Microbiome and Phageome: Key Factors in Host Organism Function and Disease Prevention in the Context of Microbiome Transplants
Abstract
1. Introduction
2. Diversity of Gut Prokaryotes and the Principles of Microbiota Transfer
2.1. Challenges in Defining the Structure of Gut Microbiota
2.2. Enterotypes of Human and Animal Guts
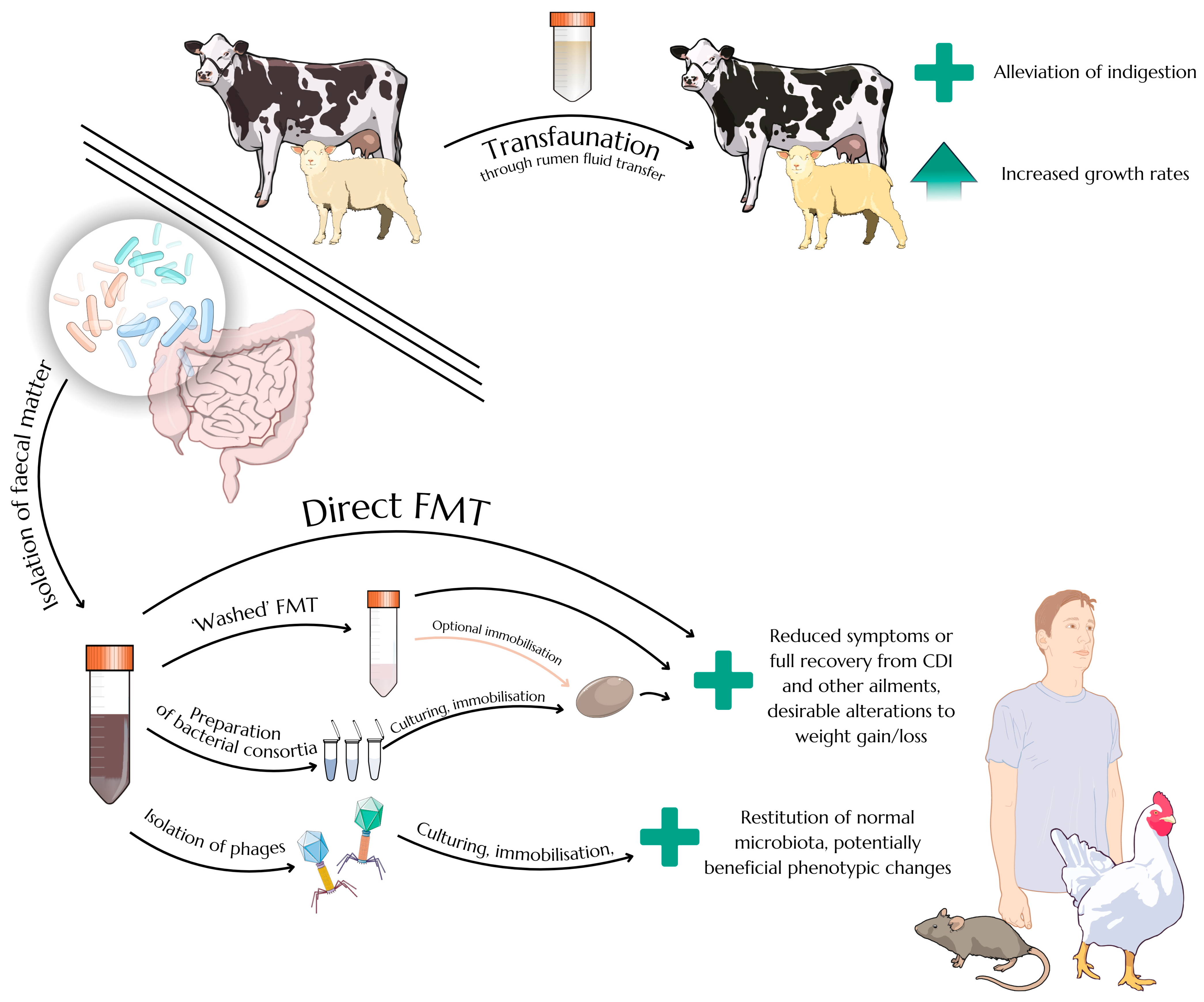
2.3. Faecal Microbiota Transplantation as an Efficient Therapeutic Approach
3. Intestinal Bacteriophages and Faecal Virome Transplants
3.1. The Phageome of the Gut and the Surprising Prevalence of crAssviridae
3.2. Advantages and Potential Benefits of Faecal Virome Transplants
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of Open Access Journals |
| FMT | Faecal microbiota transplant |
| FVT | Faecal virome transplant |
| CDI | Clostridioides difficile infection |
| IBS | Irritable bowel syndrome |
References
- Simon, J.-C.; Marchesi, J.R.; Mougel, C.; Selosse, M.-A. Host-Microbiota Interactions: From Holobiont Theory to Analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef]
- Pandey, K.; Naik, S.; Vakil, B. Probiotics, Prebiotics and Synbiotics—A Review. J. Food Sci. Technol. 2015, 52, 7577. [Google Scholar] [CrossRef]
- Hocquart, M.; Lagier, J.-C.; Cassir, N.; Saidani, N.; Eldin, C.; Kerbaj, J.; Delord, M.; Valles, C.; Brouqui, P.; Raoult, D.; et al. Early Fecal Microbiota Transplantation Improves Survival in Severe Clostridium difficile Infections. Clin. Infect. Dis. 2018, 66, 645. [Google Scholar] [CrossRef]
- Wu, D.; Liang, S.; Du, X.; Xiao, J.; Feng, H.; Ren, Z.; Yang, X.; Yang, X. Effects of Fecal Microbiota Transplantation and Fecal Virome Transplantation on LPS-Induced Intestinal Injury in Broilers. Poult. Sci. 2024, 103, 103316. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Romaní-Pérez, M.; Líebana-García, R.; Flor-Duro, A.; Bonillo-Jiménez, D.; Bullich-Vilarrubias, C.; Olivares, M.; Sanz, Y. Obesity and the Gut Microbiota: Implications of Neuroendocrine and Immune Signaling. FEBS J. 2024, 292, 1397–1420. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Dewhirst, F.E.; Olsen, I.; Fraser, G.J. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and Related Bacteria. J. Bacteriol. 1994, 176, 725. [Google Scholar] [CrossRef]
- Wallace, R.J.; McKain, N.; Broderick, G.A.; Rode, L.M.; Walker, N.D.; Newbold, C.J.; Kopecny, J. Peptidases of the Rumen Bacterium, Prevotella ruminicola. Anaerobe 1997, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-Producing Human Gut Symbiont, Clostridium butyricum, and Its Role in Health and Disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef]
- Kanai, T.; Mikami, Y.; Hayashi, A. A Breakthrough in Probiotics: Clostridium butyricum Regulates Gut Homeostasis and Anti-Inflammatory Response in Inflammatory Bowel Disease. J. Gastroenterol. 2015, 50, 928. [Google Scholar] [CrossRef]
- Hill, K.K.; Xie, G.; Foley, B.T.; Smith, T.J.; Munk, A.C.; Bruce, D.; Smith, L.A.; Brettin, T.S.; Detter, J.C. Recombination and Insertion Events Involving the Botulinum Neurotoxin Complex Genes in Clostridium botulinum Types A, B, E and F and Clostridium butyricum Type E Strains. BMC Biol. 2009, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Dykes, J.K.; Lúquez, C.; Raphael, B.H.; McCroskey, L.; Maslanka, S.E. Laboratory Investigation of the First Case of Botulism Caused by Clostridium butyricum Type e Toxin in the United States. J. Clin. Microbiol. 2015, 53, 3363. [Google Scholar] [CrossRef]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile Infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211. [Google Scholar] [CrossRef]
- Gerding, D.N.; Meyer, T.; Lee, C.; Cohen, S.H.; Murthy, U.K.; Poirier, A.; Van Schooneveld, T.C.; Pardi, D.S.; Ramos, A.; Barron, M.A.; et al. Administration of Spores of Nontoxigenic Clostridium difficile Strain M3 for Prevention of Recurrent C. difficile Infection: A Randomized Clinical Trial. JAMA 2015, 313, 1719. [Google Scholar] [CrossRef]
- Dobzhansky, T. A Critique of the Species Concept in Biology. Philos. Sci. 1935, 2, 344. [Google Scholar] [CrossRef]
- Novick, A.; Doolittle, W.F. “Species” without Species. Stud. Hist. Philos. Sci. 2021, 87, 72. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Treuren, W.V.; Fischer, C.R.; Merrill, B.D.; DeFelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A Metabolomics Pipeline for Mechanistic Interrogation of the Gut Microbiome. Nature 2021, 595, 415. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between Body Mass Index and Firmicutes/Bacteroidetes Ratio in an Adult Ukrainian Population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Tai, W.-C.; Liang, C.-M.; Wu, C.-K.; Tsai, M.-C.; Hu, W.-H.; Huang, P.-Y.; Chen, C.-H.; Kuo, Y.-H.; Yao, C.-C.; et al. Alternations of the Gut Microbiota and the Firmicutes/Bacteroidetes Ratio after Biologic Treatment in Inflammatory Bowel Disease. J. Microbiol. Immunol. Infect. 2024, 58, 62–69. [Google Scholar] [CrossRef]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic Distribution of Three Pathways for Propionate Production within the Human Gut Microbiota. ISME J. 2014, 8, 1323. [Google Scholar] [CrossRef] [PubMed]
- Fettig, N.M.; Osborne, L.C. Direct and Indirect Effects of Microbiota-Derived Metabolites on Neuroinflammation in Multiple Sclerosis. Microbes Infect. 2021, 23, 104814. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174. [Google Scholar] [CrossRef]
- Jeffery, I.B.; Claesson, M.J.; O’Toole, P.W.; Shanahan, F. Categorization of the Gut Microbiota: Enterotypes or Gradients? Nat. Rev. Microbiol. 2012, 10, 591. [Google Scholar] [CrossRef]
- Knights, D.; Ward, T.L.; McKinlay, C.E.; Miller, H.; Gonzalez, A.; McDonald, D.; Knight, R. Rethinking “Enterotypes”. Cell Host Microbe 2014, 16, 433. [Google Scholar] [CrossRef]
- Di Pierro, F. A Possible Perspective about the Compositional Models, Evolution, and Clinical Meaning of Human Enterotypes. Microorganisms 2021, 9, 2341. [Google Scholar] [CrossRef]
- Bartsch, M.; Hahn, A.; Berkemeyer, S. Bridging the Gap from Enterotypes to Personalized Dietary Recommendations: A Metabolomics Perspective on Microbiome Research. Metabolites 2023, 13, 1182. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559. [Google Scholar] [CrossRef]
- Hayashi, H.; Shibata, K.; Sakamoto, M.; Tomita, S.; Benno, Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2007, 57, 941. [Google Scholar] [CrossRef]
- Ley, R.E. Prevotella in the Gut: Choose Carefully. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Laghi, L.; Gobbetti, M.; Ercolini, D. Unusual Sub-Genus Associations of Faecal Prevotella and Bacteroides with Specific Dietary Patterns. Microbiome 2016, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2017, 3, 8. [Google Scholar] [CrossRef]
- Liang, C.; Tseng, H.-C.; Chen, H.-M.; Wang, W.-C.; Chiu, C.-M.; Chang, J.-Y.; Lu, K.-Y.; Weng, S.-L.; Chang, T.-H.; Chang, C.-H.; et al. Diversity and Enterotype in Gut Bacterial Community of Adults in Taiwan. BMC Genomics 2017, 18, 932. [Google Scholar] [CrossRef]
- Fu, T.; Pan, L.; Shang, Q.; Yu, G. Fermentation of Alginate and its Derivatives by Different Enterotypes of Human Gut Microbiota: Towards Personalized Nutrition Using Enterotype-Specific Dietary Fibers. Int. J. Biol. Macromol. 2021, 183, 1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, W.; Wang, C.; Wang, L.; He, T.; Hu, H.; Song, J.; Cui, C.; Qiao, J.; Qing, L.; et al. Enterotype Bacteroides Is Associated with a High Risk in Patients with Diabetes: A Pilot Study. J. Diabet. Res. 2020, 2020, 6047145. [Google Scholar] [CrossRef]
- Lepage, P.; Seksik, P.; Sutren, M.; de la Cochetière, M.-F.; Jian, R.; Marteau, P.; Doré, J. Biodiversity of the Mucosa-Associated Microbiota Is Stable Along the Distal Digestive Tract in Healthy Individuals and Patients With IBD. Inflamm. Bowel Dis. 2005, 11, 473. [Google Scholar] [CrossRef]
- Wang, J.; Linnenbrink, M.; Künzel, S.; Fernandes, R.; Nadeau, M.-J.; Rosenstiel, P.; Baines, J.F. Dietary History Contributes to Enterotype-like Clustering and Functional Metagenomic Content in the Intestinal Microbiome of Wild Mice. Proc. Natl. Acad. Sci. USA 2014, 111, E2703. [Google Scholar] [CrossRef]
- Li, X.; Liang, S.; Xia, Z.; Qu, J.; Liu, H.; Liu, C.; Yang, H.; Wang, J.; Madsen, L.; Hou, Y.; et al. Establishment of a Macaca Fascicularis Gut Microbiome Gene Catalog and Comparison with the Human, Pig, and Mouse Gut Microbiomes. GigaScience 2018, 7, 100. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A Catalog of the Mouse Gut Metagenome. Nat. Biotechnol. 2015, 33, 1103. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.-J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic Network Analysis Applied to Pig Gut Microbiota Identifies an Ecosystem Structure Linked with Growth Traits. ISME J. 2016, 10, 2973. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Huang, X.; Fang, S.; He, M.; Zhao, Y.; Wu, Z.; Yang, M.; Zhang, Z.; Chen, C.; Huang, L. Unraveling the Fecal Microbiota and Metagenomic Functional Capacity Associated with Feed Efficiency in Pigs. Front. Microbiol. 2017, 8, 1555. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Tiezzi, F.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Maltecca, C. Host Contributes to Longitudinal Diversity of Fecal Microbiota in Swine Selected for Lean Growth. Microbiome 2018, 6, 4. [Google Scholar] [CrossRef]
- Le Sciellour, M.; Renaudeau, D.; Zemb, O. Longitudinal Analysis of the Microbiota Composition and Enterotypes of Pigs from Post-Weaning to Finishing. Microorganisms 2019, 7, 622. [Google Scholar] [CrossRef]
- Yuan, Z.; Yan, W.; Wen, C.; Zheng, J.; Yang, N.; Sun, C. Enterotype Identification and Its Influence on Regulating the Duodenum Metabolism in Chickens. Poult. Sci. 2020, 99, 1515. [Google Scholar] [CrossRef]
- Fan, Q.; Xu, Y.; Xiao, Y.; Yang, C.; Lyu, W.; Yang, H. Linking Growth Performance and Carcass Traits with Enterotypes in Muscovy Ducks. Anim. Biosci. 2024, 37, 1213. [Google Scholar] [CrossRef] [PubMed]
- Lilli, G.; Sirot, C.; Campbell, H.; Brophy, D.; Graham, C.T.; George, I.F. Geographic Origin and Host’s Phylogeny Are Predictors of the Gut Mucosal Microbiota Diversity and Composition in Mediterranean Scorpionfishes (Scorpaena spp.). Front. Mar. Sci. 2023, 10, 1286706. [Google Scholar] [CrossRef]
- Zhang, B.; Xiao, J.; Liu, H.; Zhai, D.; Wang, Y.; Liu, S.; Xiong, F.; Xia, M. Vertical Habitat Preferences Shape the Fish Gut Microbiota in a Shallow Lake. Front. Microbiol. 2024, 15, 1341303. [Google Scholar] [CrossRef]
- Moeller, A.H.; Ochman, H. Microbiomes Are True to Type. Proc. Natl. Acad. Sci. USA 2014, 111, 9372. [Google Scholar] [CrossRef]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool Consistency Is Strongly Associated with Gut Microbiota Richness and Composition, Enterotypes and Bacterial Growth Rates. Gut 2016, 65, 57. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, W.; Shi, Y.; Fan, Z.; Ji, G. Should We Standardize the 1700-Year-Old Fecal Microbiota Transplantation? Off. J. Am. Coll. Gastroenterol.|ACG 2012, 107, 1755. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Seo, G.S. Fecal Microbiota Transplantation: Is It Safe? Clin. Endosc. 2021, 54, 157. [Google Scholar] [CrossRef]
- Marcella, C.; Cui, B.; Kelly, C.R.; Ianiro, G.; Cammarota, G.; Zhang, F. Systematic Review: The Global Incidence of Faecal Microbiota Transplantation-Related Adverse Events from 2000 to 2020. Aliment. Pharmacol. Ther. 2021, 53, 33. [Google Scholar] [CrossRef]
- Hensgens, M.P.M.; Goorhuis, A.; Dekkers, O.M.; van Benthem, B.H.B.; Kuijper, E.J. All-Cause and Disease-Specific Mortality in Hospitalized Patients With Clostridium difficile Infection: A Multicenter Cohort Study. Clin. Infect. Dis. 2013, 56, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.W.; Dharmaratne, P.; Wong, S.; Hawkey, P.; Chan, P.; Ip, M. Revisiting the Donor Screening Protocol of Faecal Microbiota Transplantation (FMT): A Systematic Review. Gut 2024, 73, 1029. [Google Scholar] [CrossRef]
- Konturek, P.C.; Dieterich, W.; Neurath, M.; Zopf, Y. Successful Therapy of Clostridium difficile Infection with Fecal Microbiota Transplantation. Gastroenterology 2017, 152, S341. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Widlak, M.; Bhala, N.; Moore, D.; Price, M.; Sharma, N.; Iqbal, T.H. Systematic Review with Meta-Analysis: The Efficacy of Faecal Microbiota Transplantation for the Treatment of Recurrent and Refractory Clostridium difficile Infection. Aliment. Pharmacol. Ther. 2017, 46, 479. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Maida, M.; Burisch, J.; Simonelli, C.; Hold, G.; Ventimiglia, M.; Gasbarrini, A.; Cammarota, G. Efficacy of Different Faecal Microbiota Transplantation Protocols for Clostridium difficile Infection: A Systematic Review and Meta-Analysis. United Eur. Gastroenterol. J. 2018, 6, 1232. [Google Scholar] [CrossRef]
- El-Salhy, M.; Hatlebakk, J.G.; Gilja, O.H.; Kristoffersen, A.B.; Hausken, T. Efficacy of Faecal Microbiota Transplantation for Patients with Irritable Bowel Syndrome in a Randomised, Double-Blind, Placebo-Controlled Study. Gut 2020, 69, 859. [Google Scholar] [CrossRef]
- He, Z.; Cui, B.-T.; Zhang, T.; Li, P.; Long, C.-Y.; Ji, G.-Z.; Zhang, F.-M. Fecal Microbiota Transplantation Cured Epilepsy in a Case with Crohn’s Disease: The First Report. World J. Gastroenterol. 2017, 23, 3565. [Google Scholar] [CrossRef]
- Cai, T.; Shi, X.; Yuan, L.; Tang, D.; Wang, F. Fecal Microbiota Transplantation in an Elderly Patient with Mental Depression. Int. Psychogeriatr. 2019, 31, 1525. [Google Scholar] [CrossRef]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy Alters Gut Ecosystem and Improves Gastrointestinal and Autism Symptoms: An Open-Label Study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- DePeters, E.J.; George, L.W. Rumen Transfaunation. Immunol. Lett. 2014, 162, 69. [Google Scholar] [CrossRef]
- Steiner, S.; Linhart, N.; Neidl, A.; Baumgartner, W.; Tichy, A.; Wittek, T. Evaluation of the Therapeutic Efficacy of Rumen Transfaunation. J. Anim. Physiol. Anim. Nutr. 2020, 104, 56. [Google Scholar] [CrossRef]
- Elrawy, A.; Mohammed, A.; Sabra, M.S.; Mahmoud, U.; Darwish, M. Impact of Rumen Juice Transfaunation on Behavioral Activities, Performance Parameters and Kidney Function in Fattening Lambs. Assiut Vet. Med. J. 2023, 69, 74. [Google Scholar] [CrossRef]
- McKinney, C.A.; Bedenice, D.; Pacheco, A.P.; Oliveira, B.C.M.; Paradis, M.-R.; Mazan, M.; Widmer, G. Assessment of Clinical and Microbiota Responses to Fecal Microbial Transplantation in Adult Horses with Diarrhea. PLoS ONE 2021, 16, e0244381. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Geng, S.; Li, Y.; Cheng, S.; Fu, X.; Yue, X.; Han, X. Exogenous Fecal Microbiota Transplantation from Local Adult Pigs to Crossbred Newborn Piglets. Front. Microbiol. 2018, 8, 02663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rodríguez, F.; Navas, M.J.; Costa-Hurtado, M.; Almagro, V.; Bosch-Camós, L.; López, E.; Cuadrado, R.; Accensi, F.; Pina-Pedrero, S.; et al. Fecal Microbiota Transplantation from Warthog to Pig Confirms the Influence of the Gut Microbiota on African Swine Fever Susceptibility. Sci. Rep. 2020, 10, 17605. [Google Scholar] [CrossRef]
- McCormack, U.M.; Curião, T.; Wilkinson, T.; Metzler-Zebeli, B.U.; Reyer, H.; Ryan, T.; Calderon-Diaz, J.A.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; et al. Fecal Microbiota Transplantation in Gestating Sows and Neonatal Offspring Alters Lifetime Intestinal Microbiota and Growth in Offspring. mSystems 2018, 3, e00134-17. [Google Scholar] [CrossRef]
- Zhang, T.; Lu, G.; Zhao, Z.; Liu, Y.; Shen, Q.; Li, P.; Chen, Y.; Yin, H.; Wang, H.; Marcella, C.; et al. Washed Microbiota Transplantation vs. Manual Fecal Microbiota Transplantation: Clinical Findings, Animal Studies and in Vitro Screening. Protein Cell 2020, 11, 251. [Google Scholar] [CrossRef]
- Varga, A.; Kocsis, B.; Sipos, D.; Kása, P.; Vigvári, S.; Pál, S.; Dembrovszky, F.; Farkas, K.; Péterfi, Z. How to Apply FMT More Effectively, Conveniently and Flexible—A Comparison of FMT Methods. Front. Cell Infect. Microbiol. 2021, 11, 657320. [Google Scholar] [CrossRef]
- Petrof, E.O.; Gloor, G.B.; Vanner, S.J.; Weese, S.J.; Carter, D.; Daigneault, M.C.; Brown, E.M.; Schroeter, K.; Allen-Vercoe, E. Stool Substitute Transplant Therapy for the Eradication of Clostridium difficile Infection: ‘RePOOPulating’ the Gut. Microbiome 2013, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Sausset, R.; Petit, M.A.; Gaboriau-Routhiau, V.; De Paepe, M. New Insights into Intestinal Phages. Mucosal Immunol. 2020, 13, 205. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Zhou, Y.; Zhao, G.; Bauer, I.K.; Droit, L.; Ndao, I.M.; Warner, B.B.; Tarr, P.I.; Wang, D.; Holtz, L.R. Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants. Nat. Med. 2015, 21, 1228. [Google Scholar] [CrossRef] [PubMed]
- Minot, S.; Bryson, A.; Chehoud, C.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Rapid Evolution of the Human Gut Virome. Proc. Natl. Acad. Sci. USA 2013, 110, 12450. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in Host-Associated Microbial Communities. NPJ Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Obeng, N.; Pratama, A.A.; Elsas, J.D. van The Significance of Mutualistic Phages for Bacterial Ecology and Evolution. Trends Microbiol. 2016, 24, 440. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy Human Gut Phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.Z.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A Highly Abundant Bacteriophage Discovered in the Unknown Sequences of Human Faecal Metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Guerin, E.; Shkoporov, A.; Stockdale, S.R.; Clooney, A.G.; Ryan, F.J.; Sutton, T.D.S.; Draper, L.A.; Gonzalez-Tortuero, E.; Ross, R.P.; Hill, C. Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut. Cell Host & Microbe 2018, 24, 653. [Google Scholar] [CrossRef]
- Chen, J.; Gissendanner, C.R.; Tikhe, C.V.; Li, H.-F.; Sun, Q.; Husseneder, C. Genomics and Geographic Diversity of Bacteriophages Associated With Endosymbionts in the Guts of Workers and Alates of Coptotermes Species (Blattodea: Rhinotermitidae). Front. Ecol. Evol. 2022, 10, 881538. [Google Scholar] [CrossRef]
- Piedade, G.J.; Schön, M.E.; Lood, C.; Fofanov, M.V.; Wesdorp, E.M.; Biggs, T.E.G.; Wu, L.; Bolhuis, H.; Fischer, M.G.; Yutin, N.; et al. Seasonal Dynamics and Diversity of Antarctic Marine Viruses Reveal a Novel Viral Seascape. Nat. Commun. 2024, 15, 9192. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 Represents the Most Abundant Bacteriophage Family in the Human Gut and Infects Bacteroides Intestinalis. Nat. Commun. 2018, 9, 4781. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Stephens, N.; Hueston, C.; Seymour, S.; Hryckowian, A.J.; Scholz, D.; Ross, R.P.; Hill, C. Long-Term Persistence of crAss-like Phage crAss001 Is Associated with Phase Variation in Bacteroides intestinalis. BMC Biol. 2021, 19, 163. [Google Scholar] [CrossRef]
- Porter, N.T.; Hryckowian, A.J.; Merrill, B.D.; Fuentes, J.J.; Gardner, J.O.; Glowacki, R.W.P.; Singh, S.; Crawford, R.D.; Snitkin, E.S.; Sonnenburg, J.L.; et al. Phase-Variable Capsular Polysaccharides and Lipoproteins Modify Bacteriophage Susceptibility in Bacteroides thetaiotaomicron. Nat. Microbiol. 2020, 5, 1170. [Google Scholar] [CrossRef]
- Guerin, E.; Shkoporov, A.N.; Stockdale, S.R.; Comas, J.C.; Khokhlova, E.V.; Clooney, A.G.; Daly, K.M.; Draper, L.A.; Stephens, N.; Scholz, D.; et al. Isolation and Characterisation of ΦcrAss002, a crAss-like Phage from the Human Gut That Infects Bacteroides Xylanisolvens. Microbiome 2021, 9, 89. [Google Scholar] [CrossRef]
- Schmidtke, D.T.; Hickey, A.S.; Liachko, I.; Sherlock, G.; Bhatt, A.S. Analysis and Culturing of the Prototypic crAssphage Reveals a Phage-Plasmid Lifestyle. bioRxiv 2024. [Google Scholar] [CrossRef]
- Ravin, N.V. N15: The Linear Phage–Plasmid. Plasmid 2011, 65, 102. [Google Scholar] [CrossRef]
- Pfeifer, E.; Moura de Sousa, J.A.; Touchon, M.; Rocha, E.P.C. Bacteria Have Numerous Distinctive Groups of Phage–Plasmids with Conserved Phage and Variable Plasmid Gene Repertoires. Nucleic Acids Res. 2021, 49, 2655. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Bonnin, R.A.; Rocha, E.P.C. Phage-Plasmids Spread Antibiotic Resistance Genes through Infection and Lysogenic Conversion. mBio 2022, 13, e01851-22. [Google Scholar] [CrossRef] [PubMed]
- Mullet, J.; Zhang, L.; Pruden, A.; Brown, C.L. Phage-Plasmid Hybrids Are Found Throughout Diverse Environments and Encode Niche-Specific Functional Traits. bioRxiv 2024. [Google Scholar] [CrossRef]
- Rasmussen, T.S.; Mentzel, C.M.J.; Danielsen, M.R.; Jakobsen, R.R.; Zachariassen, L.S.F.; Castro Mejia, J.L.; Brunse, A.; Hansen, L.H.; Hansen, C.H.F.; Hansen, A.K.; et al. Fecal Virome Transfer Improves Proliferation of Commensal Gut Akkermansia Muciniphila and Unexpectedly Enhances the Fertility Rate in Laboratory Mice. Gut Microbes 2023, 15, 2208504. [Google Scholar] [CrossRef]
- Borin, J.M.; Liu, R.; Wang, Y.; Wu, T.-C.; Chopyk, J.; Huang, L.; Kuo, P.; Ghose, C.; Meyer, J.R.; Tu, X.M.; et al. Fecal Virome Transplantation Is Sufficient to Alter Fecal Microbiota and Drive Lean and Obese Body Phenotypes in Mice. Gut Microbes 2023, 15, 2236750. [Google Scholar] [CrossRef] [PubMed]
- Draper, L.A.; Ryan, F.J.; Dalmasso, M.; Casey, P.G.; McCann, A.; Velayudhan, V.; Ross, R.P.; Hill, C. Autochthonous Faecal Viral Transfer (FVT) Impacts the Murine Microbiome after Antibiotic Perturbation. BMC Biol. 2020, 18, 173. [Google Scholar] [CrossRef]
- Selle, K.; Fletcher, J.R.; Tuson, H.; Schmitt, D.S.; McMillan, L.; Vridhambal, G.S.; Rivera, A.J.; Montgomery, S.A.; Fortier, L.-C.; Barrangou, R.; et al. In Vivo Targeting of Clostridioides difficile Using Phage-Delivered CRISPR-Cas3 Antimicrobials. mBio 2020, 11, e00019-20. [Google Scholar] [CrossRef]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Liposome-Encapsulated Bacteriophages for Enhanced Oral Phage Therapy against Salmonella spp. Appl. Environ. Microbiol. 2015, 81, 4841. [Google Scholar] [CrossRef]
- Chadha, P.; Katare, O.P.; Chhibber, S. Liposome Loaded Phage Cocktail: Enhanced Therapeutic Potential in Resolving Klebsiella pneumoniae Mediated Burn Wound Infections. Burns 2017, 43, 1532. [Google Scholar] [CrossRef]
- Śliwka, P.; Skaradziński, G.; Dusza, I.; Grzywacz, A.; Skaradzińska, A. Freeze-Drying of Encapsulated Bacteriophage T4 to Obtain Shelf-Stable Dry Preparations for Oral Application. Pharmaceutics 2023, 15, 2792. [Google Scholar] [CrossRef]
- Yazdi, Z.R.; Leaper, M.C.; Malik, D.J. The Development of Oral Solid Dosage Forms Using the Direct-Compression Tableting of Spray-Dried Bacteriophages Suitable for Targeted Delivery and Controlled Release. Processes 2023, 11, 3146. [Google Scholar] [CrossRef]
- Kawakita, R.; Leveau, J.H.J.; Jeoh, T. Optimizing Viability and Yield and Improving Stability of Gram-Negative, Non-Spore Forming Plant-Beneficial Bacteria Encapsulated by Spray-Drying. Bioprocess. Biosyst. Eng. 2021, 44, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Tufail, M.A.; Schmitz, R.A. Exploring the Probiotic Potential of Bacteroides Spp. Within One Health Paradigm. Probiotics Antimicro. Prot. 2025, 17, 681–704. [Google Scholar] [CrossRef]
- Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 2021, 22, 8937. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and Immunomodulation. Future Microbiol. 2017, 12, 905–914. [Google Scholar] [CrossRef]
- Champagne-Jorgensen, K.; Luong, T.; Darby, T.; Roach, D.R. Immunogenicity of Bacteriophages. Trends Microbiol. 2023, 31, 1058–1071. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jankowski, W.; Mizielińska, M.; Nawrotek, P. Microbiome and Phageome: Key Factors in Host Organism Function and Disease Prevention in the Context of Microbiome Transplants. Appl. Sci. 2025, 15, 5330. https://doi.org/10.3390/app15105330
Jankowski W, Mizielińska M, Nawrotek P. Microbiome and Phageome: Key Factors in Host Organism Function and Disease Prevention in the Context of Microbiome Transplants. Applied Sciences. 2025; 15(10):5330. https://doi.org/10.3390/app15105330
Chicago/Turabian StyleJankowski, Wojciech, Małgorzata Mizielińska, and Paweł Nawrotek. 2025. "Microbiome and Phageome: Key Factors in Host Organism Function and Disease Prevention in the Context of Microbiome Transplants" Applied Sciences 15, no. 10: 5330. https://doi.org/10.3390/app15105330
APA StyleJankowski, W., Mizielińska, M., & Nawrotek, P. (2025). Microbiome and Phageome: Key Factors in Host Organism Function and Disease Prevention in the Context of Microbiome Transplants. Applied Sciences, 15(10), 5330. https://doi.org/10.3390/app15105330






