The Energies of Activation and Deactivation of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase Immobilized on the Modified Nanofibrous Membrane
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of PAN/PVdF Nanofibrous Membranes by Electrospinning
2.2. Immobilization of Horseradish Peroxidase on the PAN/PVdF Nanofibrous Membranes
2.3. The Horseradish Peroxidase Activity Depending on Temperature
2.4. The Thermodynamic Parameters of Active and Deactivated Horseradish Peroxidase
2.5. Modeling of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase
3. Results
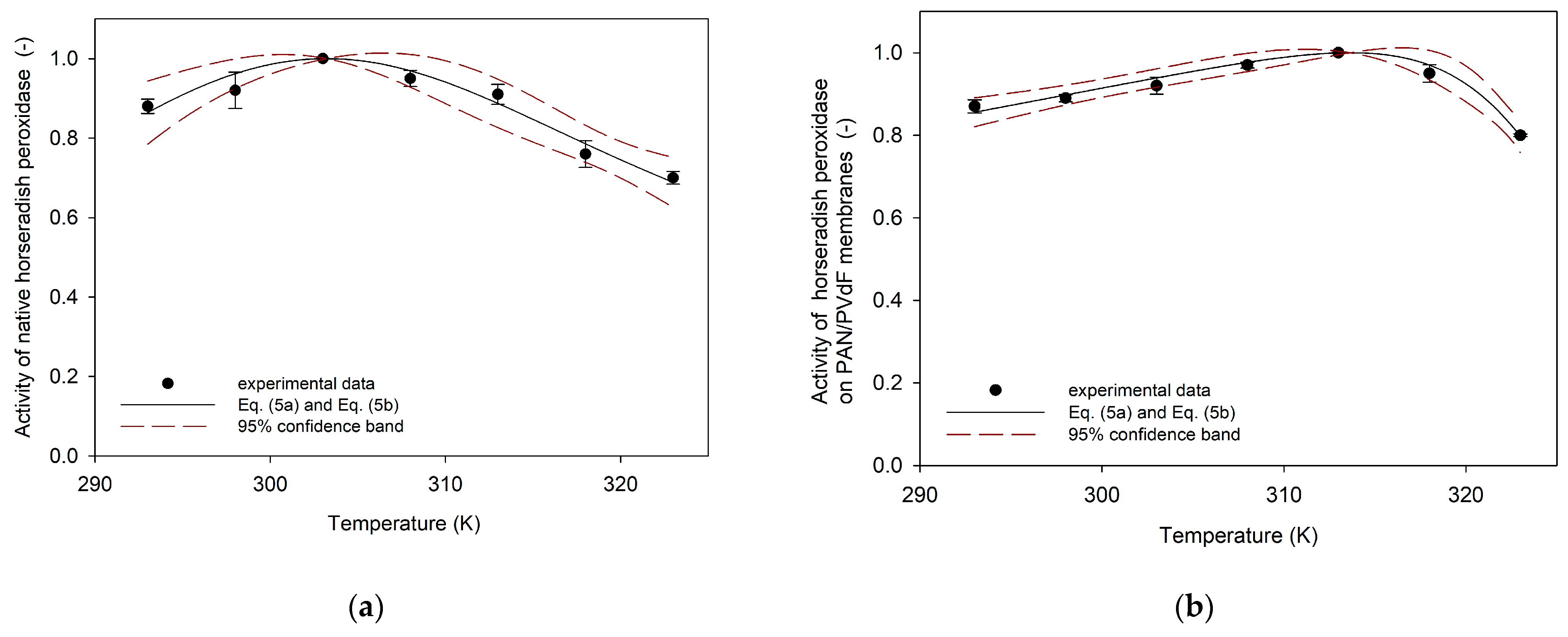
3.1. Parameters , ,
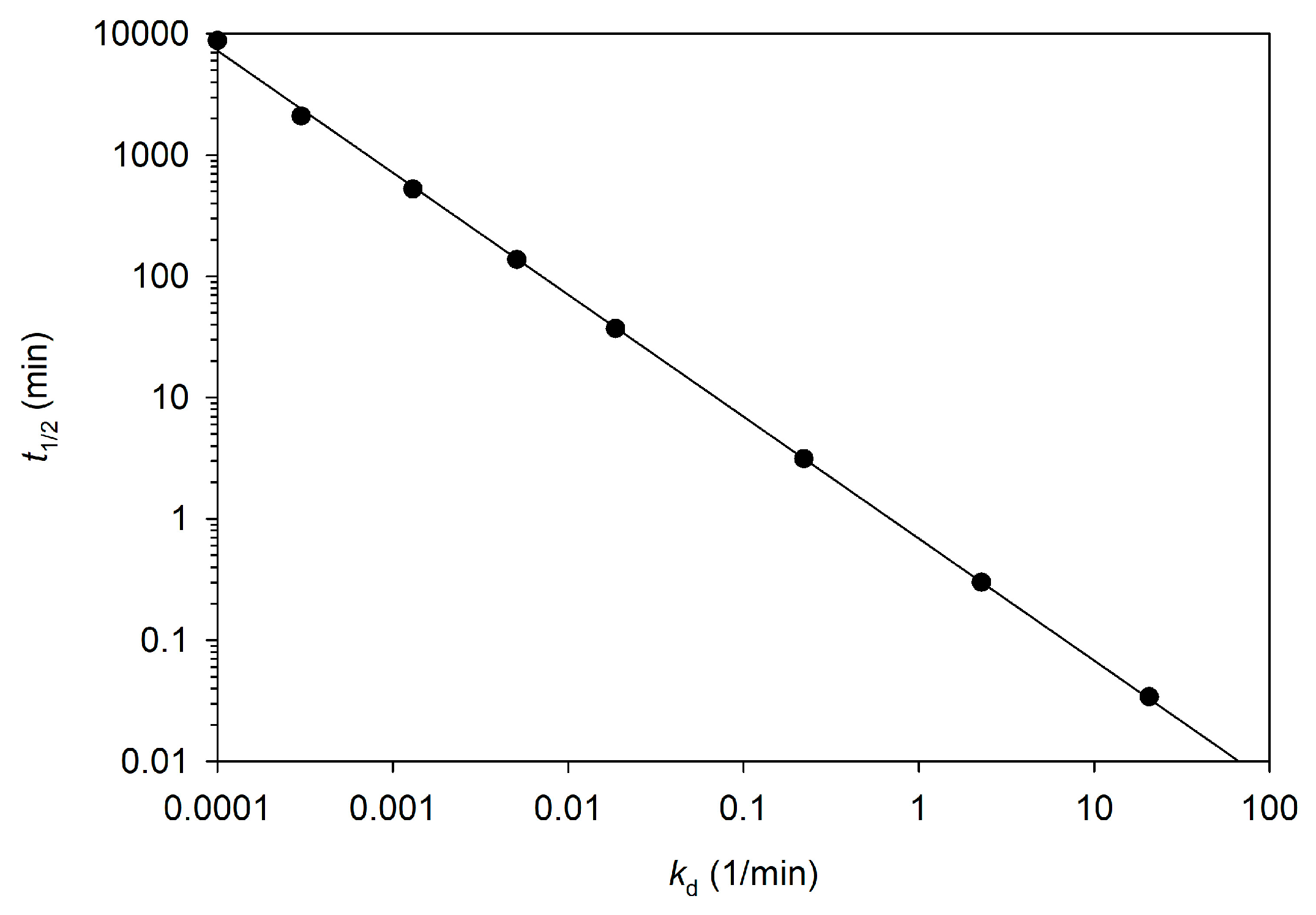
3.2. The Deactivation Constant kd and Half-Time t1/2
3.3. The Thermodynamic Parameters of Active and Deactivated Horseradish Peroxidase
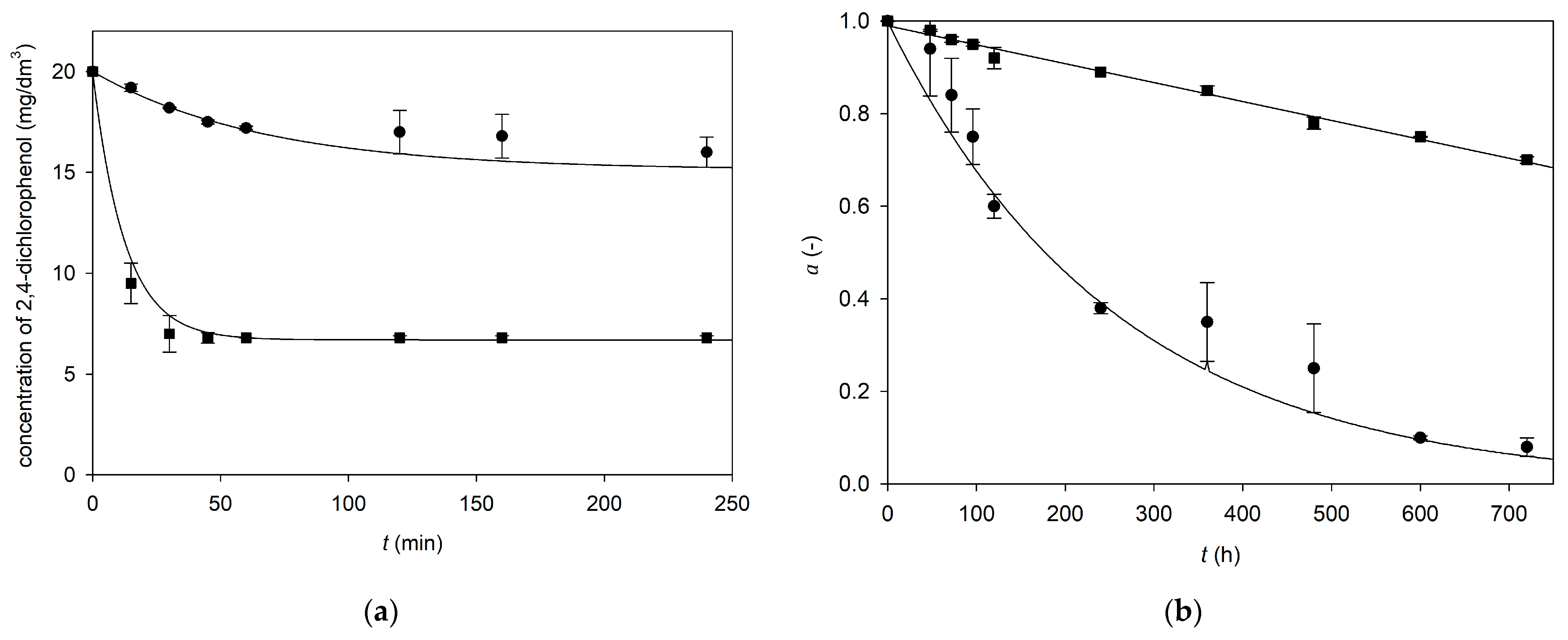
3.4. Modeling of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase
4. Discussion
4.1. The Values of the Optimum Temperature
4.2. The Values of the Activation Energy and the Deactivation Energy
4.3. The Values of the Deactivation Constant
4.4. The Values of Thermodynamic Parameters of Active and Deactivated Horseradish Peroxidase
4.5. Modeling of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antizar-Ladislao, B.; Galil, N.I. Biosorption of phenol and chlorophenols by acclimated residential biomass under bioremediation conditions in a sandy aquifer. Water Res. 2004, 38, 267–276. [Google Scholar] [CrossRef]
- Khenifi, A.; Zohra, B.; Kahina, B.; Houari, H.; Zoubir, D. Removal of 2,4–DCP from wastewater by CTAB/bentonite using one-step and two-step methods: A comparative study. Chem. Eng. J. 2009, 146, 345–354. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Igbinosa, E.O. Chlorophenols and other related derivatives of environmental concern: Properties, distribution and microbial degradation processes. Chemosphere 2011, 83, 1297–1306. [Google Scholar] [CrossRef]
- ATSDR. Comprehensive Environmental Response, Compensation, and Liability Act (CERCLA) Priority List of Hazardous Substances. 2007. Available online: https://www.atsdr.cdc.gov/spl/previous/07list (accessed on 2 January 2024).
- Czaplicka, M. Sources and transformations of chlorophenols in the natural environment. Sci. Total Environ. 2004, 322, 21–39. [Google Scholar] [CrossRef]
- EPA. Chemical Advisory and Notice of Potential Risk: Skin Exposure to Molten 2,4-Dichlorophenol Can Cause Rapid Death. 2000. Available online: https://www.epa.gov/archive/epapages/newsroom_archive/newsreleases/f4543d90482f4d8a8525688d005624d9.html (accessed on 2 January 2024).
- Aruotu, J.O.; Chikere, C.B.; Okafor, C.P.; Edamkue, I. Microbial consortium for polycyclic aromatic hydrocarbons degradation from petroleum hydrocarbon polluted soils in Rivers State, Nigeria. Appl. Sci. 2023, 13, 9335. [Google Scholar] [CrossRef]
- Liang, Y.; Chen, X.; Zeng, J.; Ye, J.; He, B.; Li, W.; Sun, J. Mesoporous polymeric ionic liquid via confined polymerization for Laccase immobilization towards efficient degradation of phenolic pollutants. Molecules 2023, 28, 2569. [Google Scholar] [CrossRef]
- Costa, R.A.; Cunha, A.S.; Peres, J.C.G.; Azzoni, A.R.; Laurenti, E.; Vianna, A.S., Jr. Enzymatic degradation of 2,4,6-trichlorophenol in a microreactor using soybean peroxidase. Symmetry 2020, 12, 1129. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, W.; Wang, S.; Xu, H.; Hu, Y. Magnetic polyethyleneimine nanoparticles fabricated via ionic liquid as bridging agents for Laccase immobilization and its application in phenolic pollutants removal. Molecules 2022, 27, 8522. [Google Scholar] [CrossRef]
- Vasileva, N.; Godjevargova, T.; Ivanova, D.; Gabrovska, K. Application of immobilized horseradish peroxidase onto modified acrylonitrile copolymer membrane in removing of phenol from water. Int. J. Biol. Macromol. 2009, 44, 190–194. [Google Scholar] [CrossRef]
- Zeyadi, M.; Almulaiky, Y.Q. A novel peroxidase from Ziziphus jujuba fruit: Purification, thermodynamics and biochemical characterization properties. Sci. Rep. 2020, 10, 8007. [Google Scholar] [CrossRef]
- Wei, N.; Xu, R.; Tang, R. Immobilization of horseradish peroxidase on modified electrospun nanofibrous membrane for 2,4-dichlorophenol removal. In Proceedings of the 2nd International Conference on Materials Chemistry and Environmental Protection, Sanya City, China, 23–25 November 2018; pp. 283–292. Available online: https://www.scitepress.org/Papers/2018/81889/81889.pdf (accessed on 2 January 2024).
- Terres, J.; Battisti, R.; Andreaus, J.; de Jesus, P.C. Decolorization and degradation of Indigo Carmine dye from aqueous solution catalyzed by horseradish peroxidase. Biocatal. Biotransform. 2014, 32, 64–73. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Ertan, H.; Poljak, A.; Bridge, W.J. Evaluating enzymatic productivity—The missing link to enzyme utility. Int. J. Mol. Sci. 2022, 23, 6908. [Google Scholar] [CrossRef]
- Ostojic, J.; Herenda, S.; Galijasevic, S.; Galic, B.; Milos, M. Inhibition of horseradish peroxidase activity by boroxine derivative, dipotassium-trioxohydroxytetrafluoro-triborate K2[B3O3F4OH]. J. Chem. 2017, 2017, 8134350. [Google Scholar] [CrossRef]
- Miłek, J. The activation energies and optimum temperatures of olive oil hydrolysis by lipase porcine pancreas. Ecol. Chem. Eng. S 2021, 28, 389–398. [Google Scholar] [CrossRef]
- Liu, C.; Tan, L.; Zhang, K.; Wang, W.; Ma, L. Immobilization of horseradish peroxidase for phenol degradation. ACS Omega 2023, 8, 26906–26915. [Google Scholar] [CrossRef]
- Xu, R.; Si, Y.; Li, F.; Zhang, B. Enzymatic removal of paracetamol from aqueous phase: Horseradish peroxidase immobilized on nanofibrous membranes. Environ. Sci. Pollut. Res. 2015, 22, 3838–3846. [Google Scholar] [CrossRef]
- Malomo, S.O.; Adeoye, R.I.; Babatunde, L.; Saheed, I.A.; Iniaghe, M.O.; Olorunniji, F.J. Suicide inactivation of horseradish peroxidase by excess hydrogen peroxide: The effects of reaction pH, buffer ion concentration, and redox mediation. Biokemistri 2011, 23, 124–128. Available online: https://www.ajol.info/index.php/biokem/article/view/77688 (accessed on 2 January 2024).
- Carvalho, C.L.S.; Gomes, F.M.; Pereira, F.M. Mathematical modeling of estrogen degradation in enzymatic membrane reactor. Ind. Biotechnol. 2020, 16, 50–55. [Google Scholar] [CrossRef]
- Miłek, J. Recombinant endo–inulinases: Determination of the activation, deactivation energies and optimum temperatures in inulin hydrolysis. J. Therm. Anal. Calorim. 2023, 148, 859–866. [Google Scholar] [CrossRef]
- Miłek, J. Determination the optimum temperatures and activation energies of inulin hydrolysis by endo-inulinase Aspergillus niger. Chem. Proc. Eng. 2020, 41, 229–236. [Google Scholar] [CrossRef]
- Miłek, J. Thermodynamics and kinetics of thermal deactivation of catalase Aspergillus niger. Pol. J. Chem. Technol. 2020, 22, 67–72. [Google Scholar] [CrossRef]
- Miłek, J. Determination of activation energies and the optimum temperatures of starch hydrolysis by α-amylase from porcine pancreas. Molecules 2021, 26, 4117. [Google Scholar] [CrossRef]
- Miłek, J. The inulin hydrolysis by recombinant exo-inulinase: Determination the optimum temperatures and activation energies. J. Therm. Anal. Calorim. 2022, 147, 8061–8067. [Google Scholar] [CrossRef]
- Kikani, B.A.; Singh, S.P. The stability and thermodynamic parameters of a very thermostable and calcium-independent α-amylase from a newly isolated bacterium, Anoxybacillus beppuensis TSSC-1. Process Biochem. 2012, 47, 1791–1798. [Google Scholar] [CrossRef]
- Jayawardena, M.B.; Yee, L.H.; Poljak, A.; Cavicchioli, R.; Kjelleberg, S.J.; Siddiqui, K.S. Enhancement of lipase stability and productivity through chemical modification and its application to latex-based polymer emulsions. Process Biochem. 2017, 57, 131–140. [Google Scholar] [CrossRef]
- Onaizi, S.A. Statistical analyses of the effect of rhamnolipid biosurfactant addition on the enzymatic removal of Bisphenol A from wastewater. Biocatal. Agric. Biotechnol. 2021, 32, 101929. [Google Scholar] [CrossRef]
- Hewson, W.D.; Dunford, H.B. Horseradish peroxidase. XVIII. The Arrhenius activation energy for the formation of compound I. Can. J. Chem. 1975, 53, 1928–1932. [Google Scholar] [CrossRef]
- Giannakopoulou, A.; Patila, M.; Spyrou, K.; Chalmpes, N.; Zarafeta, D.; Skretas, G.; Gournis, D.; Stamatis, H. Development of a four-enzyme magnetic nanobiocatalyst for multi-step cascade reactions. Catalysts 2019, 9, 995. [Google Scholar] [CrossRef]
- Grubecki, I. Optimal feed temperature for hydrogen peroxide decomposition process occurring in a bioreactor with fixed-bed of commercial catalase. A case study on thermal deactivation of the enzyme. Chem. Process Eng. 2018, 39, 491–501. [Google Scholar] [CrossRef]
- Miłek, J.; Wójcik, M.; Verschelde, W. Thermal stability for the effective use of commercial catalase. Pol. J. Chem. Technol. 2014, 16, 75–79. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Poljak, A.; De Francisci, D.; Guerriero, G.; Pilak, O.; Burg, D.; Raftery, M.J.; Parkin, D.M.; Trewhella, J.; Cavicchioli, R. A chemically modified α-amylase with a molten-globule state has entropically driven enhanced thermal stability. Protein Eng. Des. Sel. 2010, 23, 769–780. [Google Scholar] [CrossRef] [PubMed]
- De Borba, T.M.; Machado, T.B.; Brandelli, A.; Kalil, S.J. Thermal stability and catalytic properties of protease from Bacillus sp. P45 active in organic solvents and ionic liquid. Biotechnol. Prog. 2018, 34, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
| Horseradish Peroxidase | (K) | θ | (kJ/mol) | (kJ/mol) |
|---|---|---|---|---|
| Native | 303.21 ± 1.07 | 1.77 ± 0.52 | 63.35 ± 5.45 | 40.33 ± 10.72 |
| Nanofibrous membrane | 313.64 ± 1.15 | 0.06 ± 0.03 | 215.00 ± 55.04 | 6.39 ± 4.00 |
| Horseradish Peroxidase | SSE | R2 | p | F | P | ||
|---|---|---|---|---|---|---|---|
| (kJ/mol) | (K) | θ (−) | |||||
| Native | 0.0348 | 0.9538 | 0.0003 | <0.0001 | 0.0276 | 41.27 | 0.0021 |
| Nanofibrous membrane | 0.0177 | 0.9642 | 0.0174 | <0.0001 | 0.0776 | 53.92 | 0.0013 |
| Temperature (°C) | 25 | 30 | 35 | 40 | 45 | 55 | 65 | 75 |
|---|---|---|---|---|---|---|---|---|
(1/min) | 0.0001 | 0.0003 | 0.0013 | 0.0051 | 0.0186 | 0.2216 | 2.28 | 20.6 |
(min) | 8745 | 2089 | 523 | 137 | 37 | 3.13 | 0.30 | 0.034 |
| (kJ/mol) | ||||
| Native | 40.33 | 37.85 | 47.77 | −0.03 |
| Nanofibrous membrane | 63.35 | 60.87 | 47.98 | 0.04 |
| (kJ/mol) | ||||
| Native | 6.39 | 3.91 | 47.93 | −0.15 |
| Nanofibrous membrane | 215.00 | 212.52 | 64.11 | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miłek, J. The Energies of Activation and Deactivation of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase Immobilized on the Modified Nanofibrous Membrane. Appl. Sci. 2024, 14, 2423. https://doi.org/10.3390/app14062423
Miłek J. The Energies of Activation and Deactivation of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase Immobilized on the Modified Nanofibrous Membrane. Applied Sciences. 2024; 14(6):2423. https://doi.org/10.3390/app14062423
Chicago/Turabian StyleMiłek, Justyna. 2024. "The Energies of Activation and Deactivation of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase Immobilized on the Modified Nanofibrous Membrane" Applied Sciences 14, no. 6: 2423. https://doi.org/10.3390/app14062423
APA StyleMiłek, J. (2024). The Energies of Activation and Deactivation of 2,4-Dichlorophenol Degradation by Horseradish Peroxidase Immobilized on the Modified Nanofibrous Membrane. Applied Sciences, 14(6), 2423. https://doi.org/10.3390/app14062423







