Introducing an Artificial Neural Network for Virtually Increasing the Sample Size of Bioequivalence Studies
Abstract
1. Introduction
2. Materials and Methods
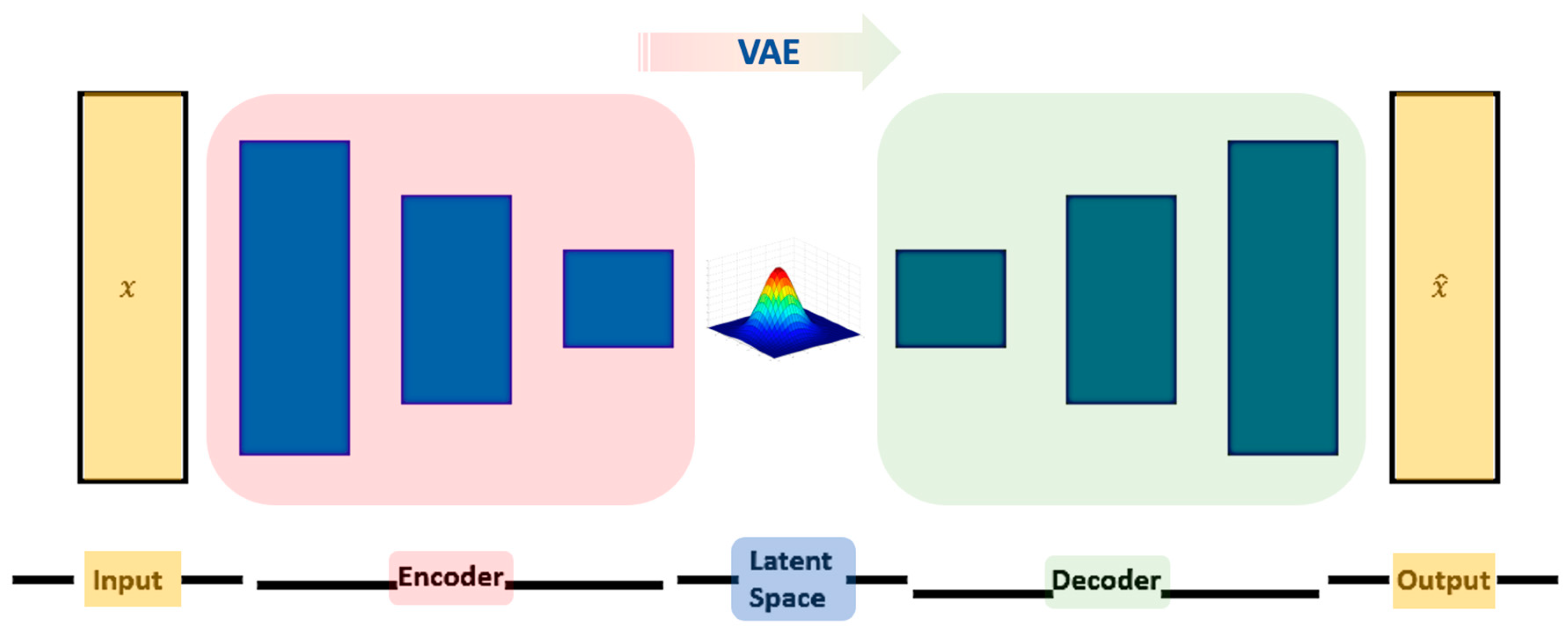
2.1. Variational Autoencoders
2.2. Tuning of Hyperparameters
2.3. Simulation Framework
- I.
- Ν individuals are randomly generated for both groups in the case of the first period:
- a.
- NT individuals for the T group for the first period, with mean μT1 and standard deviation σT1;
- b.
- NR individuals for the R group for the first period, with mean μR1 and standard deviation σR1;
- c.
- The T and R groups were set to have equal CVs;
- d.
- The sample sizes of T and R groups were assumed to be equal: NT = NR;
- e.
- Thus, the sample size of the study is: N = NT + NR.
- II.
- The N individuals from the first period are multiplied, with a randomly generated “stochastic term”, with mean μST and standard deviation σST. The stochastic term coefficient of variation (CVw) represents the “within subject variability” for each simulated volunteer between the period I and II of the crossover study [6]. It should be noted that the CVw is different to the between-subject variability (i.e., CV) discussed in step “Ic”.
- III.
- The individuals generated from steps I and II (termed as “original”) are then randomly subsampled with proportions 25%, 50%, and 75%. The so-derived groups are termed as “subsampled”.
- IV.
- Subsampled individuals are fed into an optimized VAE model to generate new individuals, termed as “generated”. The generated dataset was set to exhibit size equal to or double the “original” dataset.
- V.
- VI.
- The success (i.e., BE acceptance) or failure (i.e., non-equivalence) of the statistical test is tracked for all three datasets.
- VII.
- Steps “I–VI” are performed again for 500 repetitions in order to obtain robust values for the % BE acceptance.
- VIII.
- The results attained from step “VII” are evaluated.
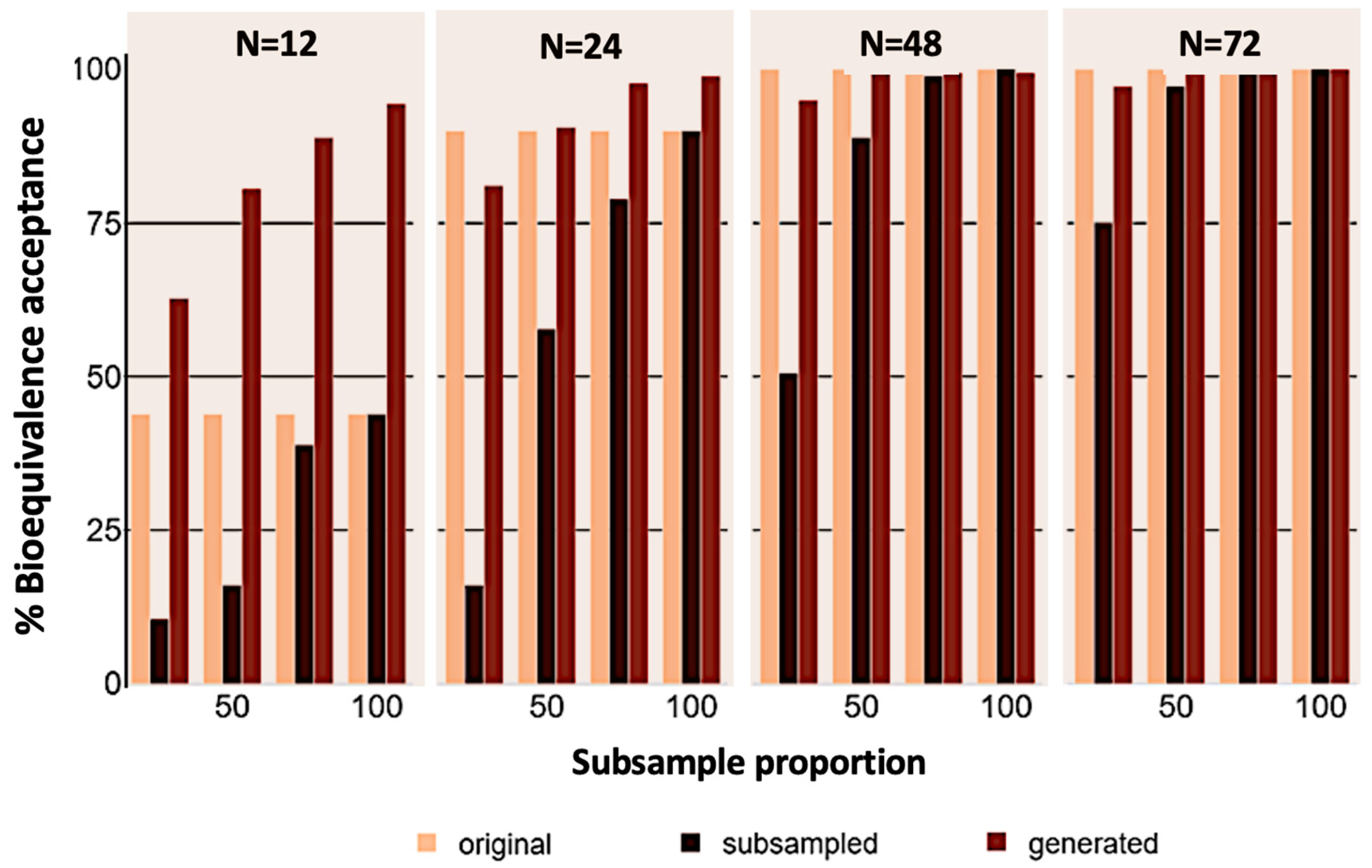
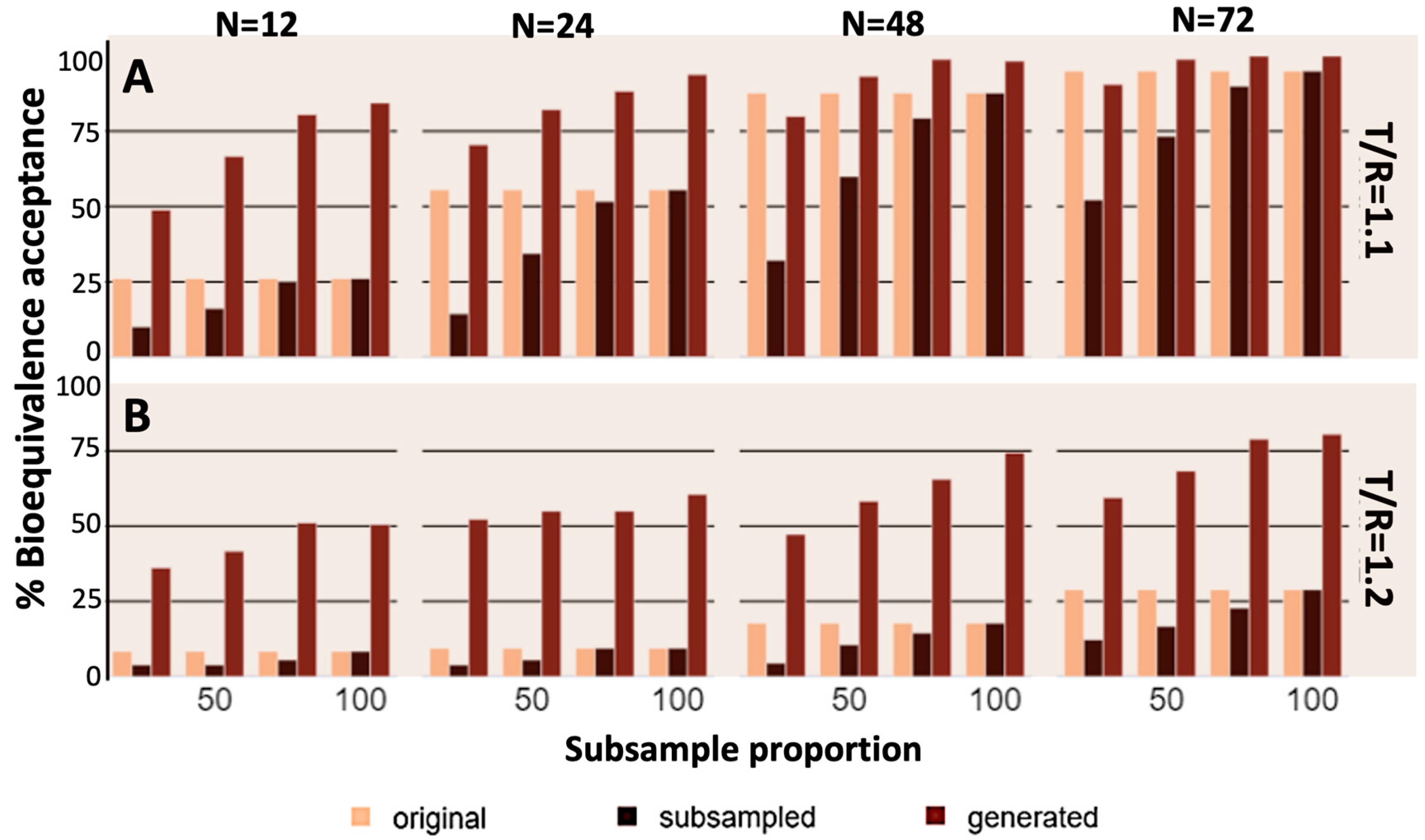
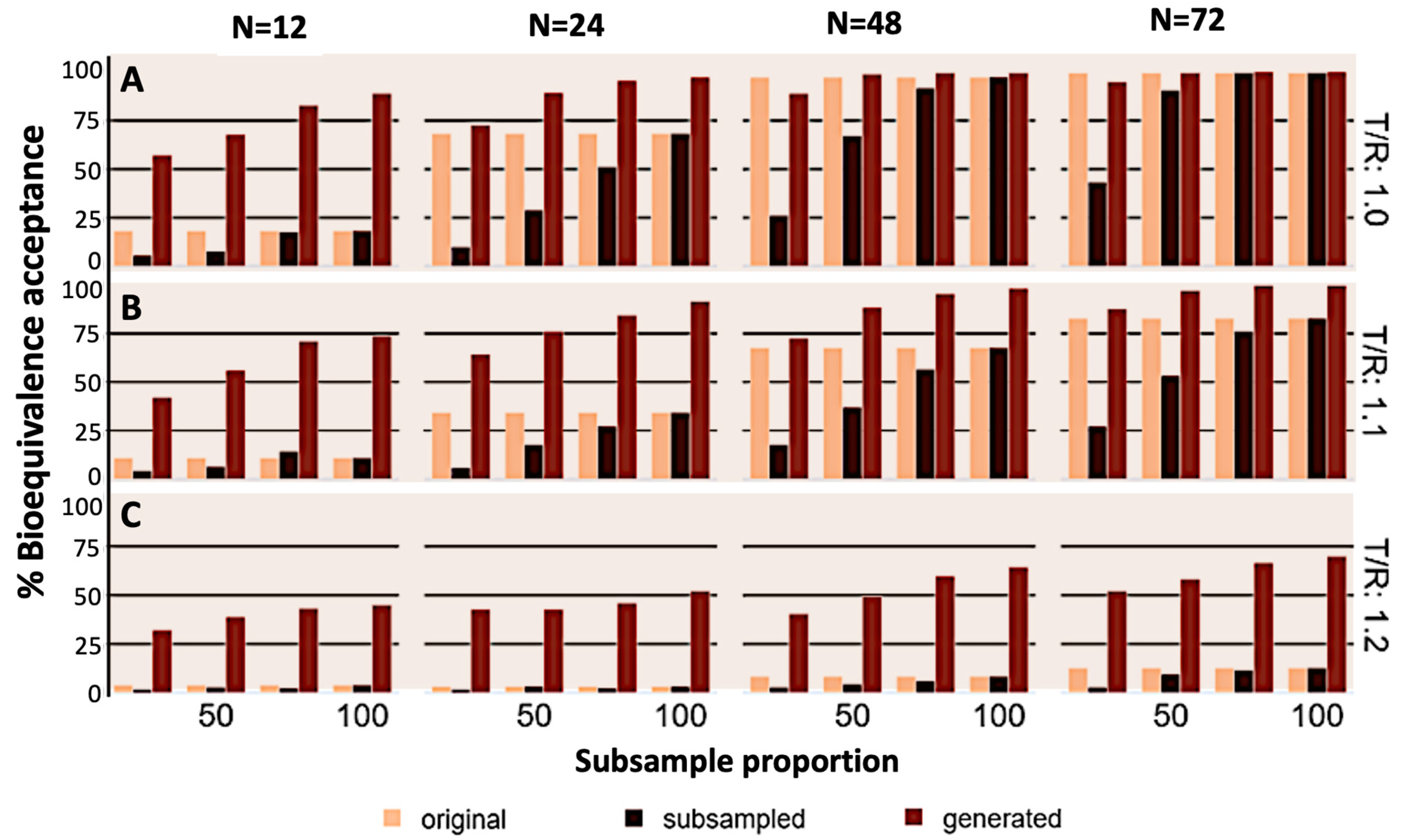
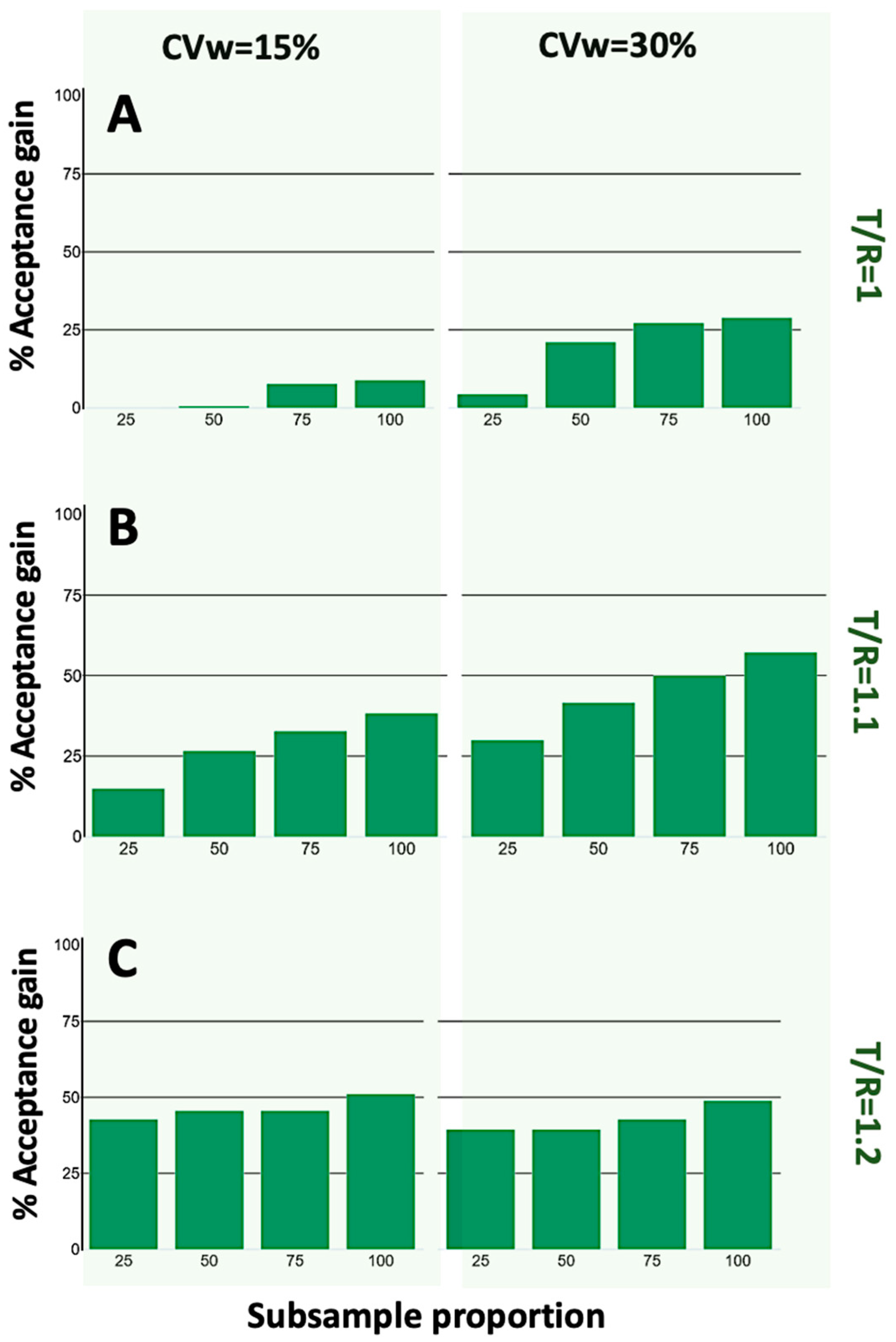
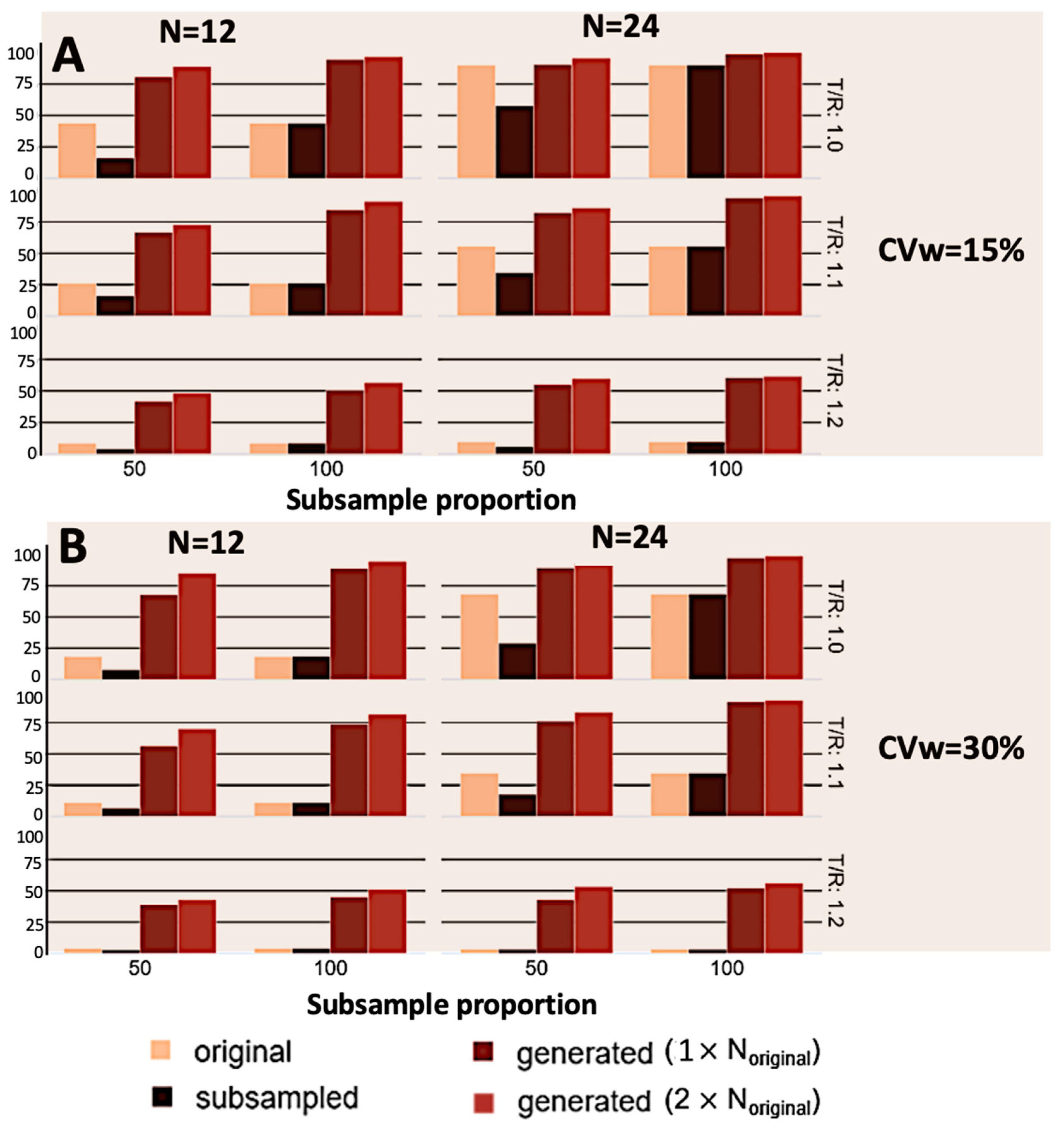
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, K.K.; Attri, J.P.; Singh, A.; Kaur, H.; Kaur, G. Basic Concepts for Sample Size Calculation: Critical Step for Any Clinical Trials. Saudi J. Anaesth. 2016, 10, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Sakpal, T.V. Sample Size Estimation in Clinical Trial. Perspect. Clin. Res. 2010, 1, 67–69. [Google Scholar] [PubMed]
- Wang, X.; Ji, X. Sample Size Estimation in Clinical Research: From Randomized Controlled Trials to Observational Studies. Chest 2020, 158, S12–S20. [Google Scholar] [CrossRef]
- European Medicines Agency; Committee for Medicinal Products for Human Use (CHMP). Guideline on the Investigation of Bioequivalence; CPMP/EWP/QWP/1401/98 Rev. 1/Corr**; Committee for Medicinal Products for Human Use (CHMP): London, UK, 20 January 2010; Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-bioequivalence-rev1_en.pdf (accessed on 23 January 2024).
- Food and Drug Administration (FDA). Guidance for Industry. Bioavailability and Bioequivalence Studies Submitted in NDAs or INDs—General Considerations. Draft Guidance. U.S. Department of Health and Human Services Food and Drug Administration. Center for Drug Evaluation and Research (CDER). December 2013. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioavailability-and-bioequivalence-studies-submitted-ndas-or-inds-general-considerations (accessed on 23 January 2024).
- Karalis, V. Modeling and Simulation in Bioequivalence. In Modeling in Biopharmaceutics, Pharmacokinetics and Pharmacodynamics. Homogeneous and Heterogeneous Approaches, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 227–255. [Google Scholar]
- Askin, S.; Burkhalter, D.; Calado, G.; El Dakrouni, S. Artificial Intelligence Applied to Clinical Trials: Opportunities and Challenges. Health Technol. 2023, 13, 203–213. [Google Scholar] [CrossRef]
- Harrer, S.; Shah, P.; Antony, B.; Hu, J. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci. 2019, 40, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Delso, G.; Cirillo, D.; Kaggie, J.D.; Valencia, A.; Metser, U.; Veit-Haibach, P. How to Design AI-Driven Clinical Trials in Nuclear Medicine. Semin. Nucl. Med. 2021, 51, 112–119. [Google Scholar] [CrossRef]
- Papadopoulos, D.; Karalis, V.D. Variational Autoencoders for Data Augmentation in Clinical Studies. Appl. Sci. 2023, 13, 8793. [Google Scholar] [CrossRef]
- Lim, C.-Y. Considerations for Crossover Design in Clinical Study. Korean J. Anesthesiol. 2021, 74, 293–299. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Z.; Su, Y.; Zhao, Q.; Li, X.; Ouyang, D. Deep Learning for in Vitro Prediction of Pharmaceutical Formulations. Acta Pharm. Sin. B 2019, 9, 177–185. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python, 2nd ed.; Manning; Simon and Schuster: New York, NY, USA, 2021. [Google Scholar]
- Atienza, R. Advanced Deep Learning with Keras: Apply Deep Learning Techniques, Autoencoders, GANs, Variational Autoencoders, Deep Reinforcement Learning, Policy Gradients, and More; Packt Publishing: Birmingham, UK, 2018. [Google Scholar]
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. Found. Trends Mach. Learn. 2019, 12, 307–392. [Google Scholar] [CrossRef]
- Endrenyi, L.; Tothfalusi, L. Bioequivalence for Highly Variable Drugs: Regulatory Agreements, Disagreements, and Harmonization. J. Pharmacokinet. Pharmacodyn. 2019, 46, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Karalis, V.D. Machine Learning in Bioequivalence: Towards Identifying an Appropriate Measure of Absorption Rate. Appl. Sci. 2022, 13, 418. [Google Scholar] [CrossRef]
- Karalis, V.D. On the Interplay between Machine Learning, Population Pharmacokinetics, and Bioequivalence to Introduce Average Slope as a New Measure for Absorption Rate. Appl. Sci. 2023, 13, 2257. [Google Scholar] [CrossRef]
- Galić, I.; Habijan, M. Deep Learning in Medical Image Analysis for Personalized Medicine. In Proceedings of the 2023 International Symposium ELMAR, Zadar, Croatia, 11–13 September 2023; IEEE: Piscataway, NJ, USA, 2023. [Google Scholar]
- Wei, Q.; Ma, S.; Tang, S.; Li, B.; Shen, J.; Xu, Y.; Fan, J. A Deep Learning-Based Recognition for Dangerous Objects Imaged in X-Ray Security Inspection Device. J. Xray Sci. Technol. 2023, 31, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Gong, E.J.; Bang, C.S.; Lee, J.J.; Baik, G.H.; Lim, H.; Jeong, J.H.; Choi, S.W.; Cho, J.; Kim, D.Y.; Lee, K.B.; et al. Deep Learning-Based Clinical Decision Support System for Gastric Neoplasms in Real-Time Endoscopy: Development and Validation Study. Endoscopy 2023, 55, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Adusumilli, N.; Friedman, A.; Patel, V. Perceptions of Artificial Intelligence Integration into Dermatology Clinical Practice: A Cross-Sectional Survey Study. J. Drugs Dermatol. 2022, 21, 135–140. [Google Scholar] [CrossRef]
- Karalis, V.D. The Integration of Artificial Intelligence into Clinical Practice. Appl. Biosci. 2024, 3, 14–44. [Google Scholar] [CrossRef]
- Galić, I.; Habijan, M.; Leventić, H.; Romić, K. Machine Learning Empowering Personalized Medicine: A Comprehensive Review of Medical Image Analysis Methods. Electronics 2023, 12, 4411. [Google Scholar] [CrossRef]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for Cardiac Contractile Dysfunction Using an Artificial Intelligence–Enabled Electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef]
- Carron, M.; Safaee Fakhr, B.; Ieppariello, G.; Foletto, M. Perioperative Care of the Obese Patient. Br. J. Surg. 2020, 107, e39–e55. [Google Scholar] [CrossRef]
- Xue, B.; Li, D.; Lu, C.; King, C.R.; Wildes, T.; Avidan, M.S.; Kannampallil, T.; Abraham, J. Use of Machine Learning to Develop and Evaluate Models Using Preoperative and Intraoperative Data to Identify Risks of Postoperative Complications. JAMA Netw. Open 2021, 4, e212240. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, R.; Requa, J.; Dao, T.; Ninh, A.; Tran, E.; Mai, D.; Lugo, M.; El-Hage Chehade, N.; Chang, K.J.; Karnes, W.E.; et al. Artificial Intelligence Using Convolutional Neural Networks for Real-Time Detection of Early Esophageal Neoplasia in Barrett’s Esophagus (with Video). Gastrointest. Endosc. 2020, 91, 1264–1271.e1. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, J.; Sun, Y.; Li, C.; Wu, W.; Jin, L.; Yang, Z.; Ni, B.; Gao, P.; Wang, P.; et al. 3D Deep Learning from CT Scans Predicts Tumor Invasiveness of Subcentimeter Pulmonary Adenocarcinomas. Cancer Res. 2018, 78, 6881–6889. [Google Scholar] [CrossRef] [PubMed]
- Bendixen, M.; Jørgensen, O.D.; Kronborg, C.; Andersen, C.; Licht, P.B. Postoperative Pain and Quality of Life after Lobectomy via Video-Assisted Thoracoscopic Surgery or Anterolateral Thoracotomy for Early Stage Lung Cancer: A Randomised Controlled Trial. Lancet Oncol. 2016, 17, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Niel, O.; Boussard, C.; Bastard, P. Artificial Intelligence Can Predict GFR Decline during the Course of ADPKD. Am. J. Kidney Dis. 2018, 71, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Cicione, A.; De Nunzio, C.; Manno, S.; Damiano, R.; Posti, A.; Lima, E.; Tubaro, A.; Balloni, F. An Update on Prostate Biopsy in the Era of Magnetic Resonance Imaging. Minerva Urol. Nephrol. 2018, 70, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.; Dinnes, J.; Chuchu, N.; Takwoingi, Y.; Bayliss, S.E.; Matin, R.N.; Jain, A.; Walter, F.M.; Williams, H.C.; Deeks, J.J. Algorithm Based Smartphone Apps to Assess Risk of Skin Cancer in Adults: Systematic Review of Diagnostic Accuracy Studies. BMJ 2020, 368, m127. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.-Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shen, Y.; Sun, H. Intelligent Algorithm-Based Analysis on Ultrasound Image Characteristics of Patients with Lower Extremity Arteriosclerosis Occlusion and Its Correlation with Diabetic Mellitus Foot. J. Healthc. Eng. 2021, 2021, 7758206. [Google Scholar] [CrossRef]
- Peng, Y.; Dharssi, S.; Chen, Q.; Keenan, T.D.; Agrón, E.; Wong, W.T.; Chew, E.Y.; Lu, Z. DeepSeeNet: A Deep Learning Model for Automated Classification of Patient-Based Age-Related Macular Degeneration Severity from Color Fundus Photographs. Ophthalmology 2019, 126, 565–575. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Fetal Brain Abnormality Classification from MRI Images of Different Gestational Age. Brain Sci. 2019, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.O.; Pedreira, C.E.; Barrena, S.; Lopez, A.; Orfao, A. A Decision-Tree Approach for the Differential Diagnosis of Chronic Lymphoid Leukemias and Peripheral B-Cell Lymphomas. Comput. Methods Programs Biomed. 2019, 178, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Quinten, V.M.; van Meurs, M.; Wolffensperger, A.E.; ter Maaten, J.C.; Ligtenberg, J.J.M. Sepsis Patients in the Emergency Department. Eur. J. Emerg. Med. 2018, 25, 328–334. [Google Scholar] [CrossRef] [PubMed]
- The Alan Turing Institute. Statistical Machine Learning for Randomised Clinical Trials (MRC CTU). Available online: https://www.turing.ac.uk/research/research-projects/statistical-machine-learning-randomised-clinical-trials-mrc-ctu (accessed on 23 January 2024).
- Fogel, D.B. Factors Associated with Clinical Trials That Fail and Opportunities for Improving the Likelihood of Success: A Review. Contemp. Clin. Trials Commun. 2018, 11, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Foster, D. (Ed.) Generative Deep Learning: Teaching Machines to Paint, Write, Compose, and Play, 2nd ed.; Karl Friston (Foreword); Oreilly & Associates Inc.: Sebastopol, CA, USA, 2023. [Google Scholar]
- Liu, C.; Gao, C.; Xia, X.; Lo, D.; Grundy, J.; Yang, X. On the Reproducibility and Replicability of Deep Learning in Software Engineering. ACM Trans. Softw. Eng. Methodol. 2022, 31, 1–46. [Google Scholar]
- Chien, J.-T. Deep Neural Network. In Source Separation and Machine Learning; Elsevier: Amsterdam, The Netherlands, 2019; pp. 259–320. [Google Scholar]
- Verma, S.; Tran, K.; Ali, Y.; Min, G. Reducing LLM Hallucinations Using Epistemic Neural Networks. arXiv 2023, arXiv:2312.15576. [Google Scholar]
- Dykstra, K.; Mehrotra, N.; Tornøe, C.W.; Kastrissios, H.; Patel, B.; Al-Huniti, N.; Jadhav, P.; Wang, Y.; Byon, W. Reporting Guidelines for Population Pharmacokinetic Analyses. J. Pharmacokinet. Pharmacodyn. 2015, 42, 301–314. [Google Scholar] [CrossRef]
- FDA. Population Pharmacokinetics Guidance for Industry. U.S. Department of Health and Human Services Food and Drug Administration. Center for Drug Evaluation and Research (CDER) Center for Biologics Evaluation and Research (CBER), 2022. Available online: https://www.fda.gov/media/128793/download (accessed on 24 March 2024).
- EMA. Guideline on Reporting the Results of Population Pharmacokinetic Analyses. Committee for Medicinal Products for Human Use (CHMP), 2007. Available online: https://www.ema.europa.eu/en/reporting-results-population-pharmacokinetic-analyses-scientific-guideline (accessed on 24 March 2024).
- Klinger, C. Bootstrapping Reality from the Limitations of Logic: Developing the Foundations of “Process Physics”, a Radical Information-Theoretic Modelling of Reality Paperback—22; VDM Publishing: Riga, Latvia, 2010. [Google Scholar]
- Eckardt, J.-N.; Hahn, W.; Röllig, C.; Stasik, S.; Platzbecker, U.; Müller-Tidow, C.; Serve, H.; Baldus, C.D.; Schliemann, C.; Schäfer-Eckart, K.; et al. Mimicking Clinical Trials with Synthetic Acute Myeloid Leukemia Patients Using Generative Artificial Intelligence. medRxiv. 2023. Available online: https://www.medrxiv.org/content/10.1101/2023.11.08.23298247v1 (accessed on 24 March 2024).
- Giuffrè, M.; Shung, D.L. Harnessing the Power of Synthetic Data in Healthcare: Innovation, Application, and Privacy. NPJ Digit. Med. 2023, 6, 186. [Google Scholar] [CrossRef]
- Lee, P.; Submitter, R.P.S.; Davis, U.C. Synthetic Data and the Future of AI. 110 Cornell Law Review. 2024. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4722162 (accessed on 24 March 2024).
- Nikolenko, S.I. Synthetic Data for Deep Learning, 1st ed.; Springer Nature: Cham, Switzerland, 2022. [Google Scholar]
- Assefa, S.A.; Dervovic, D.; Mahfouz, M.; Tillman, R.E.; Reddy, P.; Veloso, M. Generating Synthetic Data in Finance: Opportunities, Challenges and Pitfalls. In Proceedings of the First ACM International Conference on AI in Finance, New York, NY, USA, 15–16 October 2020; ACM: New York, NY, USA, 2020. [Google Scholar]
| Activation Function | Weights of Loss Function | Number of Hidden Layers | Number of Neurons in Hidden Layers | ||||
|---|---|---|---|---|---|---|---|
| Hidden Layers | Output Layer | Kullback–Leibler Part | Reconstruction Part | Encoder | Decoder | Encoder | Decoder |
| Softplus | Linear | 1 | 1 | 3 | 3 | 64-32-16 | 16-32-64 |
| 2 | 2 | ||||||
| … | … | ||||||
| 9 | 9 | ||||||
| 10 | 10 | ||||||
| Between-Subject Variability (CV) | Within-Subject Variability (CVw) | Mean Endpoint Value for the Reference | Ratio of Average Endpoints Test/Reference | Original Sample Size (N) | Subsampled Proportions | Size of Generated Data (xN) |
|---|---|---|---|---|---|---|
| 20% | 15% | 100 | 1 | 12 | 25% | 1× |
| 30% | 1.1 | 24 | 50% | 2× | ||
| 1.2 | 48 | 75% | ||||
| 72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulos, D.; Karalis, V.D. Introducing an Artificial Neural Network for Virtually Increasing the Sample Size of Bioequivalence Studies. Appl. Sci. 2024, 14, 2970. https://doi.org/10.3390/app14072970
Papadopoulos D, Karalis VD. Introducing an Artificial Neural Network for Virtually Increasing the Sample Size of Bioequivalence Studies. Applied Sciences. 2024; 14(7):2970. https://doi.org/10.3390/app14072970
Chicago/Turabian StylePapadopoulos, Dimitris, and Vangelis D. Karalis. 2024. "Introducing an Artificial Neural Network for Virtually Increasing the Sample Size of Bioequivalence Studies" Applied Sciences 14, no. 7: 2970. https://doi.org/10.3390/app14072970
APA StylePapadopoulos, D., & Karalis, V. D. (2024). Introducing an Artificial Neural Network for Virtually Increasing the Sample Size of Bioequivalence Studies. Applied Sciences, 14(7), 2970. https://doi.org/10.3390/app14072970







