Abstract
The human body contains ~1014 cells—each of which is separated by a lipid bilayer, along with its organeller. Unsaturated fatty acids are located on the external layer and, as a result, are particularly exposed to harmful factors, including xenobiotics and ionising radiation. During this activity, lipid peroxidation products are generated, e.g., 4-hydroxy-2-nonenal (HNA), 4-oxo-2(E)-nonenal (ONE), and malondialdehyde (MDA). The mentioned aldehydes can react with cytosolic 2′-deoxynucleosides via Michael addition. In this paper, the following adducts have been taken into theoretical consideration: ε-dCyt, H-ε-dAde, ε-dCyt, H-ε-dAde, H-ε-dGua, R/S-OH-PdGua, N2,3-ε-dGua, M1-dGua, N1-ε-dGua, and HNE-dGua. The presence of the above molecules can alter a cell’s antioxidant pool. With this in mind, the adiabatic ionisation potential (AIP) and vertical ionisation potential (VIP), as well as the spin and charge distributions, are discussed. For this purpose, DFT studies were performed at the M06-2x/6-31++G** level of theory in the aqueous phase (both non-equilibrated (NE) and equilibrated (EQ) solvent–solute interaction modes), together with a Hirshfeld charge and spin distribution analysis. The obtained results indicate that the AIPs of all the investigated molecules fell within a range of 5.72 and 5.98 eV, which is consistent with the reference value of 7,8-dihydro-8-oxo-2′-deoxyguanosine (OXOdGua), 5.78 eV. N2,3-ε-dGua and M1-dGua were the only exceptions, whose VIP and AIP were noted as higher. The electronic properties analysis of 2′-deoxynucleoside adducts with lipid peroxidation products reveals their potential influence on the cells’ antioxidant pool, whereby they can affect the communication process between proteins, lipids, and nucleotides.
1. Introduction
The human body contains somewhere in the region of 1014 cells, all of which are separated from each other by a lipid bilayer [1,2]. This tiny barrier, composed of proteins and sugar units, serves several functions, including protecting the cell from the outer environment, regulating the delivery of biomolecules, and playing an essential role in maintaining osmotic pressure [3]. Lipid moieties constitute around 40% of this membrane’s mass. In such abundance, phospholipids, sphingolipids, or sterols might be selected depending on their central moiety (glycerol or its derivatives) and linked moiety (such as choline, serine, or inositol). All of the above contain polyunsaturated fatty acids (PUFA) in their structure, such as linolic acid and arachidonic acid (omega-6) [4]. PUFA forms the substrate for prostaglandin, leukotrienes, and thromboxane biosynthesis, which play a signalling physiological role (as a second messenger) [5]. Each cell is continuously exposed to harmful intra- and extra-cellular factors, including ionisation radiation (IR), xenobiotics, reactive oxygen/nitrogen species (ROS/RNS), and physiological metabolites. The details of ROS and NOS activity are given in the texts of the valuable reviews of Pérez-Lebeña, Rojanasakul, and Di Meo [6,7,8]. Note that, in the cellular environment, ROS can be generated by the Haber–Weiss reaction catalysed by the transition metal ions (Fe, Cu, Co, Mn, etc.) or as a product of the respiratory cycle [9]. Among all of the ROS, H2O2 is considered relatively inert, whereas the hydroxyl radical (HO●) is the most reactive and can react with cell moieties with a diffusion-controlled rate constant of 1010 Lmol−1s−1. Its reactivity can lead to DNA damage by hydrogen atom abstraction from the sugar moiety or by the addition of HO● to the nucleobase double bond [10]. Additionally, ROS can give rise to protein modifications and trigger lipid oxidation processes that have been the subject of a valuable review article by Cocheme [11]. It should be noted that 80–90% of oxygen is consumed in the mitochondria by a four-electron reduction process [12]. PUFAs localised in the cell membrane and the mitochondrial lipid bilayer are vulnerable to ROS activity. On the other hand, genetic information is located deep within a cell as highly condensed chromatin and protected by a nucleus layer formed by lipids. Furthermore, the abundant PUFAs in a cell are susceptible to the various harmful factors mentioned above, namely malnutrition/starvation, caloric overload/obesity, and heat shock [13]. In a cell, two pathways of lipid peroxidation (LPO) formation are particularly significant: the oxidation of phospholipids by lipoxygenase (enzymatic) and the oxidation by reactive products of the reaction catalysed by transient metal ions (non-enzymatic) (Figure 1) [14].
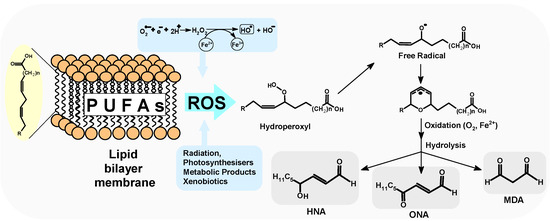
Figure 1.
A schematic representation of lipid peroxidation product formation as a result of the activity of various harmful factors. PUFAs—polyunsaturated fatty acids, ROS—reactive oxygen species, HNA—4-hydroxy-2-nonenal, ONE—4-oxo-2-nonenal, MDA—malondialdehyde. It should be pointed out that the above levels vary according to age and health.
Either process can lead to the formation of chemically reactive aldehydes in the course of LPO, which can play a significant role in a cell’s fate, causing ferroptosis, for example [15]. Like free radicals, aldehydes can react with DNA, proteins, lipids, and other cellular macromolecules, changing or impeding their function. However, they exhibit a prolonged half-life, which allows them to interact with neighbouring cells or to be delivered to other tissues in the bloodstream [16]. (Conversely, the short-lived free radicals initially formed from LPO products are perceived as secondary messengers of oxidative stress). Because of their chemical properties, aldehydes can react with exo-amino groups of nucleic base moieties that are present not only in the structure of nucleic acids but also in the nucleosides or nucleotides, thereby altering their electronic properties [17]. Free nucleosides/nucleosides are abundant molecules in the cytoplasm environment and are convenient and accessible substrates for LPO products. In this article, the ionisation potential (IP) of 4-hydroxy-2-nonenal (HNA), 4-oxo-2(E)-nonenal (ONE), and malondialdehyde (MDA) (as lipid peroxidation products (Figure 1)) and derivatives of nucleosides are theoretically investigated and compared with OXOdGua as a reference of DNA damage, as well as with canonical nucleosides. Most current studies discuss the role of LPO adducts in the context of ds-DNA (the genome). It has been found that DNA LPO product adducts can give rise to several diseases, including cancers [18]. However, they can also serve as valuable biomarkers of oxidative stress. It should be pointed out that, during radiotherapy, the level of LPO products increases, making them an indicator of the effectiveness of radiation treatment. To the author’s knowledge, no studies have been performed to determine their influence on charge transfer, especially with reference to the macromolecules present in the cytosol. The mutual communication between proteins, lipids, and nucleotides is of high scientific value and vital in cell machinery. Investigating this communication process could lead to a greater understanding of the acceleration of harmful/undesirable processes like carcinogenesis, ageing, and signal transduction. Additionally, it may result in the improved effectiveness and safety of anticancer radiotherapy/chemotherapy or combined therapies with a subsequent increase in a patient’s quality of life. Therefore, the influence of 2′-deoxynucleosides adducts with LPO products on endo-cellular charge transfer warrants future investigation.
2. Materials and Methods
The density functional theory (DFT) methodology has been applied for geometry optimisation and energy calculation of the considered nucleosides and their analogues in their neutral, positively charged forms [19]. The diastereomeric forms of the chiral centre have been assigned according to Cahn–Ingold–Prelog rules. The molecules’ ground states have been obtained using the self-consistent fields methodology with tight convergence criteria. For the nucleoside derivatives, the 2′-endo form of 2-deoxyribose has been chosen. It should be pointed out that, due to the number of freedom degrees in nucleosides, 41,472 conformers can be found [20]. All calculations were performed in the aqueous phase by DFT methodology for which Tomasi’s Conductor-like Polarised Continuum Model (CPCM) was used [21]. It should be pointed out that the solvation model with a water dielectric constant e = 78.4 cannot be directly considered for the cytoplasm [22]. However, the complexities of cytosol prohibited current quantum or DFT methodology applications limited by current supercomputer power and efficiency. For this purpose, the hybrid meta exchange–correlation functional M06-2X (54% Hartree–Fock exchange) with an augmented polarised valence double-ζ basis set 6-31++G** was used [23] (the mentioned basis set was represented as a number of atomic orbitals composed of MC,N,O/MH (4s3p1d/3s1p), consisting of 19 basis functions of C, N, O atoms and 6 basis functions of H atoms [24]). As shown previously, the M06-2X 6-31++G** is relatively cost-effective and efficient [25]. The non-equilibrium (NE) and equilibrated (EQ) solvent–solute interaction were investigated according to Sevilla’s previous studies [26]. For all studies, a charge and spin distribution was achieved using the Hirshfeld theory [27]. The Hirshfeld charge and spin population analysis is based on the topology of the electron localisation function of Becke and the molecular electrostatic potential. The differences between natural bond orbital analysis, McWeeny’s theory of electronic separability and Mulliken, Lowdin, and Morokuma population analysis are given in reference [28,29].
The following electronic properties of investigated molecules, i.e., adiabatic and vertical ionisation (AIP and VIP), are given in electron volts [eV] [30]. The following notation of the discussed states was used: Egeometrycharge. A molecule in the ground (neutral) state is described as E00, the vertical cation (VC) as E0+, and the adiabatic cation (AC) as E++. Therefore, the appropriate electronic states were defined as follows: VIPNE = E0+(NE) − E00 (vertical ionisation potential in the NE state); VIPEQ = E0+(EQ) − E00 (vertical ionisation potential in the EQ state); and AIP = E++ − E00 (adiabatic ionisation potential). All calculations were performed in the aqueous phase on the Gaussian G16 (version C.01) software package [31].
3. Results
The structure of the discussed nucleosides and their analogues in their neutral, positively charged (radical cation) forms were optimised at the M062x/6-31++G** level of theory in the aqueous phase, as presented in Figure 2.
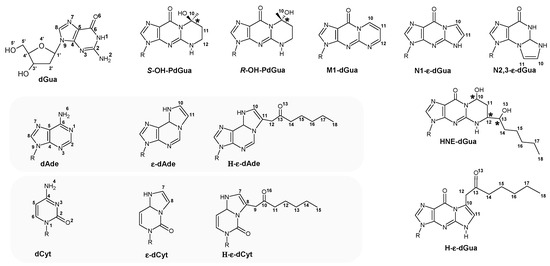
Figure 2.
A graphical representation of the investigated nucleoside adducts with atoms numbered and chiral centres indicated (*). R: 2-deoxyribose, dGua: 2′-deoxyguanosine, dAde: 2′-deoxyadenosine, dCyt: 2′deoxycytidine, ε-dCyt: 3,N4-etheno-2′-deoxycytidine, ε-dAde: 1N6-etheno-2′-deoxyadenosine, N1-ε-dGua: 1,N2-etheno-2′-deoxyguanosine, N2,3-ε-dGua: 2,N3-etheno-2′-deoxyguanosine, (R/S) 10-OH-PdGua: (R/S) 10-hydroxy-1,N2-propano-2′-deoxyguanosine, H-ε-dCyt: 8-(10-oxoheptyl)-3,N4-etheno-2′-deoxyctidine, H-ε-dAde: 11-(13-oxoheptyl)-1,N6-etheno-2′-deoxyadenosine, H-ε-dGua: 11-(13-oxoheptyl)-1,N2- etheno-2′-deoxyguanosine, HNE-dGua: 12-(13-hyroxyheptyl)-1,N2-propano-2′-deoxyguanosine, M1-dGua: malondialdehyde-2′-deoxyguanosine or 1,N3-propano-2′-deoxyguanosine.
3.1. Spatial Geometry Analysis of 2′-Deoxynucleoside Adducts with Lipid Peroxidation Products
The spatial geometries of the discussed adducts presented in Figure 2 were optimised at the M06-2x/6-31++G** level of theory in the aqueous phase using the conductor-like polarisable continuum model (CPCM) in their neutral and positively charged forms. The atom-numbering system used for canonical nucleosides with their extensions has been applied for structural cohesion and comparison with the initial heterocycles.
The comparison of geometries between the neutral and radical cation forms revealed the most significant differences in the case of N2,3-ε-dGua (0.650 [Å2]) (Table 1), as determined by the root-mean-square deviation (RMSD) calculation [32]. A similar observation was noted for the dipole moment deviation between both forms (ΔDM = 5.49 [D]) (Table 1). The above indicates that the loss of the electron (radical cation formation) forces the highest geometry distortion in the case of the N2,3-ε-dGua molecule. It should be noted that the energy of N2,3,-ε-dGua in its neutral ground state was lower by 0.9 kcal/mol than that observed for its structural analogue N1,2-ε-dGua, which makes its formation predisposed. It should also be pointed out that, in the structure of HNE-dGua, in which three chiral centres (C10, C12, C13) are present, eight diastereomeric forms are possible (Figure 2). The notation of the chirality was performed according to the atom-numbering system. Of all the diastereomeric forms, a comparison of the ground state energies of the neutral forms revealed a lower energy value for (SRR)HNE-dGua and a higher value for (SRS) (Figure 2), with the difference calculated as 2.77 kcal/mol. For 8-OH-PdG, the diastereomer C10 R was identified as slightly thermodynamically preferred by 0.02 kcal/mol (Table S1). No chiral centres were observed in the heterocycle’s derivative moiety for the remaining adducts.

Table 1.
Root-mean-square deviation (RMSD) in [Å2] of atomic positions calculated for the discussed molecules (Figure 2) in their neutral and cationic ground states (cation versus neutral); the changes in dipole moment (DM) calculated after electron loss are given in Debyes [D] (|ΔDM| = DM(E00) − DM(E0+)).
3.2. Charge and Spin Distribution Analysis
The spatial geometry of the radical cations of the molecules discussed above indicates that differences in their spin and charge distribution and that of native nucleosides should be observed. All vertical and adiabatic energies of the positively charged forms were investigated at the M06-2X/6-31++G** level of theory in the condensed (aqueous) phase. The charge and spin distributions were analysed according to Hirshfeld’s methodology [33]. The loss of the electron leads to the formation of a vertical radical cation state. The spin distribution analysis of positively charged 2′-deoxynucleoside analogues across sugar, base, and carbohydride shows that a one-electron oxidising process occurs mainly on the nucleobase moiety, i.e., more than 60% (Table 2) in all of the discussed stages (the vertical cation in non-equilibrated and equilibrated solvent–solute modes and the adiabatic cation state, i.e., after system relaxation). Exceptions were noted in the cases of N2,3-ε-dGua, ε-dCyt, and H-ε-dCyt, in which the unpaired electron was finally localised (in almost 70% of cases) on the additional etheno part derived from LPO products, i.e., HNE. For the above molecules, the spin transfer from the nucleobase to the adduct moiety after structural and solvent relaxation was noted, i.e., VCNE → VCEQ → AC.

Table 2.
The Hershfield spin distribution within ε-dCyt, ε-dAde, ε-dGua, N1-ε-dGua, N2,3-ε-dGua, (R/S) 10-OH-PdGua, H-ε-dCyt, H-ε-dAde, H-ε-dGua, HNE-dGua, and M1-dGua calculated at the M062x/6-31++G** level of theory in the condensed phase. NE: non-equilibrated solvent–solute interaction, EQ: equilibrated solvent–solute interaction. The raw data of charge and spin distribution are given in Table S2 of the Supplementary Materials. VC: vertical cation, AC: adiabatic cation, NE non-equilibrated solvent–solute interaction, EQ equilibrated solvent–solute interaction.
The spin distribution analysis of the adiabatic cation state in the atom’s course reveals that, for the discussed molecules, the atoms most prone to modification are as follows (the percentage of spin is given in parentheses): ε-dCyt: C8 (67); H-ε-dCyt: C8 (67); ε-dAde: C11 (34), N6 (30); H-ε-dAde: C11 (32), N6 (29); H-ε-dGua: N3 (16), O6 (16), C5 (15); (R)10-OH-PdGua: N3 (22), C5 (22), C8 (15); (S)10-OH-PdGua: N3 (22), C5 (22), C8 (15); M1-dGua: N3 (23), C5 (18), O6 (17), C8 (12); N1-ε-dGua: N3 (25), O6 (15), C10 (22); N2,3-ε-dGua: C10 (18), and C11 (53). In the case of HNE-dGuas, the following results were found to be dependent on the distribution of chiral centres (C10, C12, and C13): RRR: N2 (32), C5 (15), C8 (15); RRS: N2 (32), C5 (20), C8 (14); RSR: C5 (22), O6 (20), C8 (15); RSS: C5 (22), C8 (20), N3 (19); SRR: N2 (31), C5 (21), C8 (15); SRS: N2 (28), C5 (21), C8 (16); SSS: C5 (22), C8 (20), and N3 (19). It should be pointed out that the above order was noted for non-equilibrated and equilibrated vertical cations, as well as for the adiabatic cations of each of the discussed 2′-deoxynucleoside derivatives, except N2,3-ε-dGua, ε-dCyt, and H-ε-dCyt.
The charge analysis performed at the same level of theory revealed a charge accumulation mainly on the 2-ribose and aldehyde part of the 2′-deoxynucleoside adduct, leaving the base moiety almost neutral. It should be pointed out that the charge almost exclusively accumulated on the heterocyclic parts of the reference canonicals dCyt, dAdo, dGua, and the common DNA lesion OXOdGua (Table S2).
3.3. Vertical and Adiabatic Ionisation Potential (AIP) of 2′Deoxynucleosides Adducts
Under ionisation radiation or other oxidation events interacting with the cellular environment, damage to different macromolecule types can occur [34]. With this context in mind, a significant goal in biochemistry has been to determine the energy threshold necessary to ionise canonic 2′-deoxynucleosides or their analogues, such as those presented in Figure 2. Because their free forms are more susceptible to the influence of free radicals, their subcellular level can be disrupted as a result of further reactions. Looking at the issue from a different angle, modified nucleoside analogues can block radical propagation through the cytosol and protect remote regions of the cell (genome). This theory was an attempt to determine the vertical (VIP) and adiabatic ionisation potential (AIP) of nucleoside aldehyde adducts (Table 1). It should be pointed out that, directly after electron loss, the investigated “system” exists within the solvent–solute non-equilibrated (NE) interaction mode. The calculated VIPNE indicates the initial predisposition of the compound to oxidisation, therefore quenching any harmful radical in its vicinity (in the same way that antioxidants work). This state is further stabilised by the rearrangement of the condensed phase (solvent) and is described as vertical ionisation potential in the equilibrated mode (VIPEQ). Theoretically, gaseous and aqueous phase calculations were performed because it is difficult to experimentally determine the VIPNE, VIPEQ, and AIP of the molecules under investigation. In this study, the VIPNE, VIPEQ, and AIP of 2′-deoxynucleoside adducts with LPO products, as presented in Figure 2, were calculated at the M06-2x/6-31++G** level of theory and reported in Table 3. All calculations were carried out in the aqueous phase in non-equilibrated and equilibrated solvent–solute interaction modes.

Table 3.
The electronic properties, in [eV], of 2′-deoxynucleoside adducts with lipid peroxidation products: Vertical (VIP), adiabatic ionisation potential (AIP), NE: non-equilibrates, EQ: equilibrated solvent–solute interactions, calculated at the M062x/6-31++G** level of theory in the aqueous phase. The raw data are given in the Supplementary Materials, Table S1.
Among all the investigated molecules, the formation of lipid peroxidation adducts of 2′-deoxynucleoside causes a decrease in VIPNE, VIPEQ, and AIP compared to canonical 2′-deoxynucleosides. Only in the case of N2,3-ε-dGua was an increase in the ionisation potential observed, while M1-dGua showed similar properties to canonical 2′-deoxyguanosine. A comparison of the electronic properties of the discussed molecules to OXOdG (the reference DNA lesion with the lowest ionisation potential) revealed that, exclusively, the RRR and SRR diastereomers of HNE-dGua have slightly lower AIP values than 7,8-dihydro-8-oxo-2′deoxyguanosine, by 0.02 and 0.06 eV, respectively (Table 3). The resulting calculations indicate that the cells’ pool of antioxidants can increase after forming 2′-deoxynucleoside adducts with malondialdehyde, 4-hydroxyl-2-nonenal, and 4-oxo-2-nonenal.
4. Discussion
Throughout evolution, living organisms have developed protective mechanisms that eliminate unwanted R/NOS inside a cell [9]. Such mechanisms include enzymes such as superoxide dismutase (SOD), present either in prokaryotic or eukaryotic cells (its biosynthesis is controlled through the O2•− level) [35], glutathione peroxidase catalysing the H2O2 to H2O conversion [36], haem peroxidase [37], catalase [38], and many more. A different group of molecules involved in the removal of free radicals comprises so-called non-enzymatic antioxidants, which are small molecules that convert radicals into their inactive forms while becoming excited themselves (ascorbic acid, α-tocopherol, glutathione, carotenoids, and flavonoids) [39]. These defence mechanisms against reactive oxygen and nitrogen species act as guards, protecting the stability of a cell’s physiology and genetic information. It has been established that ~9 × 104 DNA damage events in each human cell are caused over 24 h [40,41]. To date, more than 80 types of DNA lesions have been identified. The human nuclear diploid genome length consists of an estimated 6.27 × 109 base pairs for male cells and 6.37 × 109 for female cells, corresponding to 6.41 pg and 6.51 pg, respectively [42]. However, the pool of nucleobases in a cell is increased by free nucleosides in the cytosol. The following concentrations, in μM, have been found for ribonucleotides: ATP: 3152, GTP: 468, UTP: 567, and CTP: 278. For 2′-deoxynucleotides, they are dATP: 24, dGTP: 5.2, dCTP: 29, dTTP: 37 [43]. Previous studies have established that guanosine exhibits the lowest ionisation potential (IP) of all the nucleobases and is easily converted by one-electron oxidation to 8-oxo-G, with subsequent decreases in IP values. The following reduction potentials have been noted: 1.29 V and 0.74 V, respectively [44,45]. This is an indication of their protective role, which is typical of antioxidants. Based on the above, this article has considered the changes in ionisation potential following DNA damage formation by PLO products. The most abundant endocellular lipid peroxidation products are HNE, MDA, and ONE. At the same time, acrolein, present in a cell in high amounts, is derived from other sources, such as smoking, fried food, etc. The following levels of aldehydes HNE, ONE and MDA, as presented in Figure 1, have been found in human plasma: 0.564 ± 0.467 nM, 0.299 ± 0.248, and >1 μM, respectively [14,46]. The local concentration of total LPO products in the microsomal and mitochondrial layer is approximately 10 mM [47], and more than 200 products have been identified to date [48]. It should be pointed out that the above level varies depending on a person’s age, health condition, and the analytical techniques used. Indeed, because of the products’ reactivity and resulting instability, ascertaining a precise measurement still poses a huge analytical challenge. However, the half-lives of HNE and ONE in physiological conditions, for example, have been estimated at 2 min [49,50]. This provides enough time for them to react with the exo-amino group of nucleobases, leading to ethano-type adducts via Michael addition [51]. It should be noted that the exo-amino groups involved in DNA base pairing are less available to LPO products, making them less abundant in the genome than OXOG, i.e., 14.6 per 106 nucleotides (nt) [52]. The levels of ε-dCyt, ε-dAde, H-ε-dGua, N2,3-ε-dGua*, (R/S) 10-OH-PdGua, H-ε-dCyt, H-ε-dAde, H-ε-dGua, HNE-dGua, and M1-dGua in calf thymus DNA have been assigned as follows (per 107 nt): 7.63, 12.4, 1.08,0.7–7 3.71, 5.81, 2.10, 2.28, 1.42, and 2.87, respectively [53,54]. The radicals formed and stabilised from nucleosides or lipids are not uniformly distributed over the cellular environment during exposure to harmful factors, such as ionisation radiation, xenobiotics, etc. It is well known that the process initiated by destructive factors, such as radiation, can create different reactive forms, such as radicals [55]. These molecules can migrate and become trapped by certain preferred molecules present in the cytosol. Both the abundant and reactive aldehyde. along with the commonly present nucleobase moieties. can trigger changes to the antioxidant potential of a cell.
Experimentally determining the ionisation potential of 2′-deoxynucleosides and their analogues presents some analytical challenges, including the preparation of intact gas-phase nucleosides, nucleotides, or nucleic acid bases and their derivatives. Therefore, little experimental data exist in the literature, most of which focuses on free bases. As a result of these limitations, IPs were computed by various theoretical methods [56]. As mentioned above, the following order of ionisation potentials was assigned: U > T > C > A > G, establishing guanosine as the most susceptible to one-electron oxidisation and radical cation trapping among all the canonical nucleosides [57].
In this study, the 2′-deoxynucleoside adducts with lipid peroxidation products, as shown in Figure 2, were taken into theoretical consideration. The spatial geometries of the neutral and radical cation forms were optimised at the M06-2x/6-31++G** level of theory in the aqueous phase. The 3D structure comparison between neutral and oxidised forms of LPO derivatives reveals the lowest RMSD value (0.156 [Å2]) for (RSS)HNE-dGua, with a ΔDM equal to 1.8 [D]. Conversely, for N2,3-OH-ε-PdGua, the highest RMSD and ΔDM values were calculated as 0.650 and 5.49, respectively. The above indicates significant structural changes forced by electron loss, making N2,3-OH-ε-PdGua less predisposed to oxidisation among all the 2′-deoxynucleosides adducts under discussion. Careful study of the spin and charge analysis obtained according to Hirshfeld’s methodology revealed that, in all the of discussed cases, the spin of positively charged molecules is predominately located on the nucleobase moiety, except for ε-dCyt, H-ε-dCyt, and N2,3-ε-dGua, as presented in Table 2. For the above 2′-deoxyncleoside adducts, unpaired electron migration was observed during solvent and solute relaxation (Table 2), i.e., in the vertical cation non-equilibrated solvent–solute state, as well as the equilibrated spin state, the unpaired electrons were found on the nucleobase moiety. In contrast, the unpaired electron migrated onto the etheno part of the adduct after structural relaxation in approximately 70% of cases. The ionisation potential is a convenient parameter for estimating the antioxidant potential of molecules. These systematic theoretical studies were performed using the Minnesota DFT functional M06-2x with a double-ζ basis set with dispersion and polarisation functions (6-31++G**) for hydrogen and heavy atoms. The ionisation potential values were calculated as vertical (VIPNE and VIPEQ) and adiabatic (AIP). The results are presented in Table 3 and show that the lowest ionisation potentials (VIPNE, VIPEQ, and AIP) in [eV] were noted in the case of 2′-deoxyadenosine adducts with LPO products for H-ε-dAdo (7.07, 6.25, 6.00, respectively). For 2′-deoxycytidine derivatives, the lowest values were noted for H-ε-dCyt (6.78, 5.96, and 5.80, respectively) and for the 2′-deoxyguanosine series. The lowest were observed for (SRR) HNE-dGua (6.80, 6.07, and 5.72, again respectively). The above values are significantly lower than those obtained for parental canonical 2′-deoxynucleosides (Table 3). It should be pointed out that, by using the same methodology for the 7,8-dihydro-8-oxo-2′-deoxyguanosine, the VIPNE, VIPEQ and AIP values were observed were as follows: 7.15, 6.15, and 5.78 [eV]. Careful analysis of the ionisation potentials showed that N2,3-ε-dGua and M1-dGua have higher values than those for dGua, while for SRR and RRR, diastereomeric forms of HNE-Gua were similar to those of OXOdG (see Table 2).
5. Conclusions
The activity of various harmful factors can lead to different DNA damage formations, which, left unrepaired, give rise to mutations and/or contribute to ageing and cancerogenesis. While DNA lesions have been thoroughly investigated [58], little light has been shed on the electronic properties of free 2′-deoxynucleosides adducts with lipid peroxidation products and their influence on a cell’s antioxidant potential.
- The results of the presented studies have been discussed on the M06-2x/6-31++G** level of theory in the aqueous phase. The non-equilibrated and equilibrated solvent–solute interaction has been considered using Tomasi’s Conductor-like Polarised Continuum Model;
- The vertical and adiabatic ionisation potentials of ε-dCyt, H-ε-dAde, ε-dCyt, H-ε-dAde, H-ε-dGua, R/S-OH-PdGua, N1-ε-dGua, HNE-dGua (8 diastereomeric forms), dCyt, dAdo, and dGuo have been calculated;
- The spatial geometry comparison between the neutral and adiabatic radical cations of discussed LPO derivatives reveals the lowest RMSD value (0.156 [Å2]) for (RSS)HNE-dGua. Conversely, for N2,3-OH-ε-PdGua, the highest RMSD value was calculated as 0.650;
- The lowest VIPNE, VIPEQ, and AIP in [eV] have been found for the following 2′-deoxynucleoside LPO adducts:H-ε-dAdo (2′-deoxyadenosine derivative) 7.07, 6.25, and 6.00, respectively,H-ε-dCyt (2′-deoxycytidine derivatives) 6.78, 5.96, and 5.80, respectively,(SRR) HNE-dGua (2′-deoxyguanosine derivatives) 6.80, 6.07, and 5.72, respectively;
- All the 2′-deoxyguanosine LPO adducts discussed in this paper have AIP values similar to OXOdGua (5.78 eV), apart from N2,3-ε-dGua and M1-dGua.
The obtained results have shown that LPO 2′-deoxynucleoside adduct formation increases the antioxidant pull of the cell, thus terminating the propagation of free radicals. When nucleoside damage, such as ε-dCyt, H-ε-dAde, ε-dCyt, H-ε-dAde, H-ε-dGua, R/S-OH-PdGua, N1-ε-dGua, and all the diastereomers of HNE-dGua become part of the cytosol, the efficiency of macromolecule lesion formation can be affected because the damage exerts a strong influence on the electron–hole transfer process.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/app15010437/s1: Table S1. The electronic state energy in Hartree and Dipole Moment (DM) in Debye was calculated on the M06-2x/6-31++G** level of theory in the condensed phase using the non-equilibrated and equilibrated solvent–solute interaction.; Table S2. Hirshfeld charge distribution calculated at the M06-2x/6-31++G** level of theory in the aqueous phase. Vertical Cation (VCNC) (NE-non-equilibrated), Vertical Cation (VCEQ) (EQ-equilibrated), Adiabatic Cation (AC).; PDB structures of discussed compounds in a zip file.; Table S3. Graphical visualisation ground state geometry and spin distribution calculated at the M06-2x/6-31++G** level of theory in the aqueous phase of the molecules discussed in the article in their Adiabatic and Vertical radical cation forms. The non-equilibrated (NE) and equilibrated (EQ) solvent–solute interaction has been taken into consideration. The optimised structures of the discussed compounds in neutral and cationic forms, such as those in *.pdb files, were compressed into a zipped PDB_structure file.
Funding
This study was supported by the Medical University of Lodz, 503/3-045-02/503-31-002.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available in Supplementary Materials.
Acknowledgments
The author gratefully acknowledges the Polish high-performance computing infrastructure PLGrid (HPC Center: ACK Cyfronet AGH) for providing computer facilities and support within computational grant no. PLG/2023/016562.
Conflicts of Interest
The author declare no conflicts of interest.
References
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Nagle, J.F.; Tristram-Nagle, S. Structure of lipid bilayers. Biochim. Biophys. Acta Rev. Biomembr. 2000, 1469, 159–195. [Google Scholar] [CrossRef]
- Rangamani, P.; Zhang, D.; Oster, G.; Shen, A.Q. Lipid tubule growth by osmotic pressure. J. R. Soc. Interface 2013, 10, 20130637. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.K.; Else, P.L.; Atkins, T.A.; Hulbert, A.J. Fatty acid composition of membrane bilayers: Importance of diet polyunsaturated fat balance. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Botta, E.; Holinstat, M. Eicosanoids in inflammation in the blood and the vessel. Front. Pharmacol. 2022, 13, 997403. [Google Scholar] [CrossRef]
- Manke, A.; Wang, L.; Rojanasakul, Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res. Int. 2013, 2013, 942916. [Google Scholar] [CrossRef]
- Juan, C.A.; de la Lastra, J.M.P.; Plou, F.J.; Pérez-Lebeña, E. The chemistry of reactive oxygen species (Ros) revisited: Outlining their role in biological macromolecules (dna, lipids and proteins) and induced pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell. Longev. 2016, 44, 1245049. [Google Scholar] [CrossRef]
- De Almeida, A.J.P.O.; De Oliveira, J.C.P.L.; Da Silva Pontes, L.V.; De Souza Júnior, J.F.; Gonçalves, T.A.F.; Dantas, S.H.; De Almeida Feitosa, M.S.; Silva, A.O.; De Medeiros, I.A. ROS: Basic Concepts, Sources, Cellular Signaling, and its Implications in Aging Pathways. Oxid. Med. Cell. Longev. 2022, 2022, 1225578. [Google Scholar] [CrossRef]
- Barry Halliwell, A.; Adhikary, M.; Dingfelder, M.D. Hydroxyl radical is a significant player in oxidative DNA damage in vivo. Chem. Soc. Rev. 2021, 50, 8355–8360. [Google Scholar] [CrossRef]
- Lennicke, C.; Cochemé, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef] [PubMed]
- Snyder, C.M.; Chandel, N.S. Mitochondrial Regulation of Cell Survival and Death During Low-Oxygen Conditions. Antioxidants Redox Signal. 2009, 11, 2673–2683. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Niki, E. Lipid peroxidation: Physiological levels and dual biological effects. Free Radic. Biol. Med. 2009, 47, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hou, L.; Guo, Z.; Wang, G.; Xu, J.; Zheng, Z.; Sun, K.; Guo, F. Lipid peroxidation in osteoarthritis: Focusing on 4-hydroxynonenal, malondialdehyde, and ferroptosis. Cell Death Discov. 2023, 9, 320. [Google Scholar] [CrossRef]
- Gentile, F.; Arcaro, A.; Pizzimenti, S.; Daga, M.; Cetrangolo, G.P.; Dianzani, C.; Lepore, A.; Graf, M.; Ames, P.R.J.; Barrera, G. DNA damage by lipid peroxidation products: Implications in cancer, inflammation and autoimmunity. AIMS Genet. 2017, 4, 103–137. [Google Scholar] [CrossRef]
- Nair, U.; Bartsch, H.; Nair, J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: A review of published adduct types and levels in humans. Free Radic. Biol. Med. 2007, 43, 1109–1120. [Google Scholar] [CrossRef]
- Voulgaridou, G.P.; Anestopoulos, I.; Franco, R.; Panayiotidis, M.I.; Pappa, A. DNA damage induced by endogenous aldehydes: Current state of knowledge. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 711, 13–27. [Google Scholar] [CrossRef]
- Bursch, M.; Mewes, J.M.; Hansen, A.; Grimme, S. Best-Practice DFT Protocols for Basic Molecular ComputationalChemistry. Angew. Chemie Int. Ed. 2022, 61, e202205735. [Google Scholar] [CrossRef]
- Evangelista, F.A.; Schaefer, H.F. Structures and energetics of adenosine radicals: (2′-dAdo—H). J. Phys. Chem. A 2004, 108, 10258–10269. [Google Scholar] [CrossRef]
- Miertus, S.; Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Markx, G.H.; Davey, C.L. The dielectric properties of biological cells at radiofrequencies: Applications in biotechnology. Enzyme Microb. Technol. 1999, 25, 161–171. [Google Scholar] [CrossRef]
- Zhao, Y.; Pu, J.; Lynch, B.J.; Truhlar, D.G. Tests of second-generation and third-generation density functionals for thermochemical kinetics. Phys. Chem. Chem. Phys. 2004, 6, 673. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Karwowski, B.T. The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds -DNA: Theoretical Studies of Clustered DNA Damage. Molecules 2023, 28, 2180. [Google Scholar] [CrossRef]
- Kumar, A.; Adhikary, A.; Sevilla, M.D.; Close, D.M. One-electron oxidation of ds(5′-GGG-3′) and ds(5′-G(8OG)G-3′) and the nature of hole distribution: A density functional theory (DFT) study. Phys. Chem. Chem. Phys. 2020, 22, 5078–5089. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Marenich, A.V.; Jerome, S.V.; Cramer, C.J.; Truhlar, D.G. Charge model 5: An extension of hirshfeld population analysis for the accurate description of molecular interactions in gaseous and condensed phases. J. Chem. Theory Comput. 2012, 8, 527–541. [Google Scholar] [CrossRef]
- Saha, S.; Roy, R.K.; Ayers, P.W. Are the Hirshfeld and Mulliken Population Analysis Schemes Consistent With Chemical Intuition? Int. J. Quantum Chem. 2009, 109, 1790–1806. [Google Scholar] [CrossRef]
- Karwowski, B.T. The influence of phosphorothioate on charge migration in single and double stranded DNA: A theoretical approach. Phys. Chem. Chem. Phys. 2015, 17, 21507–21516. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2019. [Google Scholar]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef]
- Davidson, E.R.; Chakravorty, S. A test of the Hirshfeld definition of atomic charges and moments. Theor. Chim. Acta 1992, 83, 319–330. [Google Scholar] [CrossRef]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of Ionizing Radiation on Biological Molecules—Mechanisms of Damage and Emerging Methods of Detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Meister, A.; Anderson, M.E. Glutathione. Annu Rev Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.K. Membrane peroxidases. Mol. Cell. Biochem. 1988, 128, 105–128. [Google Scholar] [CrossRef]
- Glorieux, C.; Calderon, P.B. Catalase, a remarkable enzyme: Targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017, 398, 1095–1108. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef]
- Sudhir Ambekar, S. DNA: Damage and Repair Mechanisms in Humans. Glob. J. Pharm. Pharm. Sci. 2017, 3, 7. [Google Scholar] [CrossRef]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Matter, B.; Malejka-giganti, D.; Csallany, A.S.; Tretyakova, N. Quantitative analysis of the oxidative DNA lesion, 2,2-diamino-4-(2-deoxy-β-d-erythro-pentofuranosyl)amino]-5(2H)-oxazolone (oxazolone), in vitro and in vivo by isotope dilution-capillary HPLC-ESI-MS/MS. Nucleic Acids Res 2006, 34, 5449–5460. [Google Scholar] [CrossRef]
- Harkin, C.; Cobice, D.; Watt, J.; Kurth, M.J.; Brockbank, S.; Bolton, S.; Johnston, F.; Strzelecka, A.; Lamont, J.V.; Moore, T.; et al. Analysis of reactive aldehydes in urine and plasma of type-2 diabetes mellitus patients through liquid chromatography-mass spectrometry: Reactive aldehydes as potential markers of diabetic nephropathy. Front. Nutr. 2023, 9, 997015. [Google Scholar] [CrossRef]
- Fritz, K.S.; Petersen, D.R. Free Radical Biology and Medicine An overview of the chemistry and biology of reactive aldehydes. Free Radic. Biol. Med. 2013, 59, 85–91. [Google Scholar] [CrossRef]
- Esterbauer, H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am. J. Clin. Nutr. 1993, 57, 779S–785S. [Google Scholar] [CrossRef]
- Huc, L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. Vol. 2013, 20, 1615–1630. [Google Scholar] [CrossRef]
- Lin, D.; Lee, H.; Liu, Q.; Perry, G.; Smith, M.A.; Sayre, L.M. 4-Oxo-2-nonenal Is Both More Neurotoxic and More Protein Reactive than 4-Hydroxy-2-nonenal. Chem. Res. Toxicol. 2005, 18, 1219–1231. [Google Scholar] [CrossRef]
- Hutzinger, O. The Handbook of Environmental Chemistry. Volume 2, Part A. Reactions and Processes; Springer-Verlag Berlin Heildelberg GmbH, Germany, 1980; ISBN 3540096892.
- Scanlan, L.D.; Coskun, S.H.; Jaruga, P.; Hanna, S.K.; Sims, C.M.; Almeida, J.L.; Catoe, D.; Coskun, E.; Golan, R.; Dizdaroglu, M.; et al. Measurement of Oxidatively Induced DNA Damage in Caenorhabditis elegans with High-Salt DNA Extraction and Isotope-Dilution Mass Spectrometry. Anal. Chem. 2019, 91, 12149–12155. [Google Scholar] [CrossRef]
- Chen, H.; Krishnamachari, S.; Guo, J.; Yao, L.; Murugan, P.; Weight, C.J.; Turesky, R.J. Quantitation of Lipid Peroxidation Product DNA Adducts in Human Prostate by Tandem Mass Spectrometry: A Method That Mitigates Artifacts. Chem. Res. Toxicol. 2019, 32, 1850–1862. [Google Scholar] [CrossRef]
- Swenberg, J.A.; La, D.K.; Scheller, N.A.; Wu, K. Dose-response relationships for carcinogens. Toxicol. Lett. 1995, 4274, 751–756. [Google Scholar] [CrossRef]
- Phaniendra, A.; Babu, D. Free Radicals: Properties, Sources, Targets, and Their Implication in Various Diseases. Ind. J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef]
- Close, D.M. Calculation of the Ionization Potentials of the DNA Bases in Aqueous Medium. J. Phys. Chem. A 2004, 108, 10376–10379. [Google Scholar] [CrossRef]
- Kumar, A.; Sevilla, M.D. Proton transfer induced SOMO-to-HOMO level switching in one-electron oxidized A-T and G-C base pairs: A density functional theory study. J. Phys. Chem. B 2014, 118, 5453–5458. [Google Scholar] [CrossRef] [PubMed]
- Diamantis, P.; Tavernelli, I.; Rothlisberger, U. Redox Properties of Native and Damaged DNA from Mixed Quantum Mechanical/Molecular Mechanics Molecular Dynamics Simulations. J. Chem. Theory Comput. 2020, 16, 6690–6701. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).