Abstract
This study introduces a deep-learning-based framework for detecting adversarial attacks in CT image segmentation within medical imaging. The proposed methodology includes analyzing features from various layers, particularly focusing on the first layer, and utilizing a convolutional layer-based model with specialized training. The framework is engineered to differentiate between tampered adversarial samples and authentic or noise-altered images, focusing on attack methods predominantly utilized in the medical sector. A significant aspect of the approach is employing a random forest algorithm as a binary classifier to detect attacks. This method has shown efficacy in identifying genuine samples and reducing false positives due to Gaussian noise. The contributions of this work include robust attack detection, layer-specific feature analysis, comprehensive evaluations, physician-friendly visualizations, and distinguishing between adversarial attacks and noise. This research enhances the security and reliability of CT image analysis in diagnostics.
1. Introduction
In the realm of medical imaging, especially with the widespread use of computed tomography (CT) scans for diagnostic purposes, the accuracy and integrity of the data have paramount significance. With the integration of deep learning techniques in the analysis of these scans, there emerges an opportunity for enhanced diagnostics [,,,]. However, alongside these advancements, there arises the vulnerability of adversarial attacks—meticulously crafted inputs meant to deceive deep learning models [,,]. Such perturbations, while often subtle and imperceptible to the human eye, can lead models to make incorrect predictions with potentially serious consequences in a clinical context []. Addressing this concern requires not only robust detection mechanisms but also intuitive ways for physicians to understand and counteract these adversarial inputs. By ensuring that medical professionals can visually discern genuine samples from their adversarial counterparts, it becomes possible to instill greater confidence in machine-assisted diagnoses and maintain the sanctity of patient care.
To the best of our knowledge, no research has specifically targeted the detection and visualization of adversarial attacks on medical segmentation models. However, two studies have concentrated on enhancing the robustness of deep-learning-based medical segmentation models [,]. Notably, the work by Park et al. [] is the only one that incorporates an adversarial detector, based on MagNet [], as a component to identify adversarial attacks. Although these studies aim to improve the robustness of automatic segmentation against adversarial samples, they do not focus on the direct detection of the adversarial samples themselves.
Therefore, this study is designed to address the following scenario: In a deep-learning-based computer-aided diagnosis system, suppose intruders capture and tamper with CT images. The proposed system from this work is specifically developed to detect these tampered adversarial samples, distinguishing them from genuine images or those merely affected by noise. More formally, this work introduces a framework utilizing a convolutional layer-based CT segmentation model, a specific training set, and recognized attack methods. This framework is designed to detect adversarial samples and distinguish them from genuine ones, which may only be affected by noise. Additionally, the framework provide visualization aids to assist physicians in comprehending the detection outcomes. This work places a particular emphasis on three established attack methods that are most commonly used in automated medical image diagnosis [,]: the fast gradient sign method (FGSM) [], basic iterative method (BIM) [], and stabilized medical image attack (SMIA) [].
After the comprehensive testing in this work, the experimental results confirmed the exceptional effectiveness of the proposed methodology in detecting adversarial attacks, achieving a perfect positive predictive value (PPV) and sensitivity. This is particularly evident in the first layer’s features, which outperform those from intermediate layers, underscoring their crucial role in enhancing detection capabilities. The analysis of this work, backed by empirical data, reveals that the proposed system adeptly identifies adversarial samples, surpassing various benchmark metrics. Additionally, the experiments have explored the role of high and low activation filters in distinguishing authentic from adversarial cases. While the proposed approach effectively counters Gaussian noise, often a source of Type I errors, it encounters challenges with the SMIA method, misidentifying a minority of adversarial samples as noise.
In summary, the contributions of this work are listed as follows:
- Adversarial attack detection in CT scans segmentation models: this work introduces a robust framework to detect a variety of adversarial attacks based on perturbation, including FGSM, BIM, and SMIA, when applied to CT scan images.
- Analyzing feature layers to enhance adversarial attack detection: this work highlights the significant role played by the features from the first layer in discerning genuine samples from their adversarial counterparts.
- Comparative analysis with established methods: this work conducts a comprehensive performance assessment, juxtaposing the proposed method with the existing frameworks. The proposed method from this work consistently showcases superior proficiency in detecting adversarial attacks, underscoring its efficacy and robustness in real-world CT segmentation scenarios.
- Providing visualization for physicians: recognizing the importance of interpretability in medical settings, this framework provides a visualization tailored specifically for physicians. The visualization elucidates the differences between genuine and adversarial samples, allowing for an informed decision-making process.
- Distinguishing between noise and adversarial attacks: this method effectively differentiates between images with inserted Gaussian noise—which do not degrade the performance of the segmentation model—and adversarial samples.
2. Materials and Methods
2.1. Research Objective and Problem Statement
Given a deep-learning-based automatic segmentation technique comprising convolutional filters, as well as the original medical images and their adversarial counterparts, the primary objective of this research is two-fold: (1) vulnerability identification: to pinpoint the most susceptible filter-wise components within the automatic segmentation process. By understanding the weak links, this research aims to fortify the segmentation method against adversarial perturbations. (2) Visualizing critical features of attacks for physicians: to deliver a comprehensive visual examination of the adversarial samples. This will aid physicians in understanding the subtle alterations introduced by adversarial attacks.
Specifically, the vulnerability identification can be rigorously defined as follows. Let a clean image be denoted by x and its corresponding adversarial counterpart, crafted to mislead the target classification model f, be denoted by . The features are extracted from a hidden layer within the model f, where l denotes the layer index. The primary objective is to employ an attack detection mechanism C, leveraging the feature representations and , to accurately distinguish between clean and adversarial samples. Given that the attack detection distinguishes between clean and adversarial images, the objective function is formalized using binary cross-entropy as follows:
where y denotes the true label of the i-th sample, with 0 indicating a clean image and 1 indicating an adversarial sample.
2.2. Characteristics of Features across Convolutional Layers
Convolutional filters in deep learning models are designed to learn specific features from images. Each filter within a model layer captures different features. It is widely understood that the early convolutional layers, situated closer to the input, detect basic features like edges and textures. In contrast, the deeper layers identify more abstract features, such as parts of objects []. For example, in the context of analyzing CT images that capture organs, early convolutional layers might detect basic features such as edges and textures of tissues or organ boundaries. These layers are adept at identifying simple patterns and gradients in pixel intensities, which are fundamental components of an image. Moving deeper into the network, the convolutional layers start to recognize more complex and abstract features. These could include specific structures of organs or even patterns indicative of pathological changes. In advanced layers, the network might be capable of identifying and differentiating between complex organ shapes.
Moreover, because of the nature of the convolutional operation, the initial layer closely mirrors the original input image, while the deeper layers progressively abstract the input to emphasize latent features. Therefore, as shown in Figure 1, visual analysis of the results from convolutional operations in intermediate layers can be challenging. As a result, because the perturbations in adversarial samples consist primarily of simple noise, it can be concluded that they are readily detectable by specific filters in the first layer, facilitating straightforward visual analysis.

Figure 1.
Progression of convolutional operations from the initial to the deeper layer in U-Net [].
2.3. Framework for Detection and Visualization of Adversarial Attacks
Central to the proposed approach from this work is the notion that filters in the initial layer closely reflect the input image by capturing these simple features. This characteristic is leveraged to distinguish between genuine and adversarial images, as well as allow visual analysis, providing valuable insights for physicians.
The next step is to identify the filter in the initial convolutional layer that is most activated by the adversarial sample yet least activated by the genuine sample, as highlighted by the red box and blue box, respectively, in Figure 2.
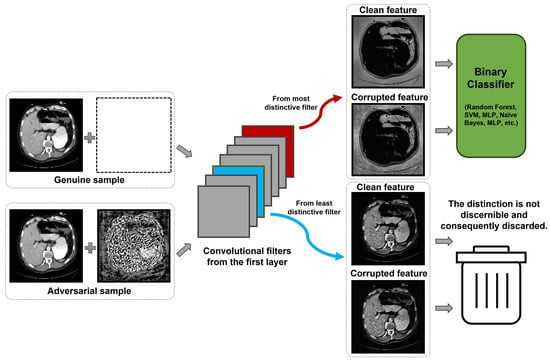
Figure 2.
Core mechanism of the proposed approach contrasting genuine and adversarial feature responses.
For genuine samples, no alterations are made, whereas for adversarial samples, perturbations are added, as shown in the left of Figure 2. Once combined, even though the adversarial sample does not explicitly distinguish between the genuine image and the perturbation, a certain convolutional filter becomes highly activated by the perturbation but remains minimally responsive to the genuine sample. This distinction is highlighted by the yellow box in the figure. After distinguishing between the genuine and adversarial samples—termed the clean feature and corrupted feature in the figure—it becomes straightforward to determine if a sample is adversarial.
The detailed process of the adversarial detection from this work is described as follows. The steps are represented by numbered circles in Figure 3.
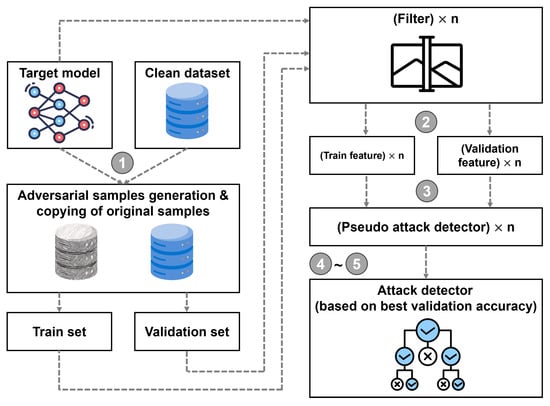
Figure 3.
Schematic representation of the attack detector derivation using the proposed approach.
- Producing adversarial samples using the target model and a clean dataset: In the beginning, the provided target model and clean dataset are utilized to create adversarial samples through a recognized attack technique. After generating these samples, they are partitioned into training and validation datasets based on subjects.
- Feature extraction from target model filters: in the next step, the training and validation datasets are processed through the target model, extracting features from each filter in its first layer. Essentially, these features serve as the input to the detection classifier, capturing unique characteristics of each image. The features are important because they contain patterns that help to differentiate between legitimate and adversarial inputs.
- Classifier training for each filter: once the feature sets are obtained, the next step is to train individual classifiers for each filter using the extracted features from the training set. This is performed to learn the mappings from features to labels (adversarial or not) for each filter. The classifiers could be any machine learning models suitable for binary classification, such as random forests, decision trees, or support vector machine.
- Identifying the most discriminative filter: after training the classifiers, the next step is to evaluate their performance on the validation set. The primary aim of this step is to identify the filter that is most effective at distinguishing between genuine and adversarial inputs. Particular interest lies in filters that produce a high mean and low variance in their classification scores across the validation set, as these filters are the most reliable and stable for detection.
- Building the final adversarial attack detector: armed with the most discriminative filter identified in the previous step, this step aims to build the final adversarial attack detector. This detector is trained using both the training and validation datasets. This comprehensive training allows the detector to generalize well to unseen data, effectively identifying adversarial attacks while minimizing false positives and negatives.
In addition, for enhanced visual analyses that would aid physicians, histogram equalization is implemented on perturbed features. Recognizing the difficulty in visually identifying minute perturbations, a post-processing technique is applied to amplify these feature values. This is achieved using histogram equalization, a technique renowned for its use of the cumulative distribution function []. Histogram equalization is formulated by
where v indicates the pixel value, and L signifies the maximum possible pixel value, typically 256 for a gray-scale image. The height and width of the input image are denoted by H and W, respectively.
Histogram equalization, as shown in Figure 4, is a method used to expand the pixel intensity values of an image. This technique is, thus, applied to the resultant features with the primary aim of highlighting any incorporated noise. Figure 4a offers a comparative view of the feature values before and after applying histogram equalization. Furthermore, Figure 4b vividly illustrates how noise becomes distinctly evident following histogram equalization (as seen at the bottom), as opposed to its more subdued presence in the original feature (shown at the top).
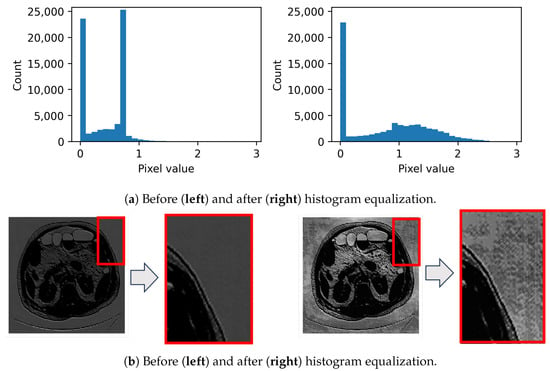
Figure 4.
Amplification of feature values via histogram equalization for enhanced perturbation visualization of a specific feature: (a) Histograms and corresponding (b) images.
3. Experiments
3.1. Data Description and Pre-Processing Methodology
3.1.1. Grayscale CT Imaging Dataset
This work sourced publicly available organ data from “Multi-Atlas Labeling Beyond the Cranial Vault—Workshop and Challenge” (BTCV) []. The BTCV dataset contains annotations for various organs, including the spleen, right and left kidneys, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, portal and splenic veins, pancreas, and both adrenal glands. It also includes images of organs without annotations.
To preprocess the CT images, pixel values are limited to a range between −135 and 215, as well as normalized. Additionally the images are resized to dimensions of 256 × 256. The dataset is allocated 75% for training and the remaining 25% for testing, based on individual patients. Given that no single image encompasses all organs, the focus was placed on those that contained seven or more organs. This resulted in a selection of 821 training images and 228 testing images.
To train the U-Net-based [] target segmentation model, the entire training dataset was utilized. For the detection of adversarial attacks, the 821 training images were partitioned evenly into a dedicated training set and a validation set. The former facilitated the training of a binary classifier tailored for adversarial attack detection, while the latter was leveraged to select the best filter to detect the adversarial sample.
3.1.2. Color Imaging Dataset of Gastrointestinal Polyp
The proposed method in this study has been initially implemented for a computed tomography (CT) segmentation model, which processes grayscale images. Additionally, the performance in detecting adversarial attacks was evaluated using a color image dataset, specifically the Kvasir SEG dataset, designed for computer-aided gastrointestinal disease detection [].
This dataset comprises 1000 images of gastrointestinal polyps, along with their corresponding ground truth masks. The images were resized to 256 × 256 pixels, and the dataset was divided into training and testing sets with a 75% to 25% split, resulting in 750 images for training and 250 for testing.
The target model, based on the U-Net architecture [], was trained using the entire training dataset. For adversarial attack detection, the training set’s polyp images were utilized in the same manner as described for CT images in the preceding section.
3.2. Implementation Details
3.2.1. Target Models
The U-Net model is employed as the target segmentation model []. The segmentation model is based on an encoder–decoder architecture enhanced with skip connections. According to the hyper-parameters defined in U-Net [], four encoders paired with an equal number of decoders are utilized. The model undergoes 200 epochs of training with a batch size of 16, relying on the AdamW optimizer [] and a learning rate set at 0.0001. As for the loss function, it aims to minimize the combined effect of cross entropy and dice loss, comparing true and predicted segmentations. Therefore, the loss function of the target model can be formulated as follows.
where and are true segmentation and predicted segmentation, respectively. Furthermore, and represents the weight of each loss component. In this work, both and are set to 0.5. Note that in this context, the terms dice loss and dice score are used interchangeably.
In addition, the structure of the target model, including the detailed hyper-parameters, is summarized in Table 1. The model structure and hyper-parameters are adopted from the original U-Net paper []. Since the target model accommodates both grayscale and color images, the input channels can be set to either 1 or 3, depending on the color scale used. Moreover, given that segmentation tasks may involve a varying number of segments, the number of output channels can also be adjusted to match the specific task at hand. For instance, in a CT segmentation model that encompasses 14 organs along with the background, the number of output channels is set to 14. Conversely, in a polyp segmentation model, which only differentiates between the polyp mask and the background, the number of output channels is reduced to 2.

Table 1.
Summary of the target model architecture based on U-Net.
3.2.2. Implemented Adversarial Attacks
The attack strategies most commonly used in automated medical image diagnosis were implemented [,]: the fast gradient sign method (FGSM) [], basic iterative method (BIM) [], and stabilized medical image attacks (SMIA) []. Below, an overview of the principles behind these implemented adversarial techniques is provided:
- FGSM: This method computes the gradients for a given input image x and its corresponding class y. These gradients are used to amplify the loss function J of the target model. By integrating this direction into the original image x, the adversarial sample is produced. The attack formulation iswhere is the step size.
- BIM: Unlike the one-step FGSM, BIM operates iteratively. This method repeatedly amplifies the loss, often for K iterations, and accumulates the gradients onto the original image. The BIM approach can be expressed aswhere the starting point corresponds to the input image x. The function serves as a clipping mechanism to ensure pixel values remain within the range to .
- SMIA: This method is tailored specifically for models in the medical domain, setting it apart from general-purpose methods like FGSM and BIM. Instead of generating noisy results like its counterparts, SMIA emphasizes noise reduction. The core concept hinges on the fact that while adversarial samples typically manifest noise, SMIA integrates a stabilization function into the loss function. This drives the noisy sample towards a blurred version obtained via a Gaussian kernel. The stabilization loss, designed for maximization in our context, is articulated aswhere W represents the Gaussian kernel used in the convolutional operation with the perturbation noise (given by ), and acts as a scalar factor balancing the loss terms.
3.2.3. Adversarial Attack Detector
The adversarial attack detectors described in this study, along with the baseline models are implemented.
- Ours: Random forest [] is utilized to distinguish between adversarial attacks based on clean and corrupted features, as shown within the green box in Figure 2. The source code of the framework proposed by this paper is available at the following URL: https://github.com/hyerica-bdml/adv_detection_ct_segmentation, accessed on 1 September 2023.
- MagNet []: This approach employs an auto-encoder to capture the distribution of genuine samples. During training, the method aims to minimize the reconstruction error between the original and reconstructed samples. Subsequently, it evaluates the distance between the input and its reconstruction to identify an adversarial sample if the reconstruction error exceeds a predetermined threshold. However this method is not specialized for medical domain but verified for MNIST [] and CIFAR []. In this work, the method is reimplemented using PyTorch [], based on the original source code, which was written in TensorFlow [] (https://github.com/Trevillie/MagNet/blob/master/defensive_models.py, accessed on 1 September 2023).
- Park []: This approach employs a deep segmentation model specialized for the medical domain to approximate the distribution of authentic samples, functioning in a manner similar to an auto-encoder. While this bears similarities to MagNet [], a key distinction lies in the input processing. Unlike MagNet, which takes raw input, this method first transforms the input into the frequency domain via discrete Fourier transform (DFT). Since the transformed image includes both real and imaginary values, the absolute value of the complex number is taken to yield a real value. Subsequently, this value is log-scaled and divided by 10 to normalize it to the range of 0 to 1. The adversarial samples are then identified by calculating the difference between the reconstructed output from the auto-encoder and the original input, using a hyper-parameterized threshold for the final decision.
- Park_spatial []: Consistent with the original approach outlined in Park [], the use of DFT has been excluded in the implementation. For both Park and Park_spatial, the hyper-parameters are sourced from the MagNet implementation, as these were not provided in Park’s original paper.
Table 2 provides an overview of the methods’ key features.

Table 2.
Key features of methods.
3.3. Evaluation Metrics
To evaluate the performance of the proposed method from this work against the baseline methods, three metrics are employed: positive predictive value (PPV), sensitivity, and accuracy. PPV quantifies the proportion of accurately predicted adversarial samples among all samples labeled as adversarial. Sensitivity, on the other hand, captures the fraction of adversarial samples that are correctly identified out of all true adversarial samples. In addition, accuracy is used to assess the capability of the binary classifier—in this context, to distinguish between genuine and adversarial samples. The formulas for PPV, sensitivity, and accuracy are as follows.
where TP, FP, TN, and FN indicate true positive, false positive, true negative, and false negative, respectively. For the attack detector, a positive label signifies an actual adversarial sample, while a negative label denotes a genuine sample. Consequently, a true positive is identified as an adversarial sample by the detector, whereas a false positive is marked as genuine, despite being an actual adversarial sample.
3.4. Experimental Results
A range of experiments was conducted, exploring different adversarial attack settings and corresponding parameters, notably the step size (EPS) and the number of iterations (NITR). For each type of attack on CT segmentation, the EPS values are set to 0.01, 0.02, and 0.1. Moreover, For each type of attack on polyp segmentation, the EPS values are configured to 0.01, 0.1, and 0.5. The number of iterations is also adjusted to 5, 10, and 15, excluding FGSM, given its nature as a one-step attack. The results of these adversarial attacks for each setting, as tested on the test set, can be found in Table 3 and Table 4. It is noteworthy that the genuine test samples achieved a dice score of 0.4524 and 0.8886 for CT segmentation and polyp segmentation performance, respectively. In the tables, a lower dice score resulting from an attack suggests that the attack is more effective.

Table 3.
Comparison of dice scores of CT segmentation for adversarial samples generated using FGSM, BIM, and SMIA, originating from a baseline dice score of 0.4524.

Table 4.
Comparison of dice scores of gastrointestinal polyp dataset for adversarial samples generated using FGSM, BIM, and SMIA, originating from a baseline dice score of 0.8886.
The subsequent sections explore both the detection efficacy of the adversarial attacks discussed in this study and provide a visual analysis to assist medical professionals. For each evaluation scenario concerning attack detection, adversarial samples are generated according to specific attack settings, matched one-to-one with the genuine samples in the test set. This results in an equal number of genuine and adversarial samples.
3.4.1. Empirical Evidence for Initial Layer Filter Selection
The choice to select filters from the initial layer is grounded in the following empirical observations using CT imaging dataset:
The first observation arises from the box plots illustrated in Figure 5. The box plots show the variability in detection accuracy across all attack methods. These are observed in the first layer, which constitutes the initial component, and in the intermediate layers corresponding to the subsequent components of the target model. The figure reveals a notable pattern: 15 out of 64 filters in the initial layer, including filters such as 2, 6, 8, and 10, consistently exhibit high detection accuracy with minimal variance. Interestingly, while the second, third, and fourth layers show remarkable stability with some filters (25 out of 64, 81 out of 128, and 54 out of 256, respectively) achieving perfect accuracy and zero variability, the fifth and sixth layers do not exhibit this trend, indicating a lack of zero variability filters and suggesting a more varied detection accuracy in these layers. However, the filters from the intermediate layers are not chosen, as elaborated in the following discussion.
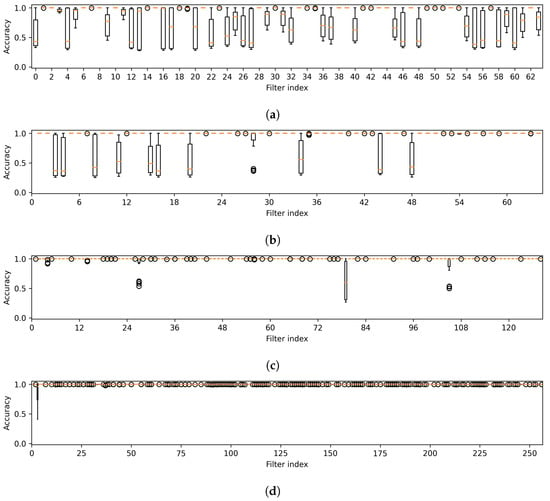
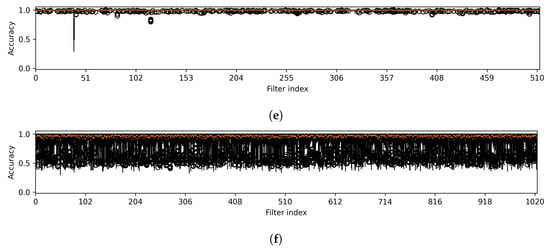
Figure 5.
Variability in accuracy across all attack methods is noted for features from the first layer (a), which is part of the first component, and from the intermediate layers (b–f), corresponding to the subsequent components of U-Net. Note that the results were obtained from a CT segmentation model.
The second consideration relates to computational overhead, where there is a significant difference in computational requirements among the layers. The initial layer requires a relatively modest computational effort, estimated at 0.06 GFLOPS. In contrast, the computation cost from the first to the fifth component increases progressively from 2.50, 6.14, 9.77, 13.40, to 17.03 GFLOPS. This indicates a substantial increase in computational load when extracting features from the deeper layers, as illustrated in Figure 6.
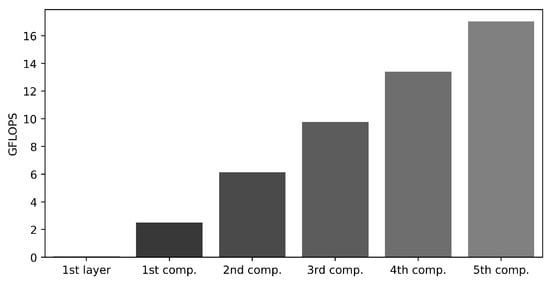
Figure 6.
GFLOPS comparison across components, illustrating the GFLOPS for the first layer within the first component and separately for each of the first to fifth components.
These empirical findings clearly underline the merits of features from the initial layer, striking a balance between detection accuracy and computational efficiency. Based on this evidence, these features are utilized to construct adversarial attack detectors tailored for various attack methodologies.
3.4.2. Comparative Analysis of Filter Efficacy in Adversarial Attack Detection
For detecting adversarial attacks within this framework, the filter that best differentiates between genuine and adversarial samples is identified based on classification accuracy as Equation (4) on validation set. As a result, Figure 5a illustrates the classification performance used for each attack method on the validation set of CT imaging dataset. In the figure, filters 2, 6, 8, and 10, among others, are typically activated by the perturbations that can be observed. In contrast, filters 0, 12, 22, 48, and so forth show less distinct activation in response to the samples. Notably, those filters with pronounced activation consistently yield high detection accuracy.
The significance of features associated with the most and least distinctive filters, specifically filter number 2 and filter number 22, is additionally examined. This analysis is conducted across FGSM, BIM, and SMIA for scenarios involving an epsilon of 0.01 and five iterations, excluding FGSM, as illustrated in Figure 7. The evaluation of feature importance is based on the mean decrease in impurity, a method used to assess how dependent variables impact prediction error in random forests []. The top 1% of features are presented according to their importance values. As indicated in the figure, the most distinctive feature predominantly concentrates on the background rather than the body, thus enabling easier distinction of adversarial samples from genuine ones. In contrast, the least distinctive feature primarily focuses on the structures surrounding the vertebrae, which may complicate the detection of attacks.
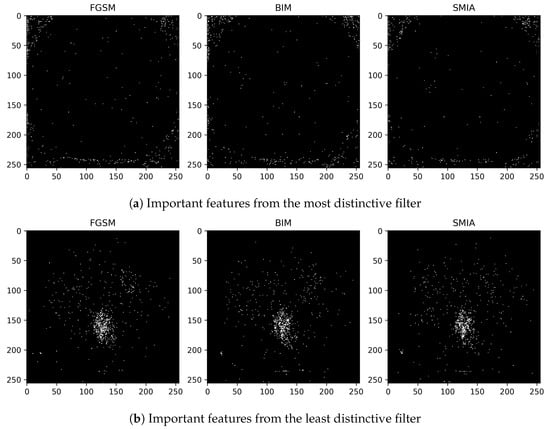
Figure 7.
Visualization of the top 1% of important features from classifiers trained using (a) the distinctive filter number 2 and (b) other than the distinctive filter number 22. These features are derived using the mean decrease in impurity from a random forest classifier for CT segmentation model.
The classification accuracy for adversarial samples, based on filters determined from the validation set results, is detailed in Table 5. This table features two distinct filters for each attack method. Evidently, filter 22, which is not underscored, displays accuracy levels spanning from 62.94% to 100%. Conversely, the underscored filter 2 in the table, denoted by its selection in our proposed framework, invariably records an impeccable accuracy of 100%, underscoring its superior distinction capability. Put simply, while filter 22 identifies features activated by both genuine and adversarial samples, filter 2 specifically pinpoints features triggered exclusively by the adversarial sample.

Table 5.
Classification accuracy of adversarial sample detection using selected filters by proposed framework in this work for CT segmentation model.
3.4.3. Comparative Evaluation of Adversarial Attack Detection of Ours against Baselines
Comparative analysis for a CT imaging dataset: As outlined in Section 3.2.3, the baseline methods were developed according to the specifications detailed in their original publications. It is important to note that these baseline methods identify adversarial samples based on the reconstruction distance; that is, an adversarial sample is detected if its reconstruction distance exceeds a certain threshold. Given that only MagNet [] provides a detailed process for determining the threshold in its original implementation, this procedure is adopted and applied uniformly across all other baseline methods to ensure consistency. Specifically, the maximum reconstruction error is calculated using the validation set, and this error value is then set as the threshold. Furthermore, the same test set was used to evaluate both the baseline methods and the proposed method in this study.
Using the test set, a comparative analysis of classification accuracies for adversarial sample detection across various methods is presented in Table 6. As shown in the table, the approach from this work, which utilizes features from filter number 2 as selected in the previous section, consistently outperforms the baseline methods. Specifically, the proposed method achieves the perfect classification accuracy under different configurations, whereas the baseline methods seldom exceed a 50% accuracy rate—with the exception of SMIA under extreme settings (i.e., epsilon = 0.1 and 15 iterations) where the accuracy reaches 82.23% and 89.69% using Park’s variations.

Table 6.
Comparative analysis of adversarial sample against CT imaging dataset detection accuracy across various methods. Note that the proposed approach in this work exclusively utilizes features from filter number 2.
In addition, a side-by-side comparison of PPV and sensitivity among various methods is presented in Table 7, which also exploits features from filter number 2. As delineated by Equation (2) for PPV and Equation (3) for sensitivity, higher values for these metrics are preferable. A low PPV suggests that the method is prone to incorrectly labeling genuine samples as adversarial, while low sensitivity implies the method may fail to identify adversarial samples. As the table reveals, the proposed approach outperforms all other methods in both PPV and sensitivity across every configuration. Notably, the proposed method achieves a flawless PPV and sensitivity scores in all scenarios.

Table 7.
Comparative analysis of PPV and sensitivity in adversarial attack detection across different methods and types of attacks using CT imaging dataset.
The confusion matrices for both the proposed method and Park_spatial, which ranked second in the SMIA tests using an epsilon of 0.1 over 15 iterations, are analyzed as depicted in Figure 8. While it is the runner-up in performance, Park_spatial fails to detect 31 out of 228 adversarial samples and incorrectly flags 16 genuine samples as adversarial.
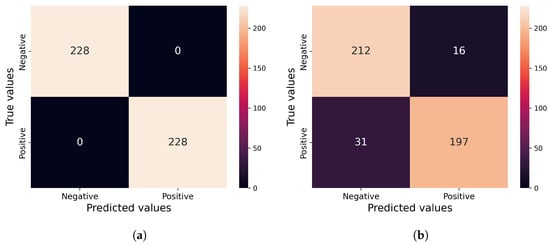
Figure 8.
(a) Proposed method in this work; (b) Baseline based on the previous work []. Confusion matrices for (a) our method and (b) Park_spatial, which achieved the second-best performance on SMIA tests with an epsilon of 0.1 and 15 iterations for CT segmentation model.
Given that the baseline methods primarily focus on capturing the reconstruction error of the input image, they perform effectively when dealing with adversarial images that exhibit extreme perturbations. This is particularly evident with images having a high step size (EPS) of 0.1 and undergoing numerous iterations, such as 15.
Comparative Analysis for a Gastrointestinal Polyp Dataset: The detection performance of the proposed method is compared to baseline approaches using a gastrointestinal polyp dataset, which utilizes a U-Net-based target model with filter 62 selected similarly to the CT imaging dataset.
Table 8 provides a comparative analysis of PPV and sensitivity between the proposed method and the baselines. It is observed that the proposed method outperforms in PPV and sensitivity in most scenarios, with the exception of FGSM attacks at epsilon values of 0.01 and 0.1. The PPVs, 0.9383 for epsilon 0.01 and 0.9881 for epsilon 0.1, stand out, despite the target model’s dice scores experiencing minimal or no decline from 0.8886 for both epsilon values. Conversely, in extreme cases, the baseline methods do not succeed in detecting adversarial samples. Particularly, at low epsilon values, all baseline methods yield a PPV of NaN, suggesting that the attack detectors from these methods classify all samples as non-adversarial.

Table 8.
Comparative analysis of PPV and sensitivity in adversarial attack detection across different methods and types of attacks using gastrointestinal polyp dataset.
3.4.4. Visualization of Comparisons of Genuine Samples and Adversarial Samples Using Histogram Equalization
Randomly selected genuine CT samples, alongside their adversarial versions, are displayed in Figure 9, Figure 10 and Figure 11. The adversarial samples were generated using FGSM, BIM, and SMIA attack methods, each with an epsilon of 0.01. While BIM and SMIA used 5 iterations, FGSM did not. These samples represent the most challenging scenarios for human visual detection. Despite the modest epsilon value, as indicated in Table 3, the adversarial attacks have a subtle but pronounced effect. Such slight modifications can lead to major diagnostic inaccuracies potentially impacting patient care.
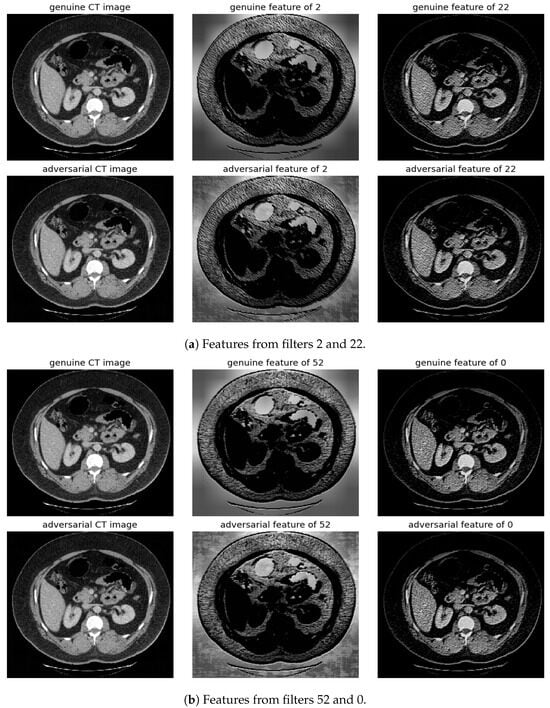
Figure 9.
Visualization under FGSM Attack: Comparison of features from the first layer between genuine and adversarial samples for the most sensitive (filters 2 and 52) and least sensitive (filters 22 and 0) filters.
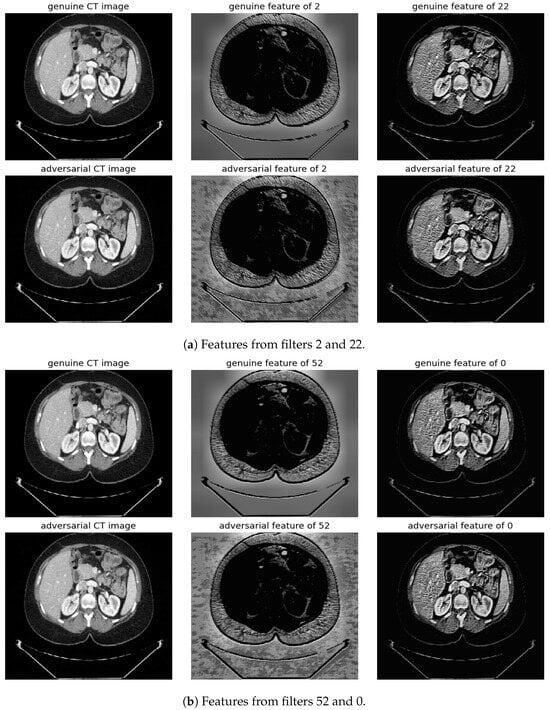
Figure 10.
Visualization under BIM Attack: Comparisons of features from the first layer between genuine and adversarial samples for the most sensitive (filters 2 and 52) and least sensitive (filters 22 and 0) filters.
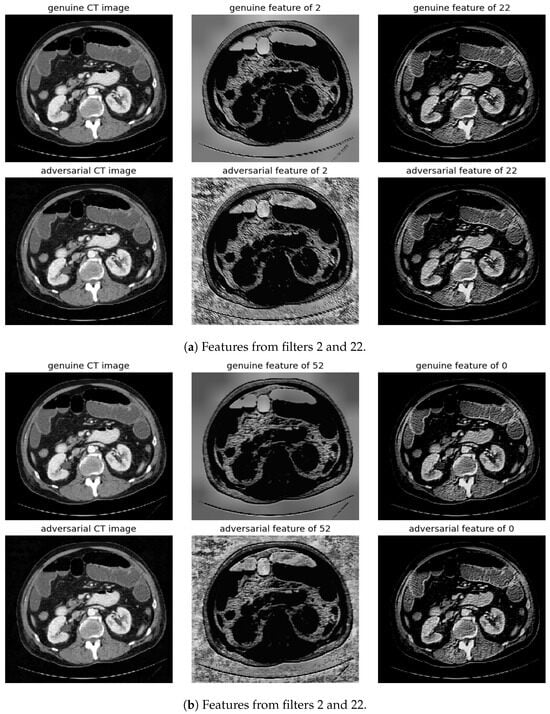
Figure 11.
Visualization under SMIA Attack: Comparison of features from the first layer between genuine and adversarial samples for the most sensitive (filters 2 and 52) and least sensitive (filter 22 and 0) filters.
To address this, histogram equalization as feature post-processing is applied. The visual representations underscore that while genuine and adversarial samples might appear similar to the naked eye, post-processing brings forth distinct feature differences when employing the most sensitive filters from 2 and 52. Perturbations, especially noticeable in the background of the adversarial samples, are frequently highlighted by these filters. Conversely, features processed with filters from 22 and 0, deemed less sensitive in prior analyses, fail to offer a stark visual contrast.
The proposed method effectively identifies adversarial attacks on gastrointestinal polyp segmentation, as illustrated in Figure 12. However, when employing histogram equalization to analyze visual features, differentiating between genuine and adversarial samples becomes difficult. This challenge is compounded in the case of gastrointestinal polyp images, which typically feature limited background areas, in contrast to CT images where adversarial perturbations in the background are more apparent and critical for attack detection.
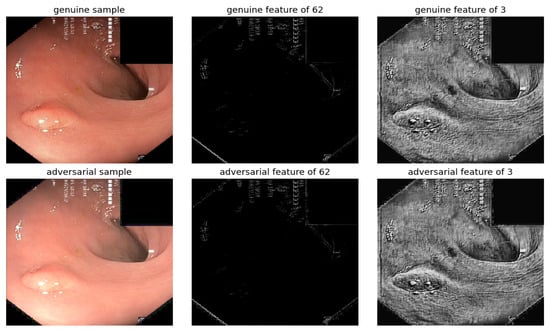
Figure 12.
Visualization under BIM attack, with EPS set to 0.01 and NITER at 15, of a gastrointestinal polyp image: Comparison of features from the first layer between genuine and adversarial samples for the most sensitive filters 62 and least sensitive filters 3.
3.5. Ablation of Binary Classifier
In this research, random forest, the binary classifier, is employed for adversarial attack detection on CT segmentation. To validate this choice and ensure robustness, an extensive ablation study encompassing a range of classifiers is conducted. These include the Gaussian process classifier, Gaussian naive Bayes, K-nearest neighbor classifier, multi-layer perceptron classifier, support vector machine, decision tree Classifier, and XGBoost. All these classifiers were implemented using their respective libraries: scikit-learn [] and XGBoost [].
Figure 13 displays box plots comparing the performance of various classifiers when subjected to adversarial attacks. Each plot aggregates results derived from 64 filters and varies according to different epsilon values and iteration counts. From the visualization, it is evident that the random forest classifier exhibits a consistent and robust performance across different attack types, particularly when compared to other classifiers. This empirical evidence from the box plots solidifies the selection of random forest as the primary binary classifier for this work, given its resilience and reliable performance against diverse adversarial challenges.
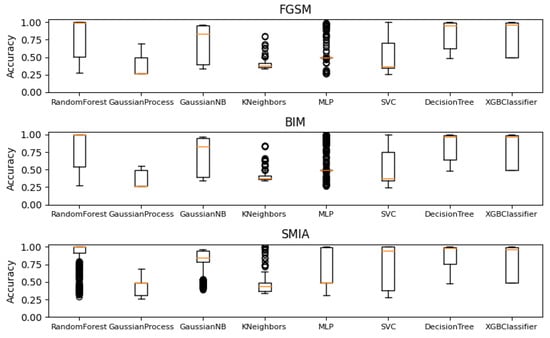
Figure 13.
Box plots illustrating the performance of various classifiers across different attack configurations, including parameters like the number of iterations and perturbation levels (EPS) for CT segmentation model.
3.6. Robustness of Distinguishing Gaussian Noise from Adversarial Attacks on CT Segmentation
In the approach targeting filters highly activated by perturbations, there was an observed tendency to misclassify certain noise types, such as Gaussian noise, as adversarial attacks. This occurs even when such noise has only a minor effect on segmentation performance. For instance, images with Gaussian noise at a variance of 0.001, which result in a reduced dice score of 0.4474 from 0.4524, are consistently identified as adversarial by our binary classifiers trained on FGSM, BIM, and SMIA attacks (on filter number 2).
To address this issue, the proposed approach is to develop noise detectors to distinct between the noise and the attack methods. Since filter number 2 is the most activated by attacks, the noise detector is trained against the adversarial samples. It showcased the highest differential accuracy between Gaussian noise and adversarial samples, achieving both a PPV and sensitivity of 1.0. In addition, out of 228 samples for each attack method, although the classifier is relatively successful in distinguishing Gaussian noise from FGSM and BIM attacks, it mislabeled 24 samples as SMIA instead of Gaussian noise. This is likely due to the objective function of SMIA incorporating a Gaussian kernel, which complicates the discrimination between the adversarial sample and Gaussian noise.
For comparative insights, other baseline detectors are also examined. MagNet identifies Gaussian noise as genuine samples with a high accuracy of 99.12% but falls behind in detecting true adversarial attacks, as shown in Table 6. The Park method also correctly identifies Gaussian noise as genuine samples with an accuracy of 96.93%, highlighting its relative effectiveness in distinguishing Gaussian noise. However, this also suggests its inconsistent performance against other types of adversarial perturbations.
3.7. Comparisons of Computation Costs across Various Methods
The inference runtimes of the proposed method were compared with those of the baseline approaches, with experiments conducted on a workstation featuring an Intel(R) Core(TM) i7-6850K CPU @ 3.60GHz, 128GB of memory, and an NVIDIA GeForce GTX 1080 Ti GPU. This experiment utilized 5000 randomly generated grayscale images, each 256 × 256 pixels in resolution. Runtimes were assessed across batch sizes from to for the 5000 data instances.
Figure 14a depicts the average runtime per batch size for each method. The figure reveals that the runtime of the proposed method experiences a moderate increase, akin to that of MagNet. However, at a batch size threshold of 256, the proposed method begins to suffer from out-of-memory errors, a problem not encountered by MagNet. This challenge is linked to the volume of model parameters processed by the GPU, as illustrated in Figure 14b. Specifically, the proposed method encompasses 704 parameters, in contrast to MagNet’s 310. Consequently, as depicted in Figure 14b, the proposed method fails to operate at batch sizes starting from 256.
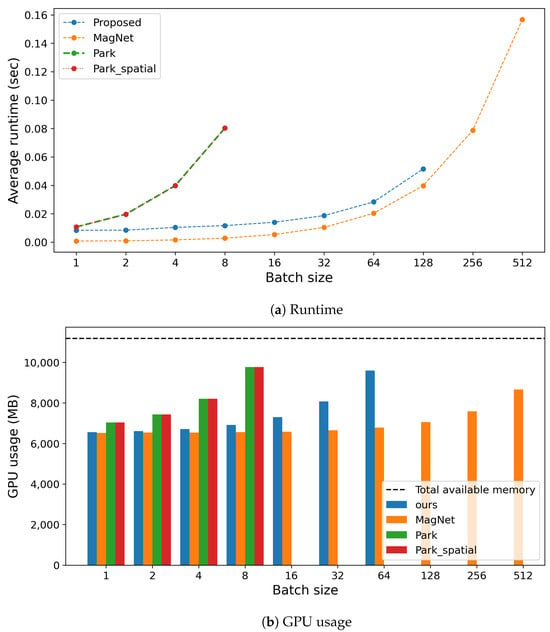
Figure 14.
Comparison of computation costs during inference, including (a) runtime and (b) GPU usage, across different methods.
Conversely, each of Park’s method iterations boasts 31,037,391 parameters, with a notable difference being Park method’s use of discrete Fourier transformation, which is omitted in the Park_spatial method. This significant parameter count causes both versions of Park’s method to experience a sharper increase in runtime compared to the proposed method and MagNet, rendering them incapable of functioning at batch sizes exceeding 16.
Both the proposed method and Park’s approach adopt the same structure for the target segmentation model to detect adversarial attacks. However, a key distinction is that the proposed method from this work relies solely on the first layer. As previously discussed in Section 3.4.1, this strategic focus on the initial layer enables our method to surpass Park’s in terms of runtime efficiency.
3.8. Limitations
This research makes a notable stride in detecting adversarial attacks on medical imaging; however, it has certain limitations worth highlighting:
- Modality-specific limitations: This study is focused on a grayscale CT dataset, which may limit its applicability to other types of medical imaging such as of skin lesions or retinal veins. For example, the visualization of features to distinguish between genuine and adversarial samples in gastrointestinal polyp images is shown to be challenging.
- Coverage of adversarial techniques: While this approach targets three attacks based on perturbations, it does not comprehensively address pixel-wise methods like DeepFool [], One-Pixel Attack [], or ASMA [] which can be used to attack specific sub-regions of the input image.
These limitations not only highlight areas of potential improvement for this work but also provide avenues for future research in the domain of medical imaging security.
3.9. Discussion
Using a real-world CT dataset, this study demonstrates the proficiency of the proposed framework in accurately detecting various adversarial attacks, including FGSM, BIM, and SMIA. Section 3.4.1 presents empirical justifications and furnishes evidence supporting the choice of initial layers for feature extraction in training the attack detector. Additionally, the findings detailed in Section 3.4.2 particularly highlight the superior stability and efficacy of first-layer features over those from the intermediate layers in discerning adversarial samples.
Further insights, using both the CT dataset and gastrointestinal polyp datasets explored in Section 3.4.3, reveal the proposed framework’s exceptional classification accuracy across various adversarial attack detection methods, consistently surpassing the baseline methods. Notably, the proposed approach demonstrates outstanding PPV and sensitivity scores across numerous configurations, indicating its robustness and reliability. A closer analysis using confusion matrices further solidifies the proposed method’s prowess, as even in the least favorable scenarios, it outperforms competing methods like Park_spatial, ensuring genuine samples remain unflagged and adversarial entities are correctly identified.
Additionally, a visualization tailored specifically for physicians is provided. Section 3.4.4 exhibits randomly selected genuine samples paired with their adversarial counterparts on CT segmentation, highlighting features after histogram equalization. This visual representation distinctly showcases the pronounced differences in features between genuine and adversarial samples. However, as illustrated in the section, the proposed method exhibits limitations in distinguishing between the features of genuine and adversarial samples within the polyp segmentation dataset.
Moreover, Section 3.5 delves into an extensive ablation study to validate the efficacy of selection of binary classifier, random forest, by contrasting its performance with a range of other classifiers. This empirical analysis reaffirms the chose classifier’s superior classification accuracy and variance, making it an ideal choice for the complex task of adversarial attack detection. While the study exposes potential vulnerabilities to Type I errors in alternative classifiers like decision tree and other baselines, random forest maintains the perfect performance, thereby substantiating its selection for our framework.
Finally, Section 3.6 demonstrates that the proposed method effectively distinguishes between Gaussian-noised images and adversarial samples using CT images, while the baseline methods are not capable. This suggests that the proposed approach provides a more nuanced and robust mechanism for the identification of different types of perturbations in medical imaging, thereby contributing to enhanced diagnostic integrity and security.
4. Conclusions
This research explored the vulnerabilities introduced by adversarial attacks in deep-learning-driven medical imaging. Utilizing real-world CT and gastrointestinal polyp datasets, the potency of early-layer features for identifying these threats was highlighted. Through exhaustive experimental results, the proposed method’s superior performance over baseline approaches is demonstrated, further substantiated by comparative evaluations with various classifiers. The proposed technique stands out in its robustness against these adversarial attempts. Additionally, visual aids equip physicians with the clarity needed to differentiate authentic scans from tampered ones. As the landscape of adversarial strategies and deep learning architectures evolves, it is crucial for the subsequent research to remain on the forefront, continuously updating our defensive strategies. We strive to reinforce the trust in deep learning’s role in medical imaging, affirming its consistent dependability in healthcare settings.
In the future work, we aim to test our approach using a variety of medical imaging techniques, including MRI and X-ray, to provide a wide-ranging safeguard in the field of medical imaging. Furthermore, we intend to mitigate the adverse activation impacts caused by adversarial samples, focusing on those filters that our framework identifies as being highly activated.
Author Contributions
Conceptualization, W.L. and Y.K.; methodology, W.L.; software, W.L.; validation, W.L.; formal analysis, W.L.; investigation, W.L.; data curation, W.L.; writing—original draft preparation, W.L.; writing—review and editing, W.L. and Y.K.; visualization, W.L.; supervision, Y.K.; project administration, Y.K.; funding acquisition, Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Seoul R&BD Program (No. RO230041). Additionally, this work was supported by the Institute of Information and communications Technology Planning and Evaluation (IITP) grant funded by the Korea government (MSIT) (No. RS-2022-00155885, Artificial Intelligence Convergence Innovation Human Resources Development (Hanyang University ERICA)). It was also supported by the BK21 FOUR (Fostering Outstanding Universities for Research) funded by the Ministry of Education (MOE, Korea) and National Research Foundation of Korea (NRF).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Restrictions apply to the availability of these data. The data were obtained from “Multi-Atlas Labeling Beyond the Cranial Vault—Workshop and Challenge”, available at https://www.synapse.org/#!Synapse:syn3193805/wiki/217789, accessed on 9 August 2022, and the “Kvasir SEG—Segmented Polyp Dataset for Computer Aided Gastrointestinal Disease Detection” accessible at https://datasets.simula.no/kvasir-seg/, accessed on 1 February 2024.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef] [PubMed]
- Choy, S.P.; Kim, B.J.; Paolino, A.; Tan, W.R.; Lim, S.M.L.; Seo, J.; Tan, S.P.; Francis, L.; Tsakok, T.; Simpson, M.; et al. Systematic review of deep learning image analyses for the diagnosis and monitoring of skin disease. NPJ Digit. Med. 2023, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, D.; Li, W.; Roth, H.R.; Landman, B.; Xu, D.; Nath, V.; Hatamizadeh, A. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 19–24 June 2022; pp. 20730–20740. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Shlens, J.; Szegedy, C. Explaining and harnessing adversarial examples. arXiv 2014, arXiv:1412.6572. [Google Scholar]
- Kurakin, A.; Goodfellow, I.J.; Bengio, S. Adversarial examples in the physical world. In Artificial Intelligence Safety and Security; Chapman and Hall/CRC: Boca Raton, FL, USA, 2018; pp. 99–112. [Google Scholar]
- Qi, G.; Lijun, G.; Song, Y.; Ma, K.; Zheng, Y. Stabilized medical image attacks. In Proceedings of the International Conference on Learning Representations, Virtual, 26 April–1 May 2020. [Google Scholar]
- Finlayson, S.G.; Bowers, J.D.; Ito, J.; Zittrain, J.L.; Beam, A.L.; Kohane, I.S. Adversarial attacks on medical machine learning. Science 2019, 363, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yang, S.; Li, G.; Li, H.; Chang, H.; Yu, Y. Non-local context encoder: Robust biomedical image segmentation against adversarial attacks. Proc. Aaai Conf. Artif. Intell. 2019, 33, 8417–8424. [Google Scholar] [CrossRef]
- Park, H.; Bayat, A.; Sabokrou, M.; Kirschke, J.S.; Menze, B.H. Robustification of segmentation models against adversarial perturbations in medical imaging. In Proceedings of the International Workshop on PRedictive Intelligence in Medicine, Lima, Peru, 8 October 2020; pp. 46–57. [Google Scholar]
- Meng, D.; Chen, H. Magnet: A two-pronged defense against adversarial examples. In Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security, Dallas, TX, USA, 30 October–3 November 2017; pp. 135–147. [Google Scholar]
- Dong, J.; Chen, J.; Xie, X.; Lai, J.; Chen, H. Adversarial Attack and Defense for Medical Image Analysis: Methods and Applications. arXiv 2023, arXiv:2303.14133. [Google Scholar]
- Muoka, G.W.; Yi, D.; Ukwuoma, C.C.; Mutale, A.; Ejiyi, C.J.; Mzee, A.K.; Gyarteng, E.S.; Alqahtani, A.; Al-antari, M.A. A comprehensive review and analysis of deep learning-based medical image adversarial attack and defense. Mathematics 2023, 11, 4272. [Google Scholar] [CrossRef]
- Olah, C.; Mordvintsev, A.; Schubert, L. Feature visualization. Distill 2017, 2, e7. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Gonzalez, R.C. Digital Image Processing; Pearson Education India: Noida, India, 2009. [Google Scholar]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Miccai multi-atlas labeling beyond the cranial vault–workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, Munich, Germany, 5–9 October 2015; Volume 5, p. 12. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In MultiMedia Modeling, Proceedings of the 26th International Conference, MMM 2020, Daejeon, Republic of Korea, 5–8 January 2020; Proceedings, Part II 26; Springer: Berlin/Heidelberg, Germany, 2020; pp. 451–462. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- LeCun, Y.; Cortes, C.; Burges, C. MNIST Handwritten Digit Database; ATT Labs: Atlanta, GA, USA, 2010; Volume 2, Available online: http://yann.lecun.com/exdb/mnist (accessed on 1 December 2023).
- Krizhevsky, A. Learning Multiple Layers of Features from Tiny Images; Technical Report; University of Toronto: Toronto, ON, Canada, 2009. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. arXiv 2015, arXiv:1603.04467. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Moosavi-Dezfooli, S.M.; Fawzi, A.; Frossard, P. Deepfool: A simple and accurate method to fool deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2574–2582. [Google Scholar]
- Su, J.; Vargas, D.V.; Sakurai, K. One pixel attack for fooling deep neural networks. IEEE Trans. Evol. Comput. 2019, 23, 828–841. [Google Scholar] [CrossRef]
- Ozbulak, U.; Van Messem, A.; De Neve, W. Impact of adversarial examples on deep learning models for biomedical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part II 22; Springer: Berlin/Heidelberg, Germany, 2019; pp. 300–308. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).