Pointwise Nonparametric Estimation of Odds Ratio Curves with R: Introducing the flexOR Package
Abstract
1. Introduction
2. The Additive Model
3. Software Description
dfgam(
response,
nl.predictors,
other.predictors = NULL,
smoother = "s",
method = "AIC",
data,
step = NULL
)
flexOR(
data,
response,
formula
)
plot(
x,
predictor,
prob = NULL,
ref.value = NULL,
conf.level = 0.95,
round.x = NULL,
ref.label = NULL,
col,
main,
xlab,
ylab,
lty,
xlim,
ylim,
xx,
ylog = TRUE,
...
)
4. Examples of Application
4.1. Acute Coronary Syndrome Data
> library ("flexOR")
> df1 <- dfgam(response="exitus",
nl.predictors=c("age","creatinine","fasting"),
other.predictors=c("anemia","sex","smoking"),
smoother="s",
method="AIC",
data = heart2)
> df1$df
df
age 8.9
creatinine 1.8
fasting 4.6
> m1 <- gam(exitus ~ s(age, 8.9) + s(creatinine, 1.8) + s(fasting, 4.6) +
anemia + sex + factor(smoking),
data=heart2,
family=binomial())
> summary(m1)
Anova for Parametric Effects
Df Sum Sq Mean Sq F value Pr(>F)
s(age, 8.9) 1.0 43.44 43.444 49.9012 3.548e-12 ***
s(creatinine, 1.8) 1.0 16.08 16.077 18.4669 1.944e-05 ***
s(fasting, 4.6) 1.0 10.71 10.709 12.3004 0.0004784 ***
anemia 1.0 15.69 15.693 18.0257 2.438e-05 ***
sex 1.0 2.71 2.713 3.1165 0.0778910 .
factor(smoking) 2.0 7.44 3.720 4.2725 0.0142708 *
Residuals 790.7 688.38 0.871
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Anova for Nonparametric Effects
Npar Df Npar Chisq P(Chi)
(Intercept)
s(age, 8.9) 7.9 21.6053 0.005358 **
s(creatinine, 1.8) 0.8 3.2991 0.050784 .
s(fasting, 4.6) 3.6 9.3431 0.040178 *
anemia
sex
factor(smoking)
> or1 <- flexOR(data = heart2, response = "exitus",
formula = ~s(age, 8.9) + s(creatinine, 1.8) + s(fasting, 4.6) +
anemia + sex + factor(smoking))
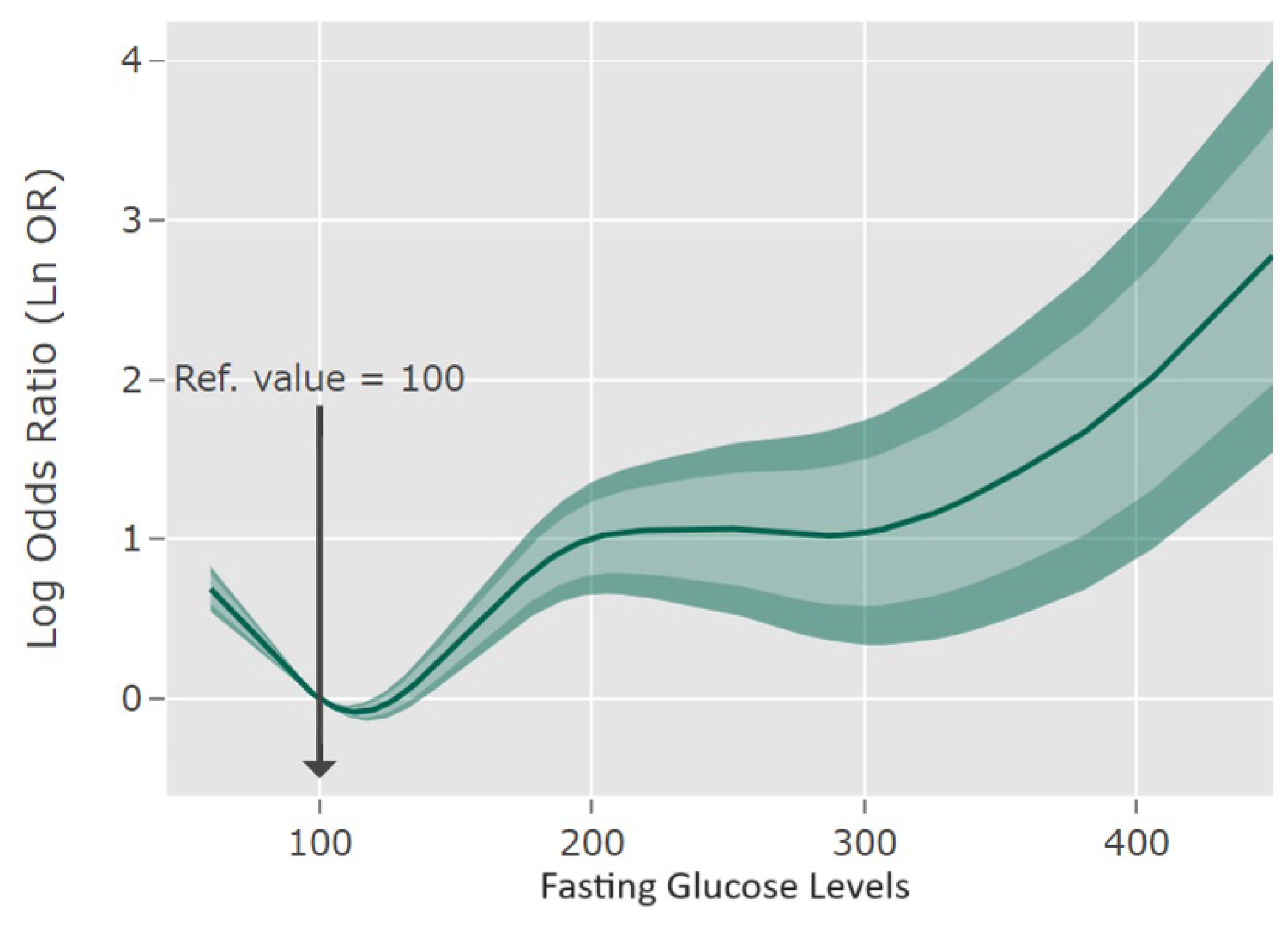
> plot(
x = or1,
predictor = "fasting",
ref.value = 100,
ref.label = "Ref. value",
col.area = c("grey75", "grey90"),
main = " ",
xlab = "Fasting blood glucose levels",
ylab = "Log Odds Ratio (Ln OR)",
lty = c(1,2,2,3,3),
ylog = TRUE,
round.x = 1,
conf.level = c(0.8, 0.95)
)
> library(plotly)
> p <- plot(
x = or1,
predictor = "fasting",
ref.value = 100,
ref.label = "Reference Label",
main = "Smooth odds ratio for Fasting blood glucose",
xlab = "Fasting blood glucose levels",
ylab = "Log Odds Ratio (Ln OR)",
lty = c(1,2,2,3,3),
xlim = c(60, 450),
round.x = 1,
conf.level = c(0.8, 0.95)
)
> tmat <- p$estimates
> xref <- p$xref
> mdata <- or1$dataset
> jj <- match(sort(unique(mdata$fasting)), mdata$fasting)
# Plotly to get shaded (two-levels) confidence bands
> fig <- plot_ly(x=mdata$fasting[jj], y=tmat[jj,5],
type = ’scatter’, mode = ’lines’,
line = list(color = ’transparent’),
showlegend = FALSE, name = ’80%UCI’)
> fig <- fig %>% add_trace(y = ~tmat[jj,3], type = ’scatter’,
mode = ’lines’,
fill = ’tonexty’, fillcolor = ’rgba(0,100,80,0.3)’,
line = list(color = ’transparent’),
showlegend = FALSE, name = ’95%UCI’)
> fig <- fig %>% add_trace(y = ~tmat[jj,2], type = ’scatter’,
mode = ’lines’,
fill = ’tonexty’, fillcolor=’rgba(0,100,80,0.3)’,
line = list(color = ’transparent’),
showlegend = FALSE, name = ’95%LCI’)
> fig <- fig %>% add_trace(y = ~tmat[jj,4], type = ’scatter’,
mode = ’lines’,
fill = ’tonexty’, fillcolor=’rgba(0,100,80,0.3)’,
line = list(color = ’transparent’),
showlegend = FALSE, name = ’80%LCI’)
> fig <- fig %>% add_trace(y = ~tmat[jj,1], type = ’scatter’,
mode = ’lines’,
line = list(color=’rgb(0,100,80)’),
showlegend = FALSE, name = ’LnOR’)
> fig <- fig %>% add_annotations( x = xref,
y = floor_to(min(tmat[jj,]), to=0.5),
xref = "x", yref = "y",
axref = "x", ayref = "y",
text = paste("Ref. value =",xref),
showarrow = T,
ax = xref,
ay = max(tmat[jj,])/2)
> fig <- fig %>% layout(#title = "",
plot_bgcolor=’rgb(229,229,229)’,
xaxis = list(title = "Fasting glucose levels",
gridcolor = ’rgb(255,255,255)’,
showgrid = TRUE,
showline = FALSE,
showticklabels = TRUE,
tickcolor = ’rgb(127,127,127)’,
ticks = ’outside’,
zeroline = FALSE),
yaxis = list(title = "Log Odds Ratio (Ln OR)",
gridcolor = ’rgb(255,255,255)’,
showgrid = TRUE,
showline = FALSE,
showticklabels = TRUE,
tickcolor = ’rgb(127,127,127)’,
ticks = ’outside’,
#range = c(-0.5,3.5),
zeroline = FALSE))
> fig
> pdval <- c (70, 80, 90, 100, 110, 120, 140, 180, 250, 400)
> predict(or1, predictor = "fasting", ref.value = 100, conf.level = 0.95,
prediction.values = pdval, ref.label = "Ref.")
Ref. LnOR lower .95 upper .95
70 0.48905582 0.3837869 0.59432470
80 0.31326688 0.2430876 0.38344614
90 0.14227425 0.1071846 0.17736388
100 0.00000000 0.0000000 0.00000000
110 -0.07323341 -0.1083230 -0.03814378
120 -0.06143541 -0.1316147 0.00874384
140 0.16943030 0.0290718 0.30978881
180 0.81032300 0.5296060 1.09104001
250 1.07569430 0.5493499 1.60203870
400 1.89278576 0.8400970 2.94547454
4.2. Pima Indians Diabetes Database
> data(PimaIndiansDiabetes2, package="mlbench")
> df2 <- dfgam(response="diabetes",
nl.predictors=c("age","mass"),
other.predictors=c("pedigree"),
smoother="s",
method="AIC",
data = PimaIndiansDiabetes2)
> df2$df
df
age 3.3
mass 4.1
> m2 <- gam(diabetes ~ s(age, df=3.3) + s(mass, df=4.1) + pedigree,
data=PimaIndiansDiabetes2, family=binomial)
> or2 <- flexOR(data = PimaIndiansDiabetes2,
response = "diabetes",
formula = ~s(age, 3.3) + s(mass, 4.1) + pedigree)
> plot(
x = or2,
predictor = "mass",
ref.value = 40,
ref.label = "Ref. value",
col.area = c("grey75", "grey90"),
main = " ",
xlab = "Body mass index",
ylab = "Log Odds Ratio (Ln OR)",
lty = c(1,2,2,3,3),
round.x = 1,
conf.level = c(0.8, 0.95)
)
> pdval <- c (20, 25, 30, 35, 40, 45, 50, 55, 60, 65)
> predict(or2, predictor = "mass", ref.value = 40, conf.level = 0.95,
prediction.values = pdval, ref.label = "Ref.")
Ref. LnOR lower .95 upper .95
20 -3.20826636 -3.7680373 -2.64849542
25 -1.61356211 -2.0333903 -1.19373390
30 -0.40263002 -0.6825155 -0.12274455
35 -0.07505977 -0.2150025 0.06488297
40 0.00000000 0.0000000 0.00000000
45 0.45600760 0.3160649 0.59595034
50 1.05087046 0.7709850 1.33075593
55 1.63284725 1.2130190 2.05267546
60 2.21353442 1.6537635 2.77330536
65 2.83405429 2.1343406 3.53376797
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Royston, P.; Altman, D.G.; Sauerbrei, W. Dichotomizing continuous predictors in multiple regression: A bad idea. Stat. Med. 2006, 25, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Barrio, I.; Rodríguez-Álvarez, M.; Arostegui, I. Categorisation of continuous variables in a logistic regression model using the R package CatPredi. In Proceedings of the MOL2NET’15, Conference on Molecular, Biomedical, and Computational & Network Science and Engineering, Athens, Greece, 5–15 December 2015; MDPI: Basel, Switzerland, 2015. [Google Scholar]
- Barrio, I.; Arostegui, I.; Rodríguez-Álvarez, M.-X.; Quintana, J.-M. A new approach to categorising continuous variables in prediction models: Proposal and validation. Stat. Methods Med. Res. 2017, 26, 2586–2602. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.J.; Tibshirani, R.J. Generalized Additive Models; Chapman & Hall/CRC: New York, NY, USA, 1990. [Google Scholar]
- Wood, S. Generalized Additive Models: An Introduction with R; Chapman & Hall/CRC: London, UK, 2017. [Google Scholar]
- Wahba, G. Spline Models for Observational Data; SIAM: Philadelphia, PA, USA, 1990. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Simonoff, J.S.; Tsai, C.L. Smoothing parameter selection in nonparametric regression using an improved akaike information criterion. J. R. Stat. Soc. Ser. B 1998, 60, 271–293. [Google Scholar] [CrossRef]
- Schwarz, G.E. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Cadarso-Suárez, C.; Meira-Machado, L.; Kneib, T.; Gude, F. Flexible hazard ratio curves for continuous predictors in multi-state models. Stat. Model. 2010, 10, 291–314. [Google Scholar] [CrossRef]
- Meira-Machado, L.; Cadarso-Suárez, C.; Araújo, A.; Gude, F. smoothHR: An R Package for Pointwise Nonparametric Estimation of Hazard Ratio Curves of Continuous Predictors. Comput. Math. Methods Med. 2013, 2013, 745742. [Google Scholar] [CrossRef] [PubMed]
- de Boor, C. A Practical Guide to Splines (Rev. Edn); Springer: New York, NY, USA, 2001. [Google Scholar]
- Figueiras, A.; Cadarso-Suárez, C. Application of nonparametric models for calculating odds-ratios and their confidence intervals for continuous exposures. Am. J. Epidemiol. 2001, 154, 264–275. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: http://www.R-project.org/ (accessed on 27 April 2024).


| Function | Description |
|---|---|
| AICc | Calculates AICc, the Akaike Information Criterion corrected for small sample sizes, for Generalized Additive Models. |
| floor_to | Takes a numeric value or vector and rounds it down to the nearest multiple of a specified base. |
| dfgam | Calculates the degrees of freedom for specified non-linear predictors in a GAM model. |
| flexOR | Computes odds ratios and CIs for predictors in GAM models. |
| plot.OR | Plots smooth odds ratios along with confidence intervals for a specified predictor. For an object of class OR. |
| predict.OR | Predicts values using a fitted OR model. |
| Covariates | AIC | GCV.Cp | REML |
|---|---|---|---|
| Age | 8.9 | 6.97 | 3.34 |
| Creatinine | 1.8 | 1.79 | 2.06 |
| Fasting | 4.6 | 4.37 | 3.47 |
| Covariates | AIC | GCV.Cp | REML |
|---|---|---|---|
| Age | 3.30 | 3.30 | 3.44 |
| Mass | 4.10 | 3.96 | 4.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azevedo, M.; Meira-Machado, L.; Gude, F.; Araújo, A. Pointwise Nonparametric Estimation of Odds Ratio Curves with R: Introducing the flexOR Package. Appl. Sci. 2024, 14, 3897. https://doi.org/10.3390/app14093897
Azevedo M, Meira-Machado L, Gude F, Araújo A. Pointwise Nonparametric Estimation of Odds Ratio Curves with R: Introducing the flexOR Package. Applied Sciences. 2024; 14(9):3897. https://doi.org/10.3390/app14093897
Chicago/Turabian StyleAzevedo, Marta, Luís Meira-Machado, Francisco Gude, and Artur Araújo. 2024. "Pointwise Nonparametric Estimation of Odds Ratio Curves with R: Introducing the flexOR Package" Applied Sciences 14, no. 9: 3897. https://doi.org/10.3390/app14093897
APA StyleAzevedo, M., Meira-Machado, L., Gude, F., & Araújo, A. (2024). Pointwise Nonparametric Estimation of Odds Ratio Curves with R: Introducing the flexOR Package. Applied Sciences, 14(9), 3897. https://doi.org/10.3390/app14093897






