A Novel Method Combining U-Net with LSTM for Three-Dimensional Soil Pore Segmentation Based on Computed Tomography Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of Soil CT Image Datasets
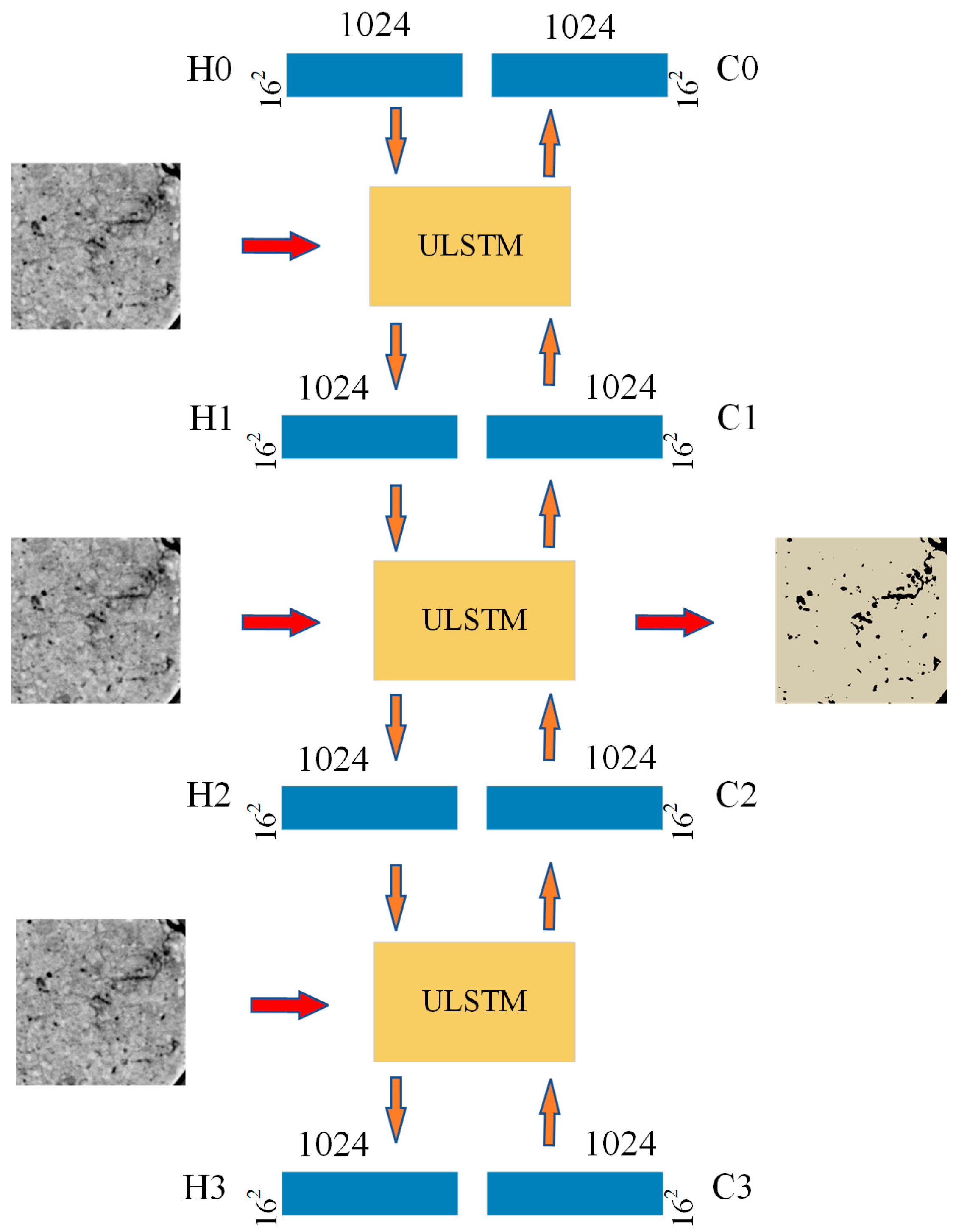
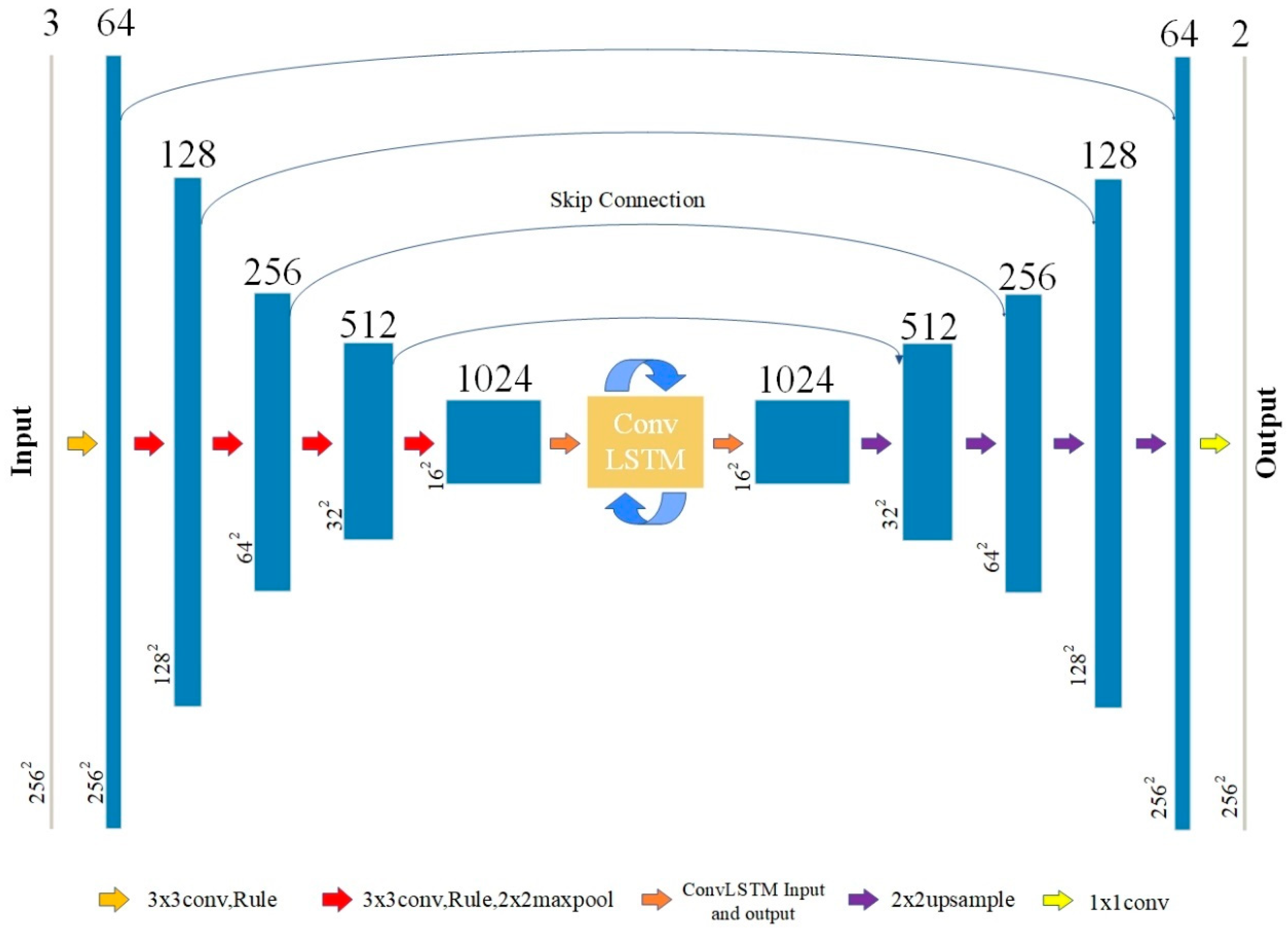
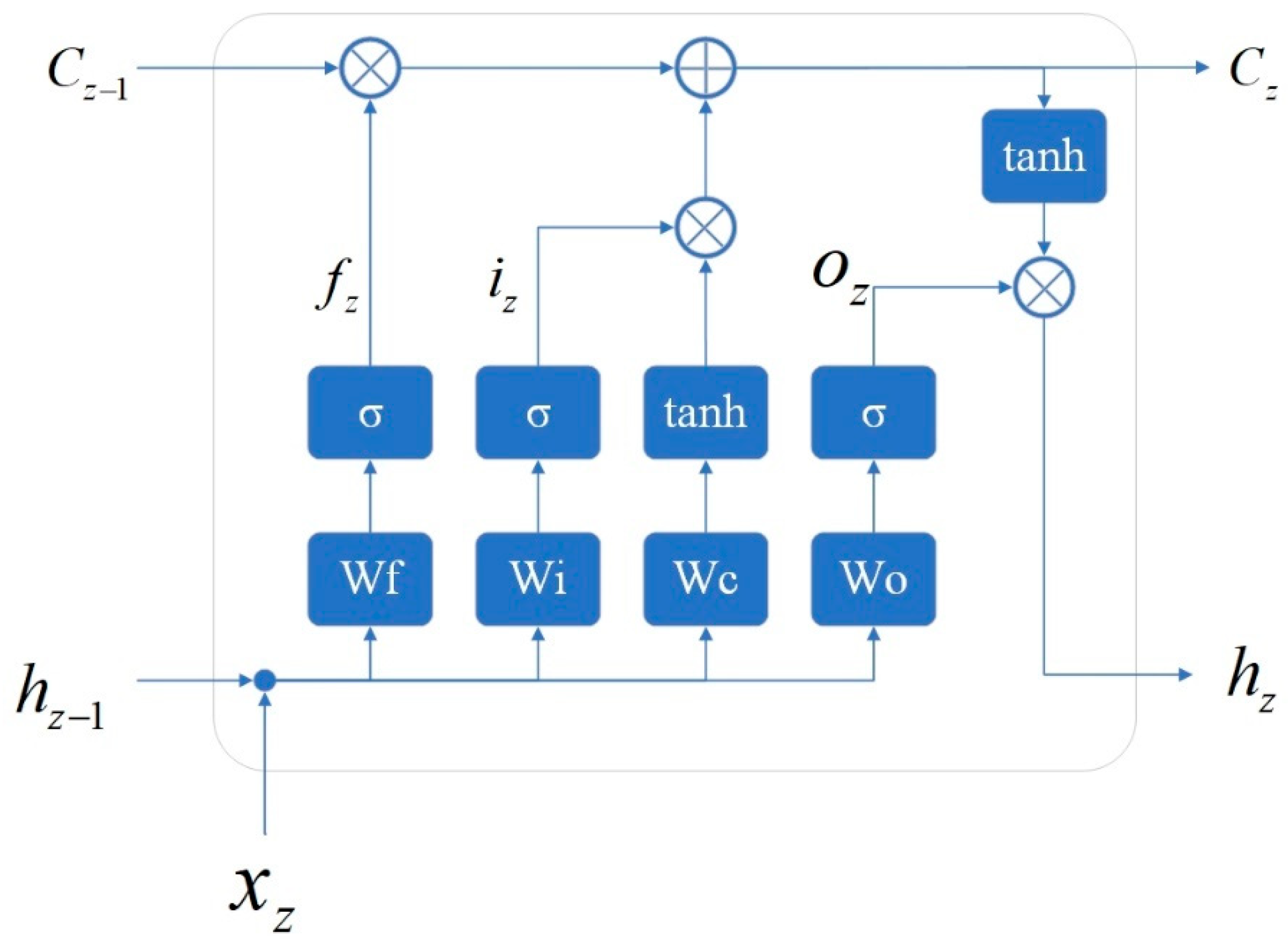
2.2. Neural Network Architecture of the Proposed Method
2.3. Evaluation Metrics
3. Results and Discussion
3.1. Experimental Details
3.2. Effects of Different Numbers of Image Slices
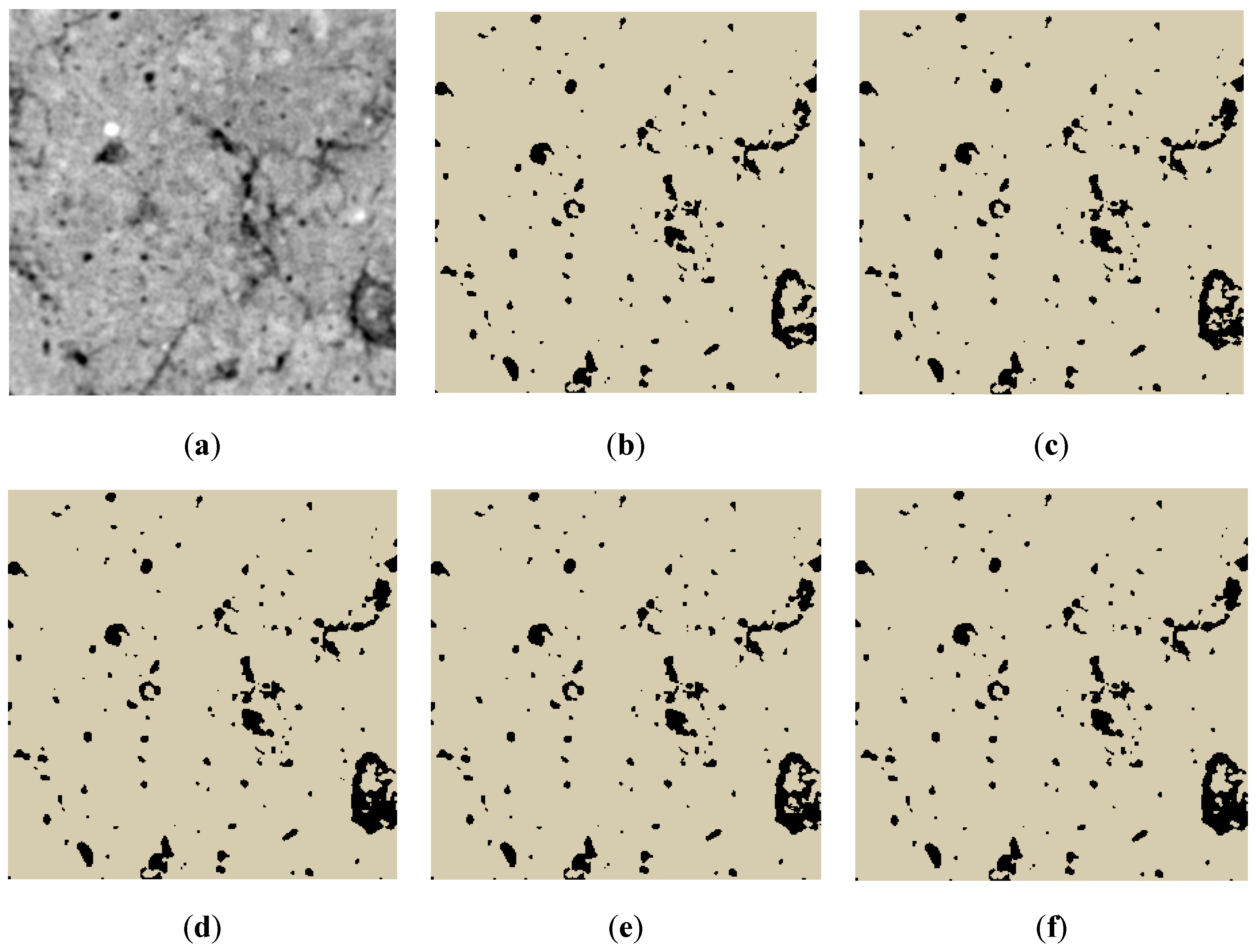
3.3. Qualitative Results
3.4. Quantitative Evaluations
3.5. Neural Network Complexity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borrelli, P.; Robinson, D.A.; Panagos, P.; Lugato, E.; Yang, J.E.; Alewell, C.; Wuepper, D.; Montanarella, L.; Ballabio, C. Land use and climate change impacts on global soil erosion by water (2015–2070). Proc. Natl. Acad. Sci. USA 2020, 117, 21994–22001. [Google Scholar] [CrossRef] [PubMed]
- Baveye, P.C.; Balseiro-Romero, M.; Bottinelli, N.; Briones, M.; Capowiez, Y.; Garnier, P.; Kravchenko, A.; Otten, W.; Pot, V.; Schlüter, S. Lessons from a landmark 1991 article on soil structure: Distinct precedence of non-destructive assessment and benefits of fresh perspectives in soil research. Soil Res. 2022, 60, 321–336. [Google Scholar] [CrossRef]
- Haubitz, B.; Prokop, M.; Döhring, W.; Ostrom, J.; Wellnhofer, P. Computed tomography of Archaeopteryx. Paleobiology 1988, 14, 206–213. [Google Scholar] [CrossRef]
- Xiong, P.; Zhang, Z.; Hallett, P.D.; Peng, X. Variable responses of maize root architecture in elite cultivars due to soil compaction and moisture. Plant Soil 2020, 455, 79–91. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Q.; Wang, X.; Ji, X.; Du, Y. A 3D reconstruction method of porous media based on improved WGAN-GP. Comput. Geosci. 2022, 165, 105151. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhou, X.-P. An integrated method for 3D reconstruction model of porous geomaterials through 2D CT images. Comput. Geosci. 2019, 123, 83–94. [Google Scholar] [CrossRef]
- Pereira, M.F.L.; Cruvinel, P.E. A model for soil computed tomography based on volumetric reconstruction, Wiener filtering and parallel processing. Comput. Electron. Agric. 2015, 111, 151–163. [Google Scholar] [CrossRef]
- Xu, J.; Ren, C.; Wang, S.; Gao, J.; Zhou, X. Permeability and microstructure of a saline intact loess after dry-wet cycles. Adv. Civil Eng. 2021, 2021, 6653697. [Google Scholar] [CrossRef]
- Zhang, X.; Neal, A.L.; Crawford, J.W.; Bacq-Labreuil, A.; Akkari, E.; Rickard, W. The effects of long-term fertilizations on soil hydraulic properties vary with scales. J. Hydrol. 2021, 593, 125890. [Google Scholar] [CrossRef] [PubMed]
- Gattullo, C.E.; Allegretta, I.; Porfido, C.; Rascio, I.; Spagnuolo, M.; Terzano, R. Assessing chromium pollution and natural stabilization processes in agricultural soils by bulk and micro X-ray analyses. Environ. Sci. Pollut. Res. 2020, 27, 22967–22979. [Google Scholar] [CrossRef] [PubMed]
- Scotson, C.P.; Duncan, S.J.; Williams, K.A.; Ruiz, S.A.; Roose, T. X-ray computed tomography imaging of solute movement through ridged and flat plant systems. Eur. J. Soil Sci. 2021, 72, 198–214. [Google Scholar] [CrossRef]
- Tang, C.-S.; Lin, L.; Cheng, Q.; Zhu, C.; Wang, D.-W.; Lin, Z.-Y.; Shi, B. Quantification and characterizing of soil microstructure features by image processing technique. Comput. Geotech. 2020, 128, 103817. [Google Scholar] [CrossRef]
- Meng, C.; Niu, J.; Yu, H.; Du, L.; Yin, Z. Research progress in influencing factors and measuring methods of three-dimensional characteristics of soil macropores. J. Beijing For. Univ. 2020, 42, 9–16. [Google Scholar]
- Karimpouli, S.; Tahmasebi, P. Segmentation of digital rock images using deep convolutional autoencoder networks. Comput. Geosci. 2019, 126, 142–150. [Google Scholar] [CrossRef]
- Tempelaere, A.; Phan, H.M.; Van De Looverbosch, T.; Verboven, P.; Nicolai, B. Non-destructive internal disorder segmentation in pear fruit by X-ray radiography and AI. Comput. Electron. Agric. 2023, 212, 108142. [Google Scholar] [CrossRef]
- Van De Looverbosch, T.; Vandenbussche, B.; Verboven, P.; Nicolaï, B. Nondestructive high-throughput sugar beet fruit analysis using X-ray CT and deep learning. Comput. Electron. Agric. 2022, 200, 107228. [Google Scholar] [CrossRef]
- Xiberta, P.; Boada, I.; Bardera, A.; Font-i-Furnols, M. A semi-automatic and an automatic segmentation algorithm to remove the internal organs from live pig CT images. Comput. Electron. Agric. 2017, 140, 290–302. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Wu, G.; Suk, H.-I. Deep learning in medical image analysis. Annu.Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Wieland, R.; Ukawa, C.; Joschko, M.; Krolczyk, A.; Fritsch, G.; Hildebrandt, T.B.; Schmidt, O.; Filser, J.; Jimenez, J.J. Use of deep learning for structural analysis of computer tomography images of soil samples. R. Soc. Open Sci. 2021, 8, 201275. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Zhang, H.; Meng, D.; Zheng, Y. InDuDoNet+: A deep unfolding dual domain network for metal artifact reduction in CT images. Med. Image Anal. 2023, 85, 102729. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Nwigbo, K.T.; Michetti, J.; Georgeot, B.; Pham, D.H.; Kouamé, D.; Basarab, A. Computed Tomography Image Restoration Using a Quantum-Based Deep Unrolled Denoiser and a Plug-and-Play Framework. In Proceedings of the 2023 31st European Signal Processing Conference (EUSIPCO), Helsinki, Finland, 4–8 September 2023; pp. 845–849. [Google Scholar]
- Saxena, N.; Day-Stirrat, R.J.; Hows, A.; Hofmann, R. Application of deep learning for semantic segmentation of sandstone thin sections. Comput. Geosci. 2021, 152, 104778. [Google Scholar] [CrossRef]
- Liang, J.; Sun, Y.; Lebedev, M.; Gurevich, B.; Nzikou, M.; Vialle, S.; Glubokovskikh, S. Multi-mineral segmentation of micro-tomographic images using a convolutional neural network. Comput. Geosci. 2022, 168, 105217. [Google Scholar] [CrossRef]
- Han, Q.; Zhao, Y.; Liu, L.; Chen, Y.; Zhao, Y. A simplified convolutional network for soil pore identification based on computed tomography imagery. Soil Sci. Soc. Am. J. 2019, 83, 1309–1318. [Google Scholar] [CrossRef]
- Bai, H.; Liu, L.; Han, Q.; Zhao, Y.; Zhao, Y. A novel UNet segmentation method based on deep learning for preferential flow in soil. Soil Till. Res. 2023, 233, 105792. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. pp. 234–241. [Google Scholar]
- Zhang, Y.; He, Z.; Jiang, R.; Liao, L.; Meng, Q. Improved Computer Vision Framework for Mesoscale Simulation of Xiyu Conglomerate Using the Discrete Element Method. Appl. Sci. 2023, 13, 13000. [Google Scholar] [CrossRef]
- Roslin, A.; Marsh, M.; Provencher, B.; Mitchell, T.; Onederra, I.; Leonardi, C. Processing of micro-CT images of granodiorite rock samples using convolutional neural networks (CNN), Part II: Semantic segmentation using a 2.5 D CNN. Miner. Eng. 2023, 195, 108027. [Google Scholar] [CrossRef]
- Phan, J.; Ruspini, L.C.; Lindseth, F. Automatic segmentation tool for 3D digital rocks by deep learning. Sci. Rep. 2021, 11, 19123. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Proceedings, Part II 19. pp. 424–432. [Google Scholar]
- Vu, M.H.; Grimbergen, G.; Simkó, A.; Nyholm, T.; Löfstedt, T. End-to-End Cascaded U-Nets with a Localization Network for Kidney Tumor Segmentation. arXiv 2019, arXiv:1910.07521. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Kitrungrotsakul, T.; Iwamoto, Y.; Han, X.-H.; Takemoto, S.; Yokota, H.; Ipponjima, S.; Nemoto, T.; Wei, X.; Chen, Y.-W. A cascade of CNN and LSTM network with 3D anchors for mitotic cell detection in 4D microscopic image. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1239–1243. [Google Scholar]
- Novikov, A.A.; Major, D.; Wimmer, M.; Lenis, D.; Bühler, K. Deep sequential segmentation of organs in volumetric medical scans. IEEE Trans. Med. Imaging 2018, 38, 1207–1215. [Google Scholar] [CrossRef]
- Stollenga, M.F.; Byeon, W.; Liwicki, M.; Schmidhuber, J. Parallel multi-dimensional LSTM, with application to fast biomedical volumetric image segmentation. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; Volume 28. [Google Scholar]
- Chen, J.; Yang, L.; Zhang, Y.; Alber, M.; Chen, D.Z. Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–12 December 2016; Volume 29. [Google Scholar]
- Ganaye, P.-A.; Sdika, M.; Triggs, B.; Benoit-Cattin, H. Removing segmentation inconsistencies with semi-supervised non-adjacency constraint. Med. Image Anal. 2019, 58, 101551. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Zhao, Y.; Xia, X.; Chen, X. Effects of freeze-thaw cycles on black soil structure at different size scales. Acta Ecol. Sin. 2014, 34, 6287–6296. [Google Scholar]
- Zhao, Y.; Han, Q.; Zhao, Y.; Liu, J. Soil pore identification with the adaptive fuzzy C-means method based on computed tomography images. J. For. Res. 2019, 30, 1043–1052. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, H.; Yeung, D.-Y.; Wong, W.-K.; Woo, W.-C. Convolutional LSTM network: A machine learning approach for precipitation nowcasting. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; Volume 28. [Google Scholar]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2013, 33, 577–590. [Google Scholar] [CrossRef] [PubMed]

| Soil Type | Number of Images | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| Primary | 3 | 96.51 ± 1.02 × 10−2 | 96.54 ± 1.55 × 10−2 | 68.02 ± 7.54 × 10−1 | 79.48 ± 2.93 × 10−1 |
| 5 | 96.47 ± 1.15 × 10−2 | 98.23 ± 9.25 × 10−3 | 66.37 ± 8.28 × 10−1 | 78.85 ± 3.53 × 10−1 | |
| 7 | 96.49 ± 1.01 × 10−2 | 96.40 ± 1.45 × 10−2 | 67.96 ± 7.64 × 10−1 | 79.38 ± 2.92 × 10−1 | |
| 9 | 96.48 ± 1.07 × 10−2 | 96.83 ± 1.31 × 10−2 | 67.51 ± 8.03 × 10−1 | 79.20 ± 3.17 × 10−1 | |
| Frozen | 3 | 98.72 ± 3.44 × 10−3 | 96.59 ± 8.08 × 10−2 | 86.80 ± 7.93 × 10−1 | 91.08 ± 1.68 × 10−1 |
| 5 | 98.88 ± 1.11 × 10−3 | 93.49 ± 2.16 × 10−1 | 92.15 ± 4.51 × 10−1 | 92.50 ± 5.08 × 10−2 | |
| 7 | 98.80 ± 9.14 × 10−4 | 92.08 ± 3.08 × 10−1 | 92.76 ± 4.05 × 10−1 | 92.07 ± 4.25 × 10−2 | |
| 9 | 98.87 ± 1.40 × 10−3 | 94.08 ± 2.29 × 10−1 | 91.48 ± 5.40 × 10−1 | 92.40 ± 6.85 × 10−2 | |
| Thawed | 3 | 98.93 ± 2.94 × 10−3 | 90.74 ± 1.90 × 10−1 | 92.68 ± 6.16 × 10−1 | 91.46 ± 1.76 × 10−1 |
| 5 | 98.89 ± 3.03 × 10−3 | 90.11 ± 1.97 × 10−1 | 92.74 ± 6.14 × 10−1 | 91.17 ± 1.80 × 10−1 | |
| 7 | 98.95 ± 3.03 × 10−3 | 90.83 ± 2.05 × 10−1 | 92.85 ± 6.09 × 10−1 | 91.59 ± 1.81 × 10−1 | |
| 9 | 98.78 ± 3.28 × 10−3 | 88.30 ± 2.28 × 10−1 | 93.02 ± 6.09 × 10−1 | 90.37 ± 1.91 × 10−1 |
| Model | Soil Type | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| CLSTM | Primary | 96.56 ± 9.35 × 10−3 | 93.40 ± 3.50 × 10−1 | 71.94 ± 1.31 | 80.45 ± 3.10 × 10−1 |
| Frozen | 97.79 ± 7.12 × 10−3 | 99.10 ± 9.05 × 10−3 | 61.92 ± 2.74 | 74.89 ± 1.61 | |
| Thawed | 97.71 ± 8.16 × 10−3 | 99.88 ± 2.80 × 10−4 | 63.79 ± 1.87 | 77.04 ± 9.82 × 10−1 | |
| U-Net | Primary | 96.57 ± 1.19 × 10−2 | 94.66 ± 4.33 × 10−1 | 70.88 ± 3.19 × 10−1 | 80.20 ± 1.48 |
| Frozen | 97.28 ± 1.85 × 10−2 | 74.79 ± 2.86 × 10−1 | 99.55 ± 6.44 × 10−1 | 85.16 ± 7.31 × 10−3 | |
| Thawed | 98.47 ± 4.59 × 10−3 | 99.94 ± 2.31 × 10−1 | 77.73 ± 4.89 × 10−4 | 87.25 ± 5.80 × 10−1 | |
| U-Net–LSTM | Primary | 96.57 ± 1.57 × 10−2 | 96.95 ± 1.37 × 10−1 | 68.86 ± 1.69 | 79.67 ± 6.32 × 10−1 |
| Frozen | 98.43 ± 1.08 × 10−2 | 98.71 ± 1.68 × 10−2 | 73.03 ± 3.88 | 82.38 ± 1.70 | |
| Thawed | 98.50 ± 7.46 × 10−3 | 99.65 ± 4.95 × 10−3 | 76.49 ± 1.92 | 85.84 ± 7.24 × 10−1 | |
| BDULSTM | Primary | 96.49 ± 1.01 × 10−2 | 96.40 ± 1.45 × 10−2 | 67.96 ± 7.64 × 10−1 | 79.38 ± 2.92 × 10−1 |
| Frozen | 98.80 ± 9.14 × 10−4 | 92.08 ± 3.08 × 10−1 | 92.76 ± 4.05 × 10−1 | 92.07 ± 4.25 × 10−2 | |
| Thawed | 98.95 ± 3.03 × 10−3 | 90.83 ± 2.05 × 10−1 | 92.85 ± 6.09 × 10−1 | 91.59 ± 1.81 × 10−1 |
| Models | Parameters (M) | FLOPs (G) | Memory (M) |
|---|---|---|---|
| CLSTM | 0.55 | 10.85 | 32.87 |
| U-Net | 7.41 | 11.60 | 202.75 |
| U-Net–LSTM | 51.90 | 38.47 | 442.50 |
| BDULSTM | 50.82 | 32.87 | 409.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Han, Q.; Zhao, Y.; Zhao, Y. A Novel Method Combining U-Net with LSTM for Three-Dimensional Soil Pore Segmentation Based on Computed Tomography Images. Appl. Sci. 2024, 14, 3352. https://doi.org/10.3390/app14083352
Liu L, Han Q, Zhao Y, Zhao Y. A Novel Method Combining U-Net with LSTM for Three-Dimensional Soil Pore Segmentation Based on Computed Tomography Images. Applied Sciences. 2024; 14(8):3352. https://doi.org/10.3390/app14083352
Chicago/Turabian StyleLiu, Lei, Qiaoling Han, Yue Zhao, and Yandong Zhao. 2024. "A Novel Method Combining U-Net with LSTM for Three-Dimensional Soil Pore Segmentation Based on Computed Tomography Images" Applied Sciences 14, no. 8: 3352. https://doi.org/10.3390/app14083352
APA StyleLiu, L., Han, Q., Zhao, Y., & Zhao, Y. (2024). A Novel Method Combining U-Net with LSTM for Three-Dimensional Soil Pore Segmentation Based on Computed Tomography Images. Applied Sciences, 14(8), 3352. https://doi.org/10.3390/app14083352






