Breast Histopathological Image Classification Based on Auto-Encoder Reconstructed Domain Adaptation
Abstract
1. Introduction
2. Related Work
3. Method
3.1. Notations
3.2. Framework
3.2.1. Deep Feature Extraction
3.2.2. High-Level Feature Reconstruction and Alignment
3.2.3. Similarity Loss
| Algorithm 1: AER-SSDA Algorithm |
| Input: BreakHis dataset, SNL dataset |
| 1. Initialize loss function weights: , and . |
| 2. Split the samples and divide into training and test sets. |
| 3. Build feature extractor, auto-encoder and classifier. |
| 4. Pre-train the feature extractor with ImageNet dataset. |
| 5. Transfer the feature extractor and corresponding parameters. |
| 6. Reconstruct the model and fine-tune it with the training dataset. |
| 7. WHILE training epochs do: |
| 8. Feed the extracted features into encoder to obtain latent features. |
| 9. Feed the latent features into decoder to reconstruct high-level reconstructed features. |
| 10. according to Equations (8) and (9). |
| 11. according to Equations (10) and (11). |
| 12. Feed the latent features into domain discriminator. |
| 13. according to Equation (13). |
| 14. Feed the latent features into sample classifier. |
| 15. according to Equations (3)–(5). |
| 16. Backpropagate with the objective function and update the parameters of the network. |
| 17. END WHILE. |
| Output: F1 score, accuracy, sensitivity and specificity of the test set |
4. Experiments
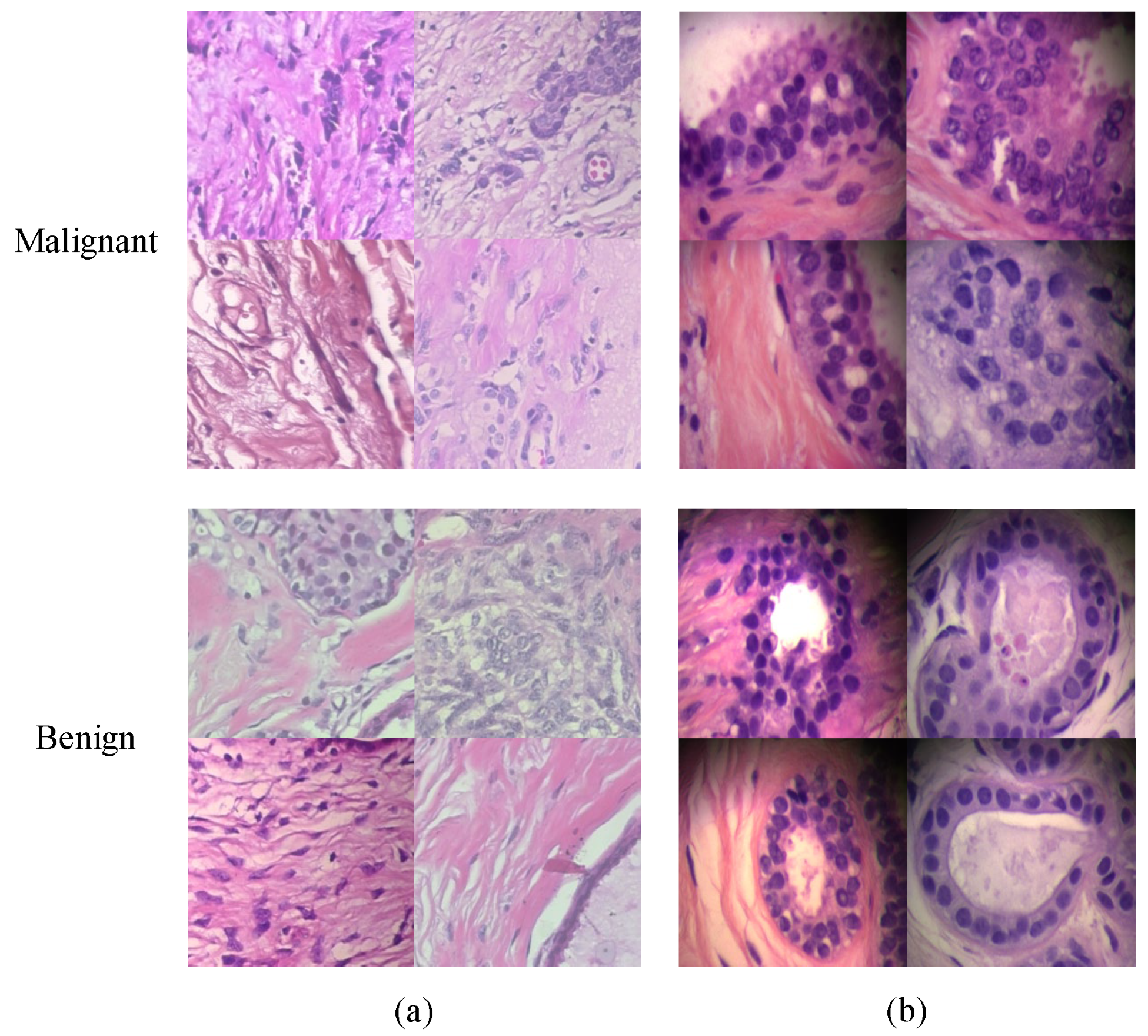
4.1. Dataset and Preprocessing
4.2. Experiment Environment and Parameter Setting
4.3. Evaluation Metrics
4.4. Result
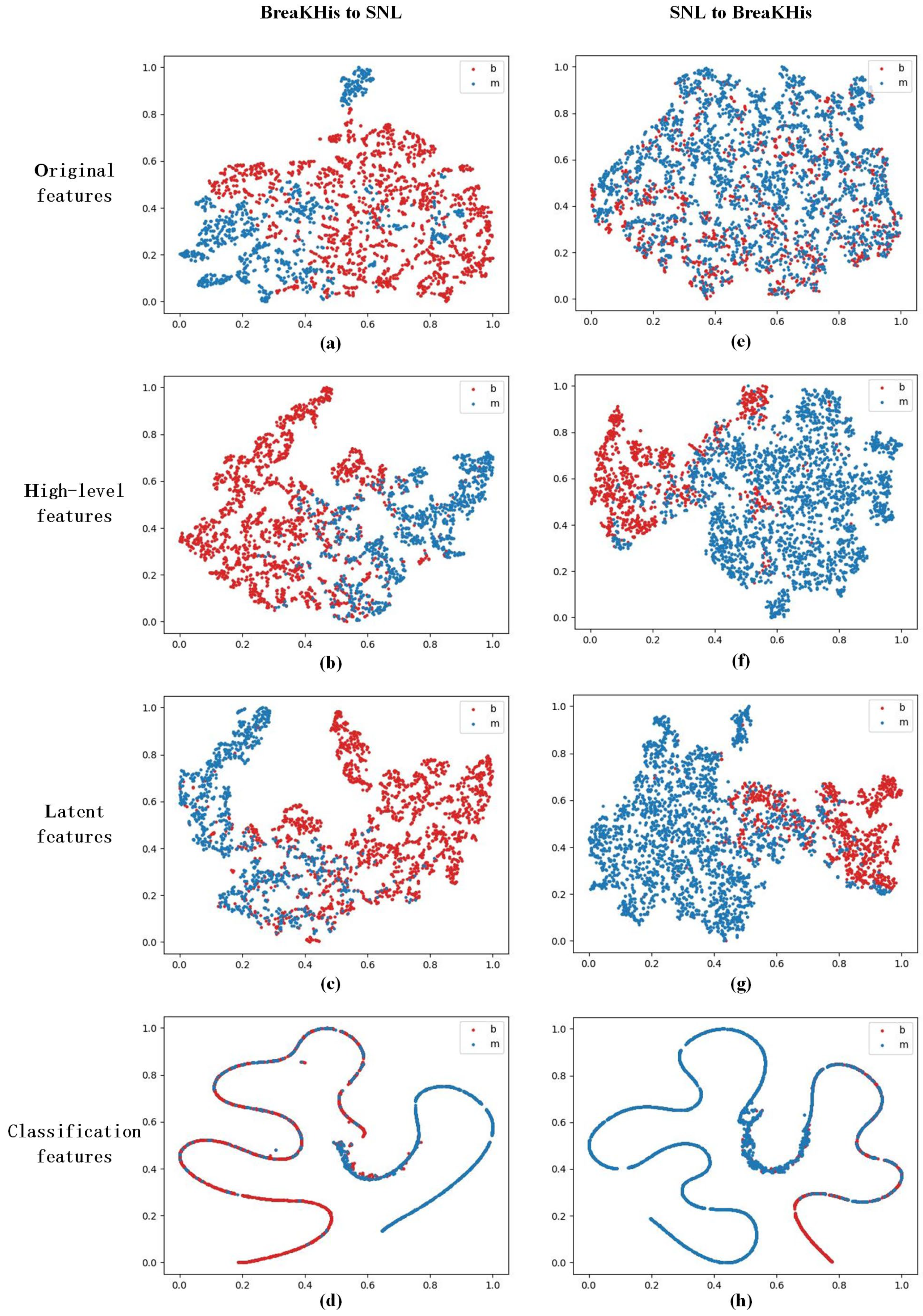
4.4.1. Feature Distribution
4.4.2. Comparative Experiment
4.4.3. Ablation Experiment
4.4.4. Discussion of Experimental Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sudharshan, P.J.; Petitjean, C.; Spanhol, F.; Oliveira, L.E.; Heutte, L.; Honeine, P. Multiple Instance Learning for Histopathological Breast Cancer Image Classification. Expert Syst. Appl. 2019, 117, 103–111. [Google Scholar] [CrossRef]
- Alirezazadeh, P.; Hejrati, B.; Monsef, A.; Fathi, A. Representation Learning-based Unsupervised Domain Adaptation for Classification of Breast Cancer Histopathology Images. Biocybern. Biomed. Eng. 2018, 38, 671–683. [Google Scholar] [CrossRef]
- Xu, B.; Liu, J.; Hou, H.; Liu, B.; Garibaldi, B.; Ellis, I.O.; Green, A.; Shen, L.; Qiu, G. Attention by Selection: A Deep Selective Attention Approach to Breast Cancer Classification. IEEE Trans. Med. Imaging 2019, 39, 1930–1941. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Sanchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen, C.; Bult, P.; Ginneken, B.; Laak, G. Deep Learning as a Tool for Increased Accuracy and Efficiency of Histopathological Diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M. Medical Image Analysis Using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Alom, M.Z.; Yakopcic, C.; Nasrin, S.; Taha, T.; Asari, V. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. MuDeRN: Multi-category Classification of Breast Histopathological Image Using Deep Residual Networks. Artif. Intell. Med. 2018, 88, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lu, Z.; Wang, J.; Ying, S.; Shi, J. A Convolutional Neural Network and Graph Convolutional Network Based Framework for Classification of Breast Histopathological Images. IEEE J. Biomed. Health Inform. 2022, 26, 3163–3173. [Google Scholar] [CrossRef] [PubMed]
- Quinones, W.R.; Ashraf, M.; Yi, M.Y. Impact of Patch Extraction Variables on Histopathological Imagery Classification using Convolution Neural Networks. In Proceedings of the 2021 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 15–17 December 2021; pp. 1176–1181. [Google Scholar]
- Li, J.; Zhang, J.; Sun, Q.; Zhang, H.; Dong, J.; Che, C.; Zhang, Q. Breast Cancer Histopathological Image Classification Based on Deep Second-order Pooling Network. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN) held as part of the IEEE World Congress on Computational Intelligence (IEEE WCCI), Glasgow, UK, 19–24 July 2020; pp. 1–7. [Google Scholar]
- Kumar, A.; Sharma, A.; Bharti, V.; Singh, A.; Singh, S.; Saxena, S. MobiHisNet: A Lightweight CNN in Mobile Edge Computing for Histopathological Image Classification. IEEE Internet Things J. 2021, 8, 17778–17789. [Google Scholar] [CrossRef]
- Gul, A.G.; Cetin, O.; Reich, C.; Flinner, N.; Prangemeier, T.; Koeppl, H. Histopathological Image Classification Based on Self-supervised Vision Transformer and Weak Labels. In Proceedings of the SPIE Journal of Medical Image 2022: Digital and Computational Pathology, San Diego, CA, USA, 20 February–28 March 2022; p. 12039. [Google Scholar]
- Huang, Y.; Zheng, H.; Liu, C.; Ding, X.; Rohde, G. Epithelium-Stroma Classification via Convolutional Neural Networks and Unsupervised Domain Adaptation in Histopathological Images. IEEE J. Biomed. Health Inform. 2017, 21, 1625–1632. [Google Scholar] [CrossRef]
- Yao, T.; Pan, Y.; Ngo, C.-W.; Li, H.; Mei, T. Semi-supervised Domain Adaptation with Subspace Learning for Visual Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 2142–2150. [Google Scholar]
- Xia, T.; Kumar, A.; Feng, D.; Kim, J. Patch-level Tumor Classification in Digital Histopathology Images with Domain Adapted Deep Learning. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Honolulu, HI, USA, 18–21 July 2018; IEEE Engineering in Medicine and Biology Society. Annual International Conference. pp. 644–647. [Google Scholar]
- Zhang, Y.; Zhang, H.; Deng, B. Semi-supervised Models are Strong Unsupervised Domain Adaptation Learners. arXiv 2021, arXiv:2106.00417. [Google Scholar]
- Wang, P.; Li, P.; Li, Y.; Xu, J.; Jiang, M. Classification of Histopathological Whole Slide Images Based on Multiple Weighted Semi-supervised Domain Adaptation. Biomed. Signal Process. Control 2022, 73, 103400. [Google Scholar] [CrossRef]
- Medela, A.; Picon, A.; Saratxaga, C.L.; Belar, O.; Cabezon, V.; Cicchi, R.; Bilbao, R.; Glover, B. Few Shot Learning in Histopathological Images: Reducing the Need of Labeled Data on Biological Datasets. In Proceedings of the 16th IEEE International Symposium on Biomedical Imaging (ISBI), Venice, Italy, 8–11 April 2019; pp. 1860–1864. [Google Scholar]
- Li, L.; Zhang, Z. Semi-Supervised Domain Adaptation by Covariance Matching. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 2724–2739. [Google Scholar] [CrossRef]
- Ahn, E.; Kumar, A.; Fulham, M.; Feng, D.; Kim, J. Unsupervised Domain Adaptation to Classify Medical Images Using Zero-Bias Convolutional Auto-Encoders and Context-Based Feature Augmentation. IEEE Trans. Med. Imaging 2020, 39, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Luo, X.; Wang, C.; Liu, W.; Lin, X. DDA-Net: Unsupervised Cross-modality Medical Image Segmentation via Dual Domain Adaptation. Comput. Methods Programs Biomed. 2021, 213, 106531. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.; Kumar, A.; Feng, D.; Fulham, M.; Kim, J. Unsupervised Deep Transfer Feature Learning for Medical Image Classification. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1915–1918. [Google Scholar]
- Zhao, H.; Ren, T.; Wang, C.; Yang, X.; Wen, Y. Multi-context Unsupervised Domain Adaption for HEp-2 Cell Classification Using Maximum Partial Classifier Discrepancy. J. Supercomput. 2022, 78, 14362–14380. [Google Scholar] [CrossRef]
- Yi, Z.; Zhang, H.; Tan, P.; Gong, M. DualGAN: Unsupervised Dual Learning for Image-to-Image Translation. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2868–2876. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [Google Scholar]
- Jue, J.; Hu, J.; Neelam, T.; Andreas, R.; Sean, B.; Joseph, D.; Harini, V. Integrating Cross-modality Hallucinated MRI with CT to Aid Mediastinal Lung Tumor Segmentation. Med. Image Comput. Comput. Assist. Interv. 2019, 11769, 221–229. [Google Scholar]
- Roels, J.; Hennies, J.; Saeys, Y.; Philips, W.; Kreshuk, A. Domain Adaptive Segmentation in Volume Electron Microscopy Imaging. In Proceedings of the 16th IEEE International Symposium on Biomedical Imaging (ISBI), Venice, Italy, 8–11 April 2019; pp. 1519–1522. [Google Scholar]
- Voigt, B.; Fischer, O.; Schilling, B.; Krumnow, C.; Herta, C. Investigation of semi-and self-supervised learning methods in the histopathological domain. J. Pathol. Inform. 2023, 14, 100305. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hu, X.; Zhang, Y.; Li, P. Semi-Supervised Representation Learning: Transfer Learning with Manifold Regularized Auto-Encoders. In Proceedings of the 9th IEEE International Conference on Big Knowledge (ICBK), Singapore, 17–18 November 2018; pp. 83–90. [Google Scholar]
- He, Y.; Carass, A.; Zuo, L.; Dewey, B.; Prince, J. Autoencoder Based Self-Supervised Test-time Adaptation for Medical Image Analysis. Med. Image Anal. 2021, 72, 102136. [Google Scholar] [CrossRef] [PubMed]
- Spanhol, A.F.; Oliveira, S.L.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Van Der Maaten, L.; Hinton, G. Visualizing Data Using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Bousmalis, K.; Silberman, N.; Krishnan, D.; Erhan, D. Domain Separation Networks. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 343–351. [Google Scholar]
- Wang, W.; Wang, H.; Zhang, Z.; Zhang, C.; Gao, Y. Semi-Supervised Domain Adaptation via Fredholm Integral Based Kernel Methods. Pattern Recognit. 2019, 85, 185–197. [Google Scholar] [CrossRef]
- Motiian, S.; Jones, Q.; Iranmanesh, S.M.; Doretto, G. Few-Shot Adversarial Domain Adaptation. In Proceedings of the 31st Annual Conference on Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 6673–6683. [Google Scholar]
- Motiian, S.; Piccirilli, M.; Adjeroh, D.A.; Doretto, G. Unified Deep Supervised Domain Adaptation and Generalization. In Proceedings of the 16th IEEE international Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 5716–5726. [Google Scholar]

| Author | Dataset | Evaluation Indicators | Model | Domain Adaptation Method |
|---|---|---|---|---|
| Alom et al. [6] | BreakHis | Accuracy | IRRCNN | None |
| Gandomkar et al. [7] | Mitosis-Atypia database | Accuracy | Based on ResNet | None |
| Li et al. [10] | BreakHis | IRR, PRR | DSoPN | None |
| Kumar et al. [11] | BreakHis | Accuracy | MobiHisNet | None |
| Wang et al. [17] | DigestPath 2019 | Accuracy, sensitivity, specificity, | HisNet | Semi-supervised domain adaptation based on manifold regularization |
| Medela et al. [18] | UMCM | Accuracy, sensitivity, specificity | Siamese Neural Network | Semi-supervised domain adaptation based on class distance metric |
| Symbol | Meaning |
|---|---|
| Annotated source domain dataset | |
| Annotated target domain dataset | |
| Unannotated target domain dataset | |
| Intermediate layer output of neural networks | |
| Activation function of neural networks | |
| Total loss of the algorithm | |
| Loss for classification task | |
| Loss for reconstruction task | |
| Domain similarity loss | |
| Domain adaptive feature alignment loss | |
| Hyperparameters of the network |
| Dataset | Class | Total Number | Patches | Train | Test |
|---|---|---|---|---|---|
| BreakHis | Malignant | 1390 | 8340 | 5838 | 2502 |
| Benign | 623 | 3738 | 2616 | 1122 | |
| SNL | Malignant | 47 | 3008 | 2048 | 960 |
| Benign | 87 | 5568 | 3840 | 1728 |
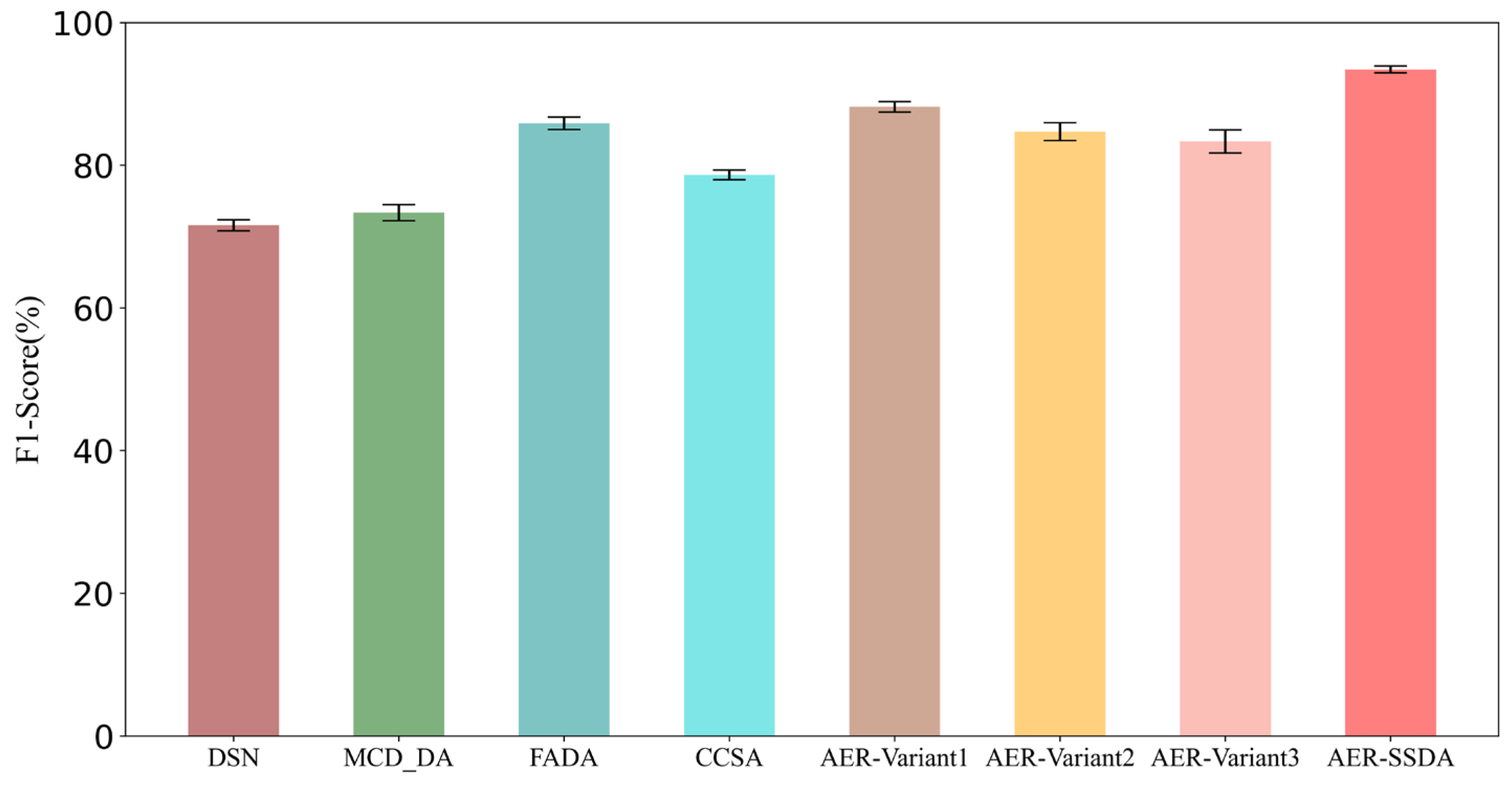
| Method | F1 Score (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| DSN [33] | 71.58 ± 0.78 | 64.87 ± 1.24 | 61.60 ± 2.31 | 73.22 ± 1.72 |
| MCD_DA [34] | 73.34 ± 1.11 | 75.03 ± 1.49 | 90.20 ± 1.76 | 66.14 ± 2.94 |
| FADA [35] | 85.86 ± 0.88 | 80.43 ± 0.60 | 82.65 ± 0.92 | 74.78 ± 0.89 |
| CCSA [36] | 78.63 ± 0.69 | 78.07 ± 0.73 | 80.12 ± 1.08 | 76.35 ± 1.43 |
| AER-SSDA | 93.40 ± 0.48 | 95.24 ± 0.32 | 94.66 ± 0.87 | 95.56 ± 1.27 |
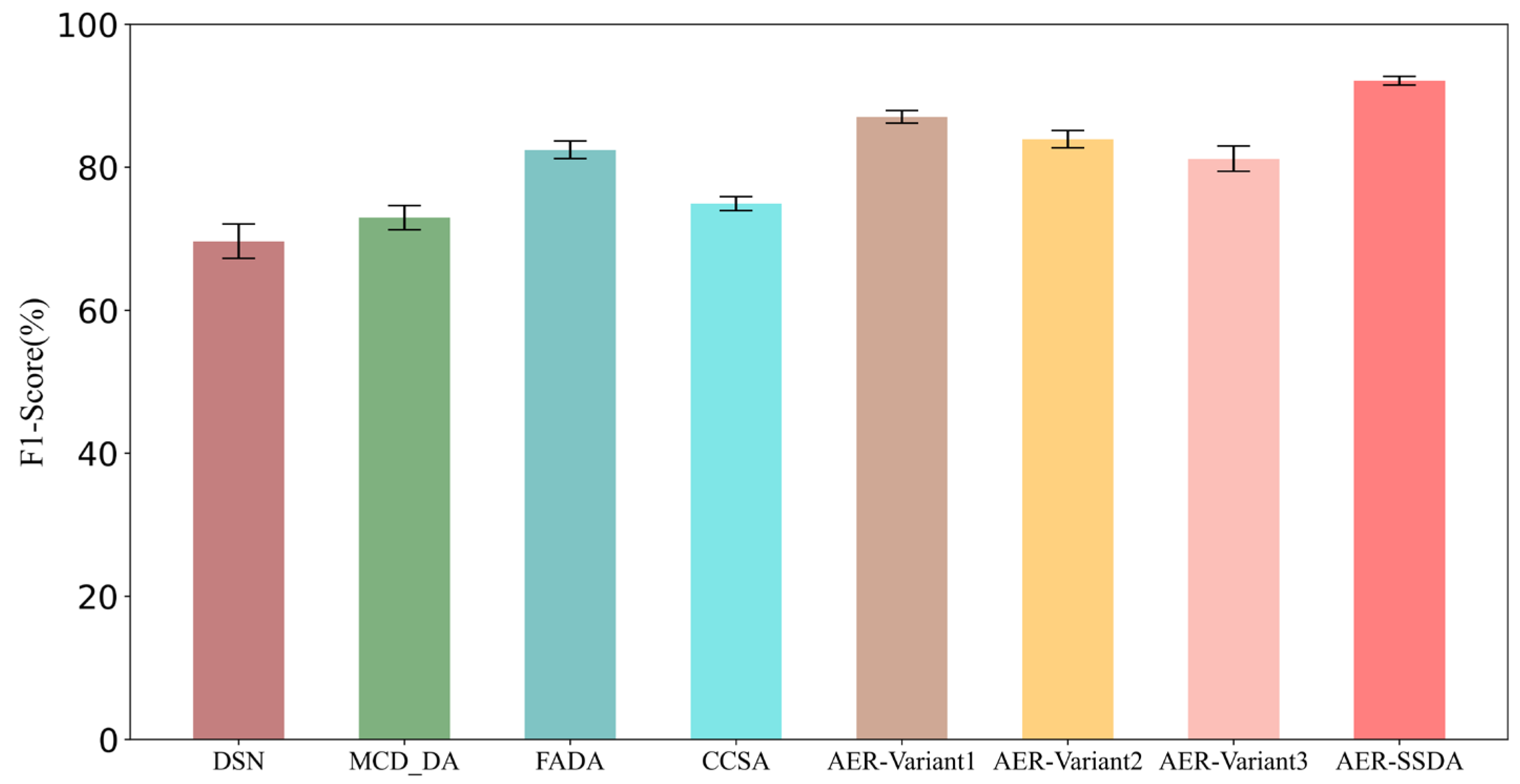
| Method | F1 Score (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| DSN [33] | 69.64 ± 2.39 | 77.78 ± 2.35 | 71.11 ± 3.17 | 81.48 ± 2.83 |
| MCD_DA [34] | 72.95 ± 1.68 | 74.74 ± 1.46 | 95.17 ± 1.27 | 63.38 ± 4.58 |
| FADA [35] | 82.42 ± 1.23 | 78.25 ± 1.07 | 79.88 ± 1.64 | 73.46 ± 2.04 |
| CCSA [36] | 74.91 ± 0.98 | 74.65 ± 0.85 | 74.58 ± 1.67 | 75.45 ± 1.52 |
| AER-SSDA | 92.08 ± 0.59 | 89.47 ± 0.47 | 88.87 ± 1.19 | 89.14 ± 1.22 |
| Method | Loss | F1 Score (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|---|---|---|
| AER-Variant1 | ✓ | ✓ | 88.17 ± 0.71 | 90.96 ± 0.67 | 93.33 ± 1.21 | 89.63 ± 1.37 | |
| AER-Variant2 | ✓ | ✓ | 84.70 ± 1.25 | 88.57 ± 1.01 | 87.98 ± 2.12 | 88.89 ± 1.46 | |
| AER-Variant3 | ✓ | ✓ | 83.32 ± 1.63 | 87.62 ± 2.09 | 85.33 ± 2.75 | 88.86 ± 1.94 | |
| AER-SSDA | ✓ | ✓ | ✓ | 93.40 ± 0.48 | 95.24 ± 0.32 | 94.66 ± 0.87 | 95.56 ± 1.27 |
| Method | Loss | F1 Score (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|---|---|---|
| AER-Variant1 | ✓ | ✓ | 87.02 ± 0.86 | 84.17 ± 0.95 | 82.40 ± 1.43 | 88.13 ± 1.37 | |
| AER-Variant2 | ✓ | ✓ | 83.89 ± 1.21 | 81.23 ± 1.41 | 75.53 ± 2.38 | 87.80 ± 1.89 | |
| AER-Variant3 | ✓ | ✓ | 81.17 ± 1.76 | 77.26 ± 1.83 | 68.71 ± 3.41 | 88.75 ± 1.53 | |
| AER-SSDA | ✓ | ✓ | ✓ | 92.08 ± 0.59 | 89.47 ± 0.47 | 88.87 ± 1.19 | 89.14 ± 1.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Zhang, J.; Li, Y.; Guo, Y.; Li, P. Breast Histopathological Image Classification Based on Auto-Encoder Reconstructed Domain Adaptation. Appl. Sci. 2024, 14, 11802. https://doi.org/10.3390/app142411802
Wang P, Zhang J, Li Y, Guo Y, Li P. Breast Histopathological Image Classification Based on Auto-Encoder Reconstructed Domain Adaptation. Applied Sciences. 2024; 14(24):11802. https://doi.org/10.3390/app142411802
Chicago/Turabian StyleWang, Pin, Jinhua Zhang, Yongming Li, Yurou Guo, and Pufei Li. 2024. "Breast Histopathological Image Classification Based on Auto-Encoder Reconstructed Domain Adaptation" Applied Sciences 14, no. 24: 11802. https://doi.org/10.3390/app142411802
APA StyleWang, P., Zhang, J., Li, Y., Guo, Y., & Li, P. (2024). Breast Histopathological Image Classification Based on Auto-Encoder Reconstructed Domain Adaptation. Applied Sciences, 14(24), 11802. https://doi.org/10.3390/app142411802







