combines a compression algorithm with the tag index construction algorithm to improve storage efficiency.
2.2.2. Preprocessing of Protein Database
STIP allows users to specify the maximum amount of memory to be used for constructing the tag index. Before generating the tag index, STIP predicts the size of the tag index and partitions the tag index into blocks. The partitioning results are used to optimize the tag index construction process, allowing for batch construction of the tag index.
Firstly, traverse the input protein database and construct two basic index structures: the protein index and the enzyme digestion site index. The protein index stores all protein information in the database, including protein ID, protein name, detailed description, and amino acid sequences. The enzyme digestion site index records the specific enzyme digestion sites for each protein. The construction process of the protein index and specific enzymatic digestion site index is shown in Figure 3.
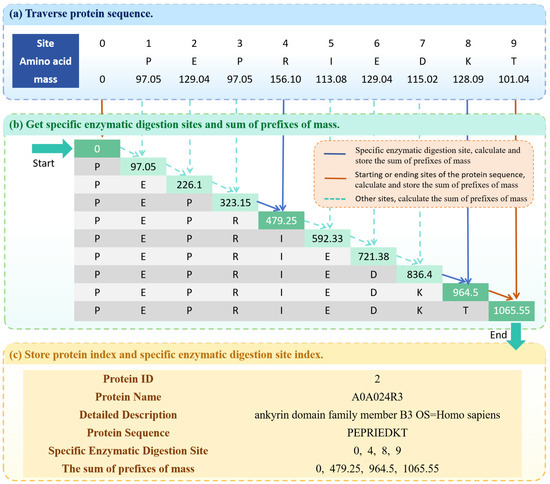
Figure 3.
The overview of generating protein index. (a) Traverse protein sequence. (b) Obtain specific enzymatic digestion sites and sum of prefixes of mass. (c) Store protein index and specific enzymatic digestion site index.
To generate the tag index for large-scale databases under memory constraints, STIP uses a greedy algorithm to partition the tag index into blocks. We generated protein databases of different sizes using random sampling from NCBI-nr as input data to construct tag indexes and calculate space consumption. We predict the index space based on the fitting results of the protein database and index space consumption. The specific process is as follows. The database is traversed to generate a set of t-mer tags, which are tags composed of k amino acids, and calculate the frequency of occurrence for each t-mer tag. For each tag , the storage cost S of its tag index can be estimated based on the frequency of its appearance in the protein database; the formula is as follows:
where represents the frequency of appearance of tags in protein databases. The tags are sorted in lexicographical order to form an ordered tag set. Based on the memory threshold M set by users, the greedy algorithm is applied to determine the start and end tags for each tag index block. Finally, the tag index blocks are initialized according to the partitioning results.
2.2.3. Generate Sequence Tag Index
Firstly, for a given protein database , where M is the total number of proteins, and represents the i-th protein sequence. For sequence , with length , all subsequences of length k are generated using the Rabin–Karp algorithm, and these subsequences are the tags. For example, for sequence , where , , five 5-mer tags can be generated: , PTIDE, TIDEK }. During the index construction process, any sequence containing anomalous characters is detected and flagged to prevent any impact on data integrity in order to ensure the system can handle such scenarios correctly.
The second step is tag encoding. The traditional method encodes the 26 uppercase letters, while the improved method proposed in this paper encodes only the characters corresponding to the 20 amino acids that make up proteins, making it more suitable for practical applications. There are 20 common amino acids, and in protein databases, protein sequences are generally composed of 20 types of uppercase letters. In the NCBI-nr dataset, besides the 20 common amino acid types corresponding to characters, other characters account for less than 0.03%. For any t-mer tag , the corresponding key in the tag index is as follows:
Each sequence tag in the tag index has a unique key corresponding to this encoding strategy. Since the number of keys in the hash table is , covering all possible t-mer tags, the time complexity of searching is .
Generate the t-mer tag index using tags obtained by the Rabin–Karp algorithm. Sort all t-mer tags in ascending lexicographic order to form an ordered set T_sorted , where N is the number of t-mer tags in the protein database. Record the protein ID and starting position of the tag, then generate the tag index. Its formula is as follows:
where represents the unique identifier of the i -th protein in the database, Positioni represents the starting position of the tag in the protein sequence, and m represents the number of occurrences of the tag in the protein database.
2.2.4. Compress Sequence Tag Index
In order to optimize the storage efficiency of large-scale protein databases, STIP designed a compression algorithm specifically for tag indexing, using a three-layer compression mechanism to reduce the storage cost of the tag index. The compression methods employed in this paper are lossless compression techniques, including delta encoding, index dimensionality reduction, and dynamic bit-width encoding.
The first step of compression is delta encoding, which stores the difference between adjacent numbers in the index rather than storing the numbers themselves. This is highly effective for storing sequences of incrementing numbers. Since the tag index generation process traverses the protein information in sequence, for each inverted list of the tag index, the protein IDs exhibit a monotonically increasing property. Therefore, the differential encoding strategy is applied to compress the protein IDs. For example, if the protein IDs in the tag index are [2, 5, 8, 10], after applying delta encoding, only the differences [2, 3, 3, 2] need to be stored, and these differences can be represented using fewer bits. The specific implementation process is shown in Formula (5). In each inverted list, the first protein ID is used as the base value and remains unchanged, while the differences between each pair of adjacent protein IDs are calculated and stored in the tag index instead of the actual protein IDs. The formulas are as follows:
where represents the unique identifier of the protein. The differential encoding strategy not only compresses the data scale within linear time complexity but also transforms the protein IDs into low-value data. Based on statistics from the human proteomics dataset downloaded from the UniProt official website, which includes human protein sequences, this conversion can improve the compression effect of subsequent dynamic bit-width encoding, compressing the original 32-bit integer protein ID into an average of 6.2 bits per value.
The second step is to generate a two-level hierarchical index which consists of a tag index array. The tag index obtained in the previous step is reduced to one dimension. The starting positions of different types of tag in the one-dimensional index form the auxiliary index. This step involves the linear reconstruction of the high-dimensional tag index. During retrieval, the auxiliary index allows the tag positioning information in the tag index array to be accessed in time complexity.
The third step is to use dynamic bit-width encoding to compress the tag index array and the auxiliary index. First, the protein IDs and starting positions in the tag index are stored separately, generating two tag index arrays: one for the protein IDs and one for the starting positions. STIP utilizes the local similarity of the data within the tag index array to divide the array into fixed-size blocks, and each block by default stores 256 data entries. Each block is compressed independently, using dynamic bit-width encoding to store different values. Specifically, STIP calculates the minimum number of bits (denoted as b) required for 80% of the values in each block. Regular data is encoded using b bits, while values exceeding are treated as outliers. A patch mechanism is used to store these outliers in a separate index, recording the position of each outlier. Extra space is used to store the values of the outliers, preventing the bit-width for regular data from being inflated due to the presence of outliers.
After compression, the tag index consists of three parts: the protein ID array, the starting position array, and the auxiliary index. The compressed tag index is then stored on the disk, making it easy to retrieve directly during search. The core advantage of STIP is that it significantly reduces storage cost through delta encoding, dimensionality reduction, and dynamic bit-width compression, while maintaining constant-time retrieval complexity.
2.3. STIP-Search
The scale of high-throughput mass spectrometry data is rapidly increasing, and existing peptide retrieval algorithms are encountering the challenge of long retrieval times. This paper proposes a peptide sequence retrieval algorithm, named STIP-Search, which utilizes both protein and tag indexes to narrow the search scope, thereby achieving fast tag localization and improved peptide sequence retrieval efficiency.
2.3.1. Overall Workflow of STIP-Search
STIP-Search is a peptide retrieval algorithm using a tag index. Figure 4 is the overall workflow of the STIP-Search algorithm. In this example, the quality window is set to [−350 Da, 350 Da], and the threshold for missed enzymatic digestion sites is set to 2. First, the tag index is used to quickly locate the tags onto the protein sequences based on the protein IDs and starting positions. In protein databases, the N-terminus typically represents the left end of the sequence, while the C-terminus represents the right end of the sequence. Starting from the N-terminus and C-terminus of the tag, specific enzymatic digestion sites are traversed towards both sides, resulting in candidate peptide sequences within the quality window. Then, the number of missed cleavage sites is checked, and peptides exceeding the missed cleavage threshold are removed, ultimately yielding 5 peptide sequences.
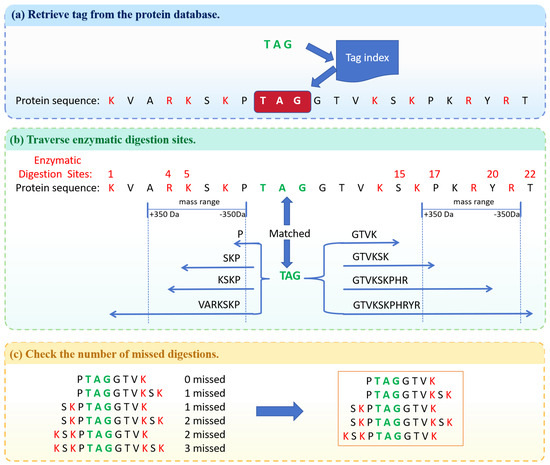
Figure 4.
The overall workflow of the STIP-Search algorithm. (a) Retrieve tag from the protein database. (b) Traverse enzymatic digestion sites. (c) Check the number of missed digestions.
2.3.2. Retrieve Tag Positioning Information from Tag Index
In traditional protein retrieval algorithms based on the inverted index, a single tag index can only support the retrieval of tags of a fixed length. When retrieving tags of different lengths, corresponding tags must be reconstructed. To overcome the limitations of single-length tag retrieval in traditional methods, we designed a variable-length sequence tag retrieval algorithm that is compatible with the STIP algorithm, enabling efficient retrieval of t-mer tags using a k-mer tag index.
When the length of the tag to be retrieved is the same as the index length (), the corresponding tag can be directly retrieved using the tag index. The auxiliary index is used to obtain the range where the tag information is stored. Within the tag index, the tags are arranged in lexicographical order, and the information corresponding to each tag is stored together. During the search, only the two endpoints of the index range need to be obtained:
When the length of the tag to be retrieved is less than the length of the tag index , traditional methods use tag sequence extension to retrieve tags. Specifically, by enumerating amino acid sequences, t-mer tags are supplemented with k-mer tags, and then the k-mer tag index can be used for retrieval. This method of expanding the amino acid sequence has resulted in a significant amount of time cost. STIP Search utilizes the orderliness of tag index to design a more efficient retrieval strategy. Through the two-level hierarchical index structure, the starting and ending positions of each tag in the index are determined to quickly retrieve tags. STIP Search utilizes the orderliness and continuity of data within the tag index to locate tags within time complexity.
2.3.3. Retrieve Peptides from the Protein Database
Once the tag is located within the protein sequence, the corresponding peptides are retrieved based on the tag’s positioning information. The process of recalling specific enzyme cleavage candidate peptides is as follows. Firstly, all specific enzymatic digestion site information is read from the protein index, and the tag is used as the center to traverse the specific enzymatic digestion sites on both sides. The residue mass information in the specific enzymatic digestion site index is used to quickly calculate the sum of the continuous residue masses. If the peptide segment corresponding to the current site meets the user’s set number of missing cleavage sites and the precursor mass window, then the site is the endpoint of the candidate peptide sequence. When the sum of residue masses exceeds the quality tolerance range of search or the number of missing enzymatic digestion sites reaches the upper limit, the traversal is ended and each specific enzyme cleavage candidate peptide is retrieved. This algorithm transforms the task of traversing the entire protein sequence into a task of only traversing specific enzymatic digestion sites and residue masses by utilizing an index of these digestion sites, thus reducing retrieval time.
After obtaining specific enzymatic digestion candidate peptides through STIP, non-specific enzymatic digestion candidate peptides are obtained by detecting the mass of the tag’s two ends. The sequence tags are mapped back to the candidate peptides of the specific enzymatic digestion. The tags closest to the N-terminus and C-terminus of the peptide sequence are called the N-terminal judgment tag and the C-terminal judgment tag, respectively. For the offset quality of the N-terminal judgment tag and C-terminal judgment tag, we first check whether it is lower than the minimum amino acid quality. If it is lower than the minimum amino acid quality, the corresponding site is considered as a candidate peptide endpoint, and the amino acids beyond the break point are removed to obtain a non-specific candidate peptide sequence. The retrieval process of non-specific enzymatic digested peptides is shown in Figure 5:
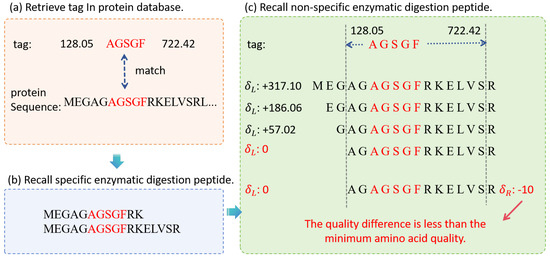
Figure 5.
The overall workflow of non-specific peptide retrieval. (a) Retrieve tag in protein database. (b) Recall specific enzymatic digestion peptide. (c) Recall non-specific enzymatic digestion peptide.
3. Results
To evaluate the storage and time cost of STIP, this study compares with four mainstream tag index generation and retrieval methods used in Open-pFind, MODplus, TIIP and PIPI2. Since Open-pFind, MODplus, and PIPI2 do not provide open source code or independent tag index generation modules, this paper reconstructs the index generation and retrieval algorithms based on the method frameworks described in their original publications. This paper strictly follows the optimal parameter settings recommended by the authors and uses Python 3.9 to implement the core architecture.
3.1. Performance of Tag Index Generation Algorithm
In this experiment, five tag index generation algorithms are compared. Protein databases of 10 species are selected for testing. Table 2 shows the storage cost constructed by different methods on protein databases of ten species. STIP has the lowest storage cost of indexes, achieving optimal space performance. Compared with Open-pFind, STIP reduces index space consumption by 58.88% to 78.18%. Compared with MODplus, STIP reduces storage cost by 38.51% to 74.09%. Compared with TIIP, STIP reduces the index space consumption by 73.87% to 83.16%. The storage cost of the 5-mer tag index generated by the five methods is shown in Figure 6. Compared with PIPI2, the index space consumption constructed by STIP on the Danio rerio dataset increased by 2.38%. The reason for this result is that the Danio rerio dataset has fewer tag types and a smaller database size. On the other nine species datasets, the spatial performance of STIP is significantly higher than that of PIPI2, verifying that STIP has obvious advantages in protein databases with larger scale.

Table 2.
Comparison of storage cost of index constructed by Open-pFind, MODplus, TIIP, PIPI2, and STIP on protein databases of ten species.
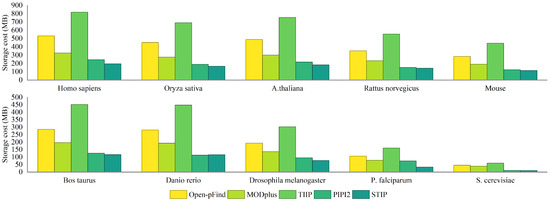
Figure 6.
Storage cost of tag index generated by Open-pFind, MODplus, TIIP, PIPI2, and STIP on FASTA files from ten species.
To test the performance of STIP in constructing indexes on protein databases of different sizes, we used a random sampling method to select proteins from the NCBI nr dataset and construct protein databases of different sizes. We set the tag length from 1 to 6, and the space consumption of the index is shown in Figure 7.
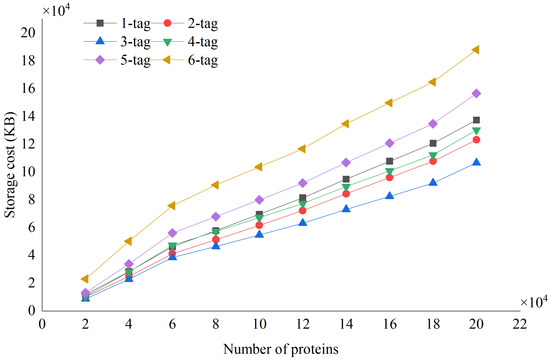
Figure 7.
The index storage cost of STIP on 10 protein databases of different sizes, with 6 colors representing the indexes corresponding to tags from the 1-mer tag to the 6-mer tag.
Analyzing the experimental results, it can be found that the index space corresponding to 3-mer tags is the smallest. The larger index space corresponding to shorter tags is because short tags often appear more frequently in protein databases, so it needs to be recorded in tag indexes. The larger index space corresponding to longer tags is due to the greater number of types of long tags. The 3-mer tag, composed of 20 amino acids, has 8000 types, while the 6-mer tag has as many as types, resulting in a higher consumption of tag index space for the latter. In addition, STIP successfully constructed an index for the NCBI nr protein database, with an index storage space of 179.8 GB and a compression rate of 24.8%. This is a task that existing protein database index construction algorithms cannot accomplish, and this result verifies the ability of STIP on large-scale protein databases.
To investigate the robustness and reliability of the STIP algorithm, we have conducted a series of ablation experiments. The experimental results are shown in Figure 8. After removing tag sequence encoding, index dimensionality reduction, delta encoding, and dynamic bit width encoding from STIP, compared with the tag index construction algorithm that removes all compression methods, they, respectively, save 15.36%, 20.09%, 58.77%, and 62.16% of storage cost. Among them, the optimization effect of index dimensionality reduction is not significant on large-scale protein databases, but the optimization effect on spatial performance is significant on small-scale databases. On the Homo sapiens dataset, index dimensionality reduction reduces space consumption by 1.71%. On the S.cerevisiae dataset, this method reduced space consumption by 56.43%. The reason for this result is that small-scale protein databases generate fewer types of sequence tag, and there are many empty index items in the index structure based on the inverted index. So, it causes a lot of additional space consumption. After dimensionality reduction of the index, the invalid index items are removed, greatly reducing space consumption.
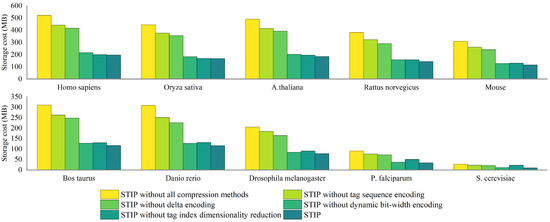
Figure 8.
Ablation experiment on FASTA files from ten species. Storage cost of index generation algorithms after removing tag sequence encoding, index dimensionality reduction, delta encoding, dynamic bit width encoding, and all compression methods from STIP.
3.2. Performance of Tag Index Partitioning Algorithm
Before performing tag index block partitioning, it is necessary to pre-calculate the storage space requirements of each index block based on the number distribution of different tag types. This experiment is based on NCBI-nr. To generate a test dataset, we obtain sub-databases with gradient tag quantities through systematic sampling; the 5-mer tag index is generated, and its storage space occupancy is measured. The purpose of this experiment is to verify the effectiveness of estimating index storage cost using the number of tags. Figure 9 shows the actual storage cost and the relative error between the actual and theoretical index space for different tag counts. The error is caused by the fact that the size of the tag index is not only related to the number of tags, but also to the numerical size of the information within the tag index.
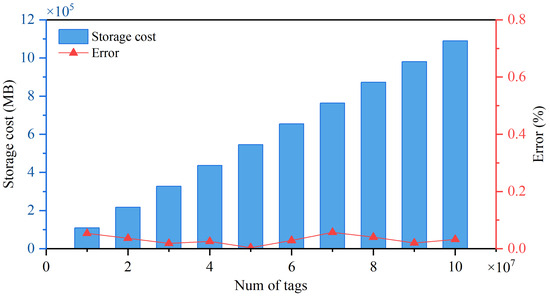
Figure 9.
Performance of tag index storage cost estimations on 10 protein databases of different sizes.
The results show that there is a significant linear correlation between storage space occupancy and tag quantity, and the trend of change closely matches the theoretical prediction, with the maximum relative error being 0.065%. The experimental results effectively validated the mathematical validity and engineering applicability of estimating tag index storage cost using the number of tags, providing reliable index space prediction results for tag index partitioning algorithms.
In addition to the tag index partitioning method based on greedy algorithm used in this article, there is also a method that divides the tag index by evenly dividing the number of tag types. In order to facilitate differentiation in subsequent comparative experiments, we call the former Greedy Load Balancing Partitioning (GLP) and the latter Uniform Lexicographical Partitioning (ULP). To verify indexing block performance of GLP for large-scale databases, we selected the NCBI-nr database as a test database. Since 5-mer tags are the most used in practical applications, this experiment generates a 5-mer tag index and compares it with an equal dictionary sequence partitioning (ULP). ULP partitions the tag index by evenly distributing the types of tag within the index.
Block utilization rate refers to the degree of effective usage of a storage block. It measures how much of the available space within a block is actually used to store data. A higher block utilization rate indicates that a larger portion of the block is being used, while a lower rate suggests that the block has a significant amount of unused space, potentially leading to inefficiencies. The formula is as follows:
where x is the amount of data actually stored in the block, and M is the total available capacity of the block.
Figure 10 shows the performance of ULP and GLP. The GLP strategy fully utilizes memory resources while adhering to the set memory threshold. A total of 95.83% of the index blocks in the GLP strategy achieve a saturation rate of 99.98% or higher. Additionally, the GLP strategy generates 24 blocks, which is a 54% reduction compared to the equal dictionary sequence partitioning strategy. This results in more concentrated index information, making subsequent retrieval easier. The different partitioning strategies may affect retrieval time, primarily because multiple calls to the tag index blocks are made during retrieval. Figure 11 shows the I/O operations during the retrieval process for the label indices generated using the two strategies. The GLP strategy reduces operations I/O by 54% compared to ULP. This experiment validates the necessity of the GLP algorithm in large-scale index generation and provides empirical support for developing memory-adaptive index architectures.
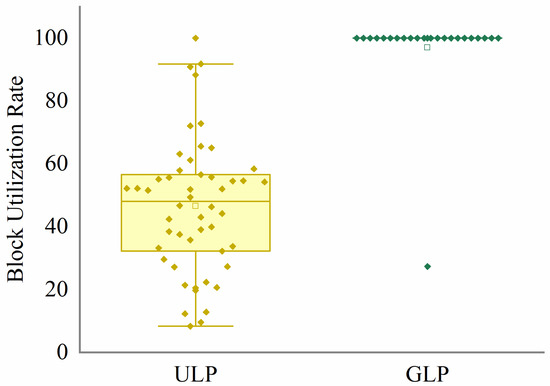
Figure 10.
Block utilization rate comparison of ULP and GLP on NCBI-nr. The points represent the block saturation rate for each index, and the boxes represent the interquartile range of ULP and GLP.
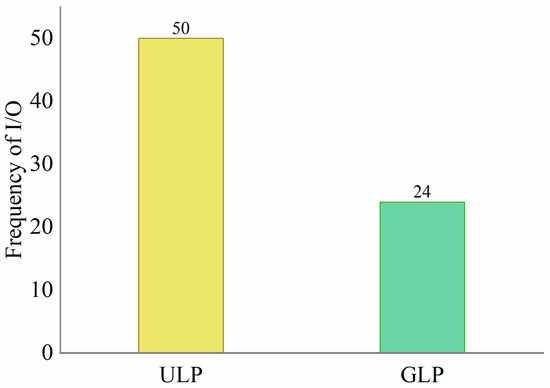
Figure 11.
Block utilization rate comparison of ULP and GLP on NCBI-nr.
3.3. Performance of STIP-Search Algorithm
Time cost is an important metric for evaluating the retrieval performance of protein sequence database search engines. We measured the retrieval time for recalling candidate peptides using 3-mer, 4-mer, and 5-mer tags. The experimental results are shown in Figure 12.
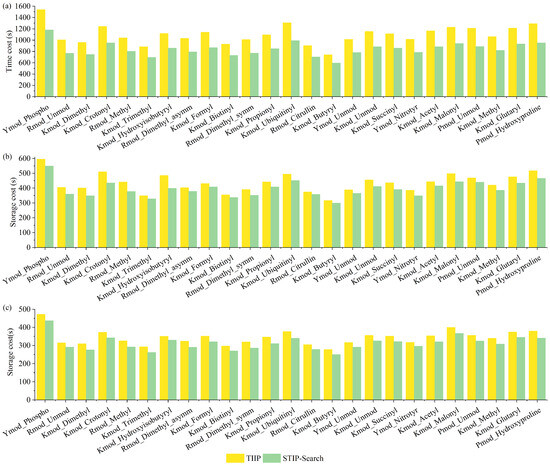
Figure 12.
Time cost of retrieving peptide sequence on PXD009449. (a) Time cost comparisons of peptide identification using 3-mer tag between TIIP and STIP-Search on PXD009449. (b) Time cost comparisons of peptide identification using 4-mer tag between TIIP and STIP-Search on PXD009449. (c) Time cost comparisons of peptide identification using 5-mer tag between TIIP and STIP-Search on PXD009449.
Currently, algorithms that use variable-length tags to recall candidate peptides include PIPI2 and TIIP. TIIP accelerates the retrieval process by precomputing specific enzymatic digestion sites, resulting in the lowest time complexity. In this experiment, TIIP was chosen for comparison to test the time performance of STIP-Search in recalling candidate peptides with variable length tags. The results demonstrate that, compared to TIIP, STIP-Search reduces the peptide recall time by 14.01% to 23.31%, validating the speed advantage of STIP-Search in practical applications of variable length tag retrieval.
4. Discussion
This paper proposes a new tag index generation algorithm named STIP, which combines tag index construction with data compression techniques. While maintaining high retrieval speed, it successfully reduces the storage cost of tag indexes by 76.2%. STIP uses a greedy algorithm to partition the tag index, optimizing the index construction process and providing a new technical approach for constructing the tag indexes in large-scale protein databases.
Compared with mainstream protein data index construction algorithms such as Open-pFind, MODplus, TIIP, and PIPI2, STIP combines the index construction algorithm and the data compression algorithm, and constructs the index on datasets of ten common species with the lowest storage cost. In addition, STIP can construct a protein index and tag index for large-scale protein databases such as NCBI-nr, which is a task that other algorithms participating in the testing cannot complete, verifying the applicability of this algorithm in a large-scale protein database.
Under the condition of not exceeding the set memory threshold, STIP, using greedy load-balanced partitioning, reduces the frequency of I/O by 52% and improves block saturation by 108.33% compared to ULP. The experimental validation on PXD009449 datasets demonstrates the superior search speed of STIP-Search. Tag indexing is widely used in the field of proteomics, and to address the issues of large index size and high memory demands, compressing tag index will be an important research direction.
5. Conclusions
In order to reduce the storage cost during peptide sequence retrieval, this paper proposes a protein index algorithm and a tag index construction algorithm. Firstly, a protein index construction method is designed to calculate the specific enzyme cleavage site and residue masses, then store them in the protein index to avoid redundant calculation of relevant information during the retrieval stage. Secondly, to solve the bottleneck problem of high memory requirement in the traditional tag index construction process, a tag index blocking algorithm is designed. Tags are grouped before tag index construction based on greedy algorithm, and the tag index is constructed in batches based on the grouping results. Finally, in order to reduce storage cost, we designed a tag index construction and compression algorithm. It utilizes delta encoding, index reduction, and dynamic bit width encoding to compress the tag index, reducing storage cost by 76.2%. Additionally, we design a protein sequence database retrieval algorithm, STIP-Search, based on the tag index constructed by STIP. STIP-Search achieves rapid tag localization and peptide sequence identification.
However, as data size continues to grow, further improving the compression ratio while maintaining high retrieval efficiency remains a key challenge. Future research could focus on developing more efficient compression algorithms, particularly those based on deep learning techniques. We believe that the methods presented in this paper will play a significant role in protein identification, driving further progress of proteomics.
Author Contributions
Conceptualization, H.W.; methodology, H.W., P.Z. and X.X.; software, X.X.; validation, H.W. and X.X.; formal analysis, H.W., P.Z. and X.X.; investigation, X.X. and Y.F.; resources, H.W.; data curation, X.X., Y.F., D.Z. and L.Y.; writing—original draft preparation, X.X.; writing—review and editing, H.W.; visualization, X.X.; supervision, H.W.; project administration, H.W.; funding acquisition, H.W. All authors have read and agreed to the published version of the manuscript.
Funding
Support Program for Outstanding Youth Innovation Teams in Higher Educational Institutions of Shandong Province (2019KJN048).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets can be downloaded from UniProt (link: https://www.uniprot.org/ (accessed on 9 November 2024)).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation; in the writing of the manuscript; or in the decision to publish the results.
References
- Mann, M.; Wilm, M. Error-Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal. Chem. 1994, 66, 4390–4399. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; Hoopmann, M.R.; Jahan, T.A.; Egertson, J.D.; Noble, W.S.; MacCoss, M.J. A deeper look into Comet–implementation and features. J. Am. Soc. Mass Spectrom. 2015, 26, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Xu, S.; Zhang, X.; Ling, T.; Dong, N.; Ouyang, W.; Gao, Z.; Chang, C.; Sun, S. ContraNovo: A Contrastive Learning Approach to Enhance De Novo Peptide Sequencing. AAAI 2024, 38, 144–152. [Google Scholar] [CrossRef]
- Zhou, R.; Zhao, H.; Zhong, J.; Duan, G. PepGPL: A Multi-Task Framework for Identifying Peptide-Protein Interactions and Corresponding Binding Residues. In Proceedings of the 15th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, New York, NY, USA, 16 December 2024. [Google Scholar]
- Lu, B.; Chen, T. A suffix tree approach to the interpretation of tandem mass spectra: Applications to peptides of non-specific digestion and post-translational modifications. Bioinformatics 2003, 19, ii113–ii121. [Google Scholar] [CrossRef]
- Devabhaktuni, A.; Lin, S.; Zhang, L.; Swaminathan, K.; Gonzalez, C.G.; Olsson, N.; Pearlman, S.M.; Rawson, K.; Elias, J.E. TagGraph reveals vast protein modification landscapes from large tandem mass spectrometry datasets. Nat. Biotechnol. 2019, 37, 469–479. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Lai, S.; Zhao, P.; Zhou, C.; Li, N.; Yu, W. PIPI2: Sensitive Tag-Based Database Search to Identify Peptides with Multiple Post-translational Modifications. J. Proteome Res. 2024, 23, 1960–1969. [Google Scholar] [CrossRef]
- Li, D.; Gao, W.; Ling, C.X.; Wang, X.; Sun, R.; He, S. IndexToolkit: An open source toolbox to index protein databases for high-throughput proteomics. Bioinformatics 2006, 22, 2572–2573. [Google Scholar] [CrossRef][Green Version]
- Zhou, C.; Chi, H.; Wang, L.-H.; Li, Y.; Wu, Y.-J.; Fu, Y.; Sun, R.-X.; He, S.-M. Speeding up tandem mass spectrometry-based database searching by longest common prefix. BMC Bioinform. 2010, 11, 577. [Google Scholar] [CrossRef]
- Li, Y.; Chi, H.; Wang, L.-H.; Wang, H.-P.; Fu, Y.; Yuan, Z.-F.; Li, S.-J.; Liu, Y.-S.; Sun, R.-X.; Zeng, R.; et al. Speeding up tandem mass spectrometry based database searching by peptide and spectrum indexing. Rapid Commun. Mass Spectrom. 2010, 24, 807–814. [Google Scholar] [CrossRef]
- Chi, H.; He, K.; Yang, B.; Chen, Z.; Sun, R.-X.; Fan, S.-B.; Zhang, K.; Liu, C.; Yuan, Z.-F.; Wang, Q.-H.; et al. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J. Proteom. 2015, 125, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Liu, C.; Yang, H.; Zeng, W.-F.; Wu, L.; Zhou, W.-J.; Wang, R.-M.; Niu, X.-N.; Ding, Y.-H.; Zhang, Y.; et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018, 36, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Na, S.; Kim, J.; Paek, E. MODplus: Robust and Unrestrictive Identification of Post-Translational Modifications Using Mass Spectrometry. Anal. Chem. 2019, 91, 11324–11333. [Google Scholar] [CrossRef]
- Na, S.; Bandeira, N.; Paek, E. Fast Multi-blind Modification Search through Tandem Mass Spectrometry. Mol. Cell. Proteom. 2012, 11, M111.010199. [Google Scholar] [CrossRef]
- Zhou, P.; Hou, X.; Wang, H. A New Tag Index Scheme Enables Fast Peptide Retrieval for Protein Identification. J. Comput. Chem. 2022, 10, 14–23. [Google Scholar] [CrossRef]
- d’Acierno, A. IsAProteinDB: An Indexed Database of Trypsinized Proteins for Fast Peptide Mass Fingerprinting. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 1195–1201. [Google Scholar] [CrossRef]
- Maabreh, M.; Gupta, A.; Saeed, F. A parallel peptide indexer and decoy generator for crux tide using OpenMP. In Proceedings of the 2016 International Conference on High Performance Computing & Simulation (HPCS), Innsbruck, Austria, 18–22 July 2016; IEEE: Innsbruck, Austria, 2016. [Google Scholar]
- Haseeb, M.; Saeed, F. Efficient Shared Peak Counting in Database Peptide Search Using Compact Data Structure for Fragment-Ion Index. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: San Diego, CA, USA, 2019. [Google Scholar]
- Acquaye, N.A.F.L.; Kertesz-Farkas, A.; Noble, W.S. Efficient indexing of peptides for database search using Tide. J. Proteome Res. 2023, 22, 577–584. [Google Scholar] [CrossRef]
- Diament, B.J.; Noble, W.S. Faster SEQUEST searching for peptide identification from tandem mass spectra. J. Proteome Res. 2011, 10, 3871–3879. [Google Scholar] [CrossRef]
- Kim, H.; Mirdita, M.; Steinegger, M. Foldcomp: A library and format for compressing and indexing large protein structure sets. Bioinformatics 2023, 39, btad153. [Google Scholar] [CrossRef]
- Zolg, D.P.; Wilhelm, M.; Schmidt, T.; Médard, G.; Zerweck, J.; Knaute, T.; Wenschuh, H.; Reimer, U.; Schnatbaum, K.; Kuster, B. ProteomeTools: Systematic Characterization of 21 Post-translational Protein Modifications by Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Using Synthetic Peptides. Mol. Cell. Proteom. 2018, 17, 1850–1863. [Google Scholar] [CrossRef] [PubMed]
- Grumbach, S.; Tahi, F. Compression of DNA sequences. In Proceedings of the Proceedings DCC93: Data Compression Conference, Snowbird, UT, USA, 30 March–1 April 1993; IEEE: Snowbird, UT, USA, 1993; pp. 340–350. [Google Scholar]
- Ma, B.; Tromp, J.; Li, M. PatternHunter: Faster and more sensitive homology search. Bioinformatics 2002, 18, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.J.; Cox, A.J.; Rosone, G. Lightweight algorithms for constructing and inverting the BWT of string collections. Theor. Comput. Sci. 2013, 483, 134–148. [Google Scholar] [CrossRef]
- Navarro, G. Technical perspective: The compression power of the BWT. Commun. ACM 2022, 65, 90. [Google Scholar] [CrossRef]
- Hang, Z.; Pan, X.; Sun, J.; Teng, L.; Jiang, J. Application of improved LZW compression algorithm based on CZ-BWT in power message. Comput. Integr. Manuf. Syst. 2024, 30, 1570–1574. [Google Scholar]
- Hong, A.; Oliva, M.; Köppl, D.; Bannai, H.; Boucher, C.; Gagie, T. Pfp-fm: An accelerated FM-index. Algorithms Mol. Biol. 2024, 19, 15–28. [Google Scholar] [CrossRef]
- Anderson, T.; Wheeler, T.J. An optimized FM-index library for nucleotide and amino acid search. Algorithms Mol. Biol. 2021, 16, 25–42. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y. Accelerating spliced alignment of long RNA sequencing reads using parallel maximal exact match retrieval. Comput. Biol. Med. 2024, 175, 108542. [Google Scholar] [CrossRef]
- Herruzo, J.M.; Fernandez, I.; González-Navarro, S.; Plata, O. Enabling fast and energy-efficient FM-index exact matching using processing-near-memory. J. Supercomput. 2021, 77, 10226–10251. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).