1. Introduction
Symbolic regression (SR) is a task where we aim to find a closed-form mathematical equation which describes linear and nonlinear dependencies in data without making prior assumptions. The goal is to make predictions for unseen data by training a mathematical expression with a finite set of existing observations. While there is a huge variety of methods for solving regression problems, starting from linear models over decision trees [
1] up to neural networks [
2], SR methods deliver non-linear and human-readable closed-form expressions as models with smooth and differentiable outputs. Therefore, SR is commonly applied in situations where we cannot rely on black-box models like neural networks, as they require comprehensible and traceable results for throughout model verification.
Bias and variance are properties of a machine learning algorithm that affect the interpretability of its models. The variance of an algorithm describes, how the outputs of its models change when there are differences in the used training data. SR algorithms are considered as high-variance algorithms, which means that even slightly different training data can lead to very dissimilar models [
3]. However, high variance of an algorithm implies that it is capable to fit a model, to a certain extent, perfectly to training data. It is often necessary to limit the variance in order to perform well on unseen data, without restricting it too much, which would prevent highly accurate models and cause so-called bias. Balancing both properties is called the bias/variance trade-off [
1].
From a practitioner’s point of view, perturbations caused by variance do not spark trustworthiness for practitioners, as we would expect small changes in training data to have little effect on the overall results. Therefore, many algorithms use, e.g., statistical tools to gracefully deal with bias and variance or ignore this property as the primary focus is accuracy regardless of the model structure [
4]. However, bias and variance have specific implications to SR. SR is known for its human-readable and potentially interpretable white-box model structure, but high variance limits this feature. On the other hand, the complexity of models is limited for the sake of interpretability, which causes bias. Algorithmic aspects of SR amplify these effects: First, the stochastic nature of a Genetic Programming (GP)-based [
5] SR algorithm results in differences in models between multiple SR runs even when the training data do not change at all. Second, since there is no guarantee for optimality in an SR search space [
6], models trained in different SR runs might even provide very similar accuracy despite being completely different mathematical expressions due to, e.g., bloat [
5] or over-parameterization [
7] that increase the size of a model without affecting its accuracy.
1.1. Research Question
Controlling the variance while still achieving high accuracy is an ambient goal in SR research and both directly and indirectly targeted by new algorithms. In this work, we examine the bias and the variance of contemporary SR algorithms. We show which algorithms perform most consistent despite perturbations in training data. The results are put into perspective with their achieved accuracy and parsimony. We analyze which algorithms in recent SR research have been the most promising regarding robustness and reliability by mitigating variance while still providing high accuracy. This work complements and builds upon the existing results of the SRBench benchmark [
8], which is a solid benchmark suite for SR algorithms. SRBench compares the accuracy of several SR algorithms for problems from the PMLB benchmark suite [
9]. The results, which are available at
cavalab.org/srbench (accessed on 21 November 2024), also include the trained models for each algorithm and data set. We reuse these published results and analyze the bias and variance of the already published models.
1.2. Related Work
Although most algorithmic advancements in SR algorithms affect their bias and variance, actual measurements and analyses of these properties are sparse. One analyses of bias and variance of SR algorithms was done by Keijzer and Babovic [
10]. They calculated bias and variance measurements but only for few data sets and only for ensemble bagging of standard GP-based SR. Kammerer et al. [
11] analyzed the variance for two different GP-based SR variants and compared them with Random Forest regression [
12] and linear regression on few data sets. Highly related to bias and variance is the work by de Franca et al. [
13], who provided a successor of SRBench. In this benchmark, they tested specific properties of algorithms for a few specific data sets, instead of only the accuracy and model size for a wide range of different data sets. Those tests included, e.g., whether algorithms identified the ground truth on a symbolic level instead of just approximating it with an expression of any structure. In our work, we use the first version of SRBench as we focus on the semantic of models on a broad range of data sets. More research about bias and variance was performed on other algorithms, most recently for neural networks by Neal et al. [
4] and Yang et al. [
14]. They evaluated the relation between bias/variance to model structure and number of model parameters. Belkin et al. [
15] questioned the classical understanding of the relationship between bias and variance and their dependence on the dimensionality of the problem for very large black-box models.
2. Bias/Variance Decomposition
In supervised machine learning tasks, we want to identify a function
, which approximates the unknown ground truth
for any
x drawn from a problem-specific distribution
P.
x is a vector of so-called features and
y the scalar target. The target values contain randomly distributed noise
. In this work, we define
with
being a problem-specific standard deviation, as it is done in SRBench [
8]. Therefore, our data-generating function is
. To find an approximation
, we use a training set
with
and their corresponding
y values. The goal of machine learning is to learn a prediction model
which minimizes a predefined loss function
on a training set
D. This means that the output of a machine learning model depends on the used algorithm, the features
x and the set of training samples
D.
Bias and variance of an algorithm occur due to changes in the training data
D and have a direct effect on the error and loss of models. We use the mean squared error as the loss function. For this loss function, Hastie et al. [
1] describe that a model’s expected error on previously unseen data consists of bias, variance and irreducible noise
. However, Domingos [
16] describes how the decomposition of the error into bias and variance is also possible for other loss functions.
The distribution of training sets
D that contain values drawn from
P also leads to a distribution of model outputs
for a single fixed feature vector
. The bias is the difference between the ground truth value
and the expected value over
D over the
of model outputs
, as depicted in Equation (
1). The bias describes, how far our estimation is “off” on average from the truth.
Variance defines, how far the outputs of estimators spread on a specific point
when they were trained on different data sets. It is independent of the ground truth. It is defined in Equation (
2) as the expected squared difference between the output of models and the average output of those model on a specific point
. Both properties are not mutually exclusive. However, algorithms with high variance tend to have lower bias and vice versa, so practitioners need to find an algorithm setting that minimizes both properties. This is called the bias-variance trade-off [
1].
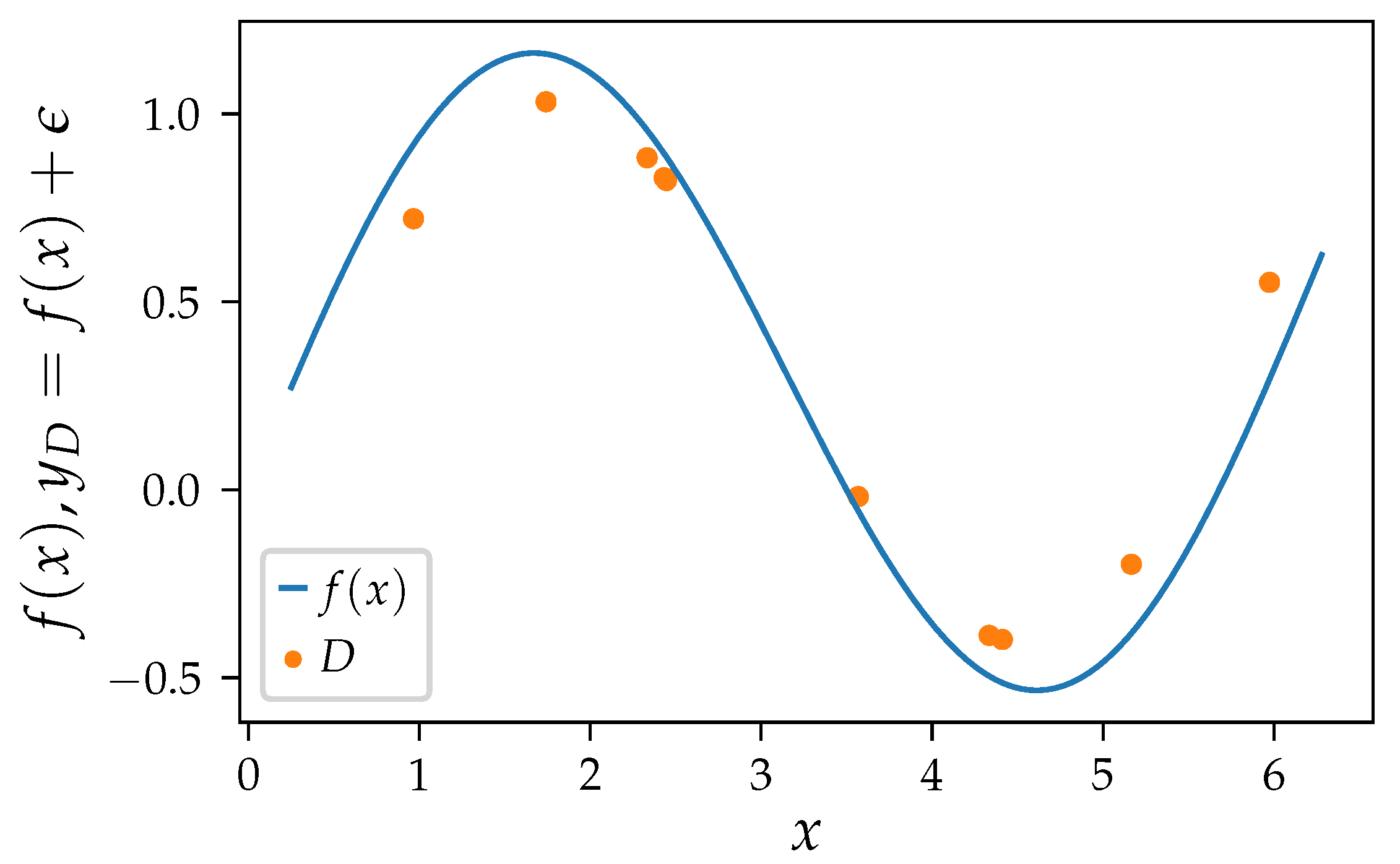
An example inspired by Geman et al. [
17] for high bias, high variance and an optimal trade-off between both is given in
Figure 1 and
Figure 2, where we want to approximate an oscillating function with polynomial regression. We use polynomial regression as the results nicely translates to SR as its search space is a subset of SR methods with an arithmetic function set. We add normal distributed noise to each target value
y, so
with
and
. Each set of samples
D consists of ten randomly sampled observations
with
.
Figure 1 shows the ground truth and one training set of samples. We get a different model with every different set of samples
D. Depending on the algorithm settings, those models behave more or less similar, resulting in different bias and variance measures. We expect the error of a perfect model to be equally distributed as the irreducible noise
.
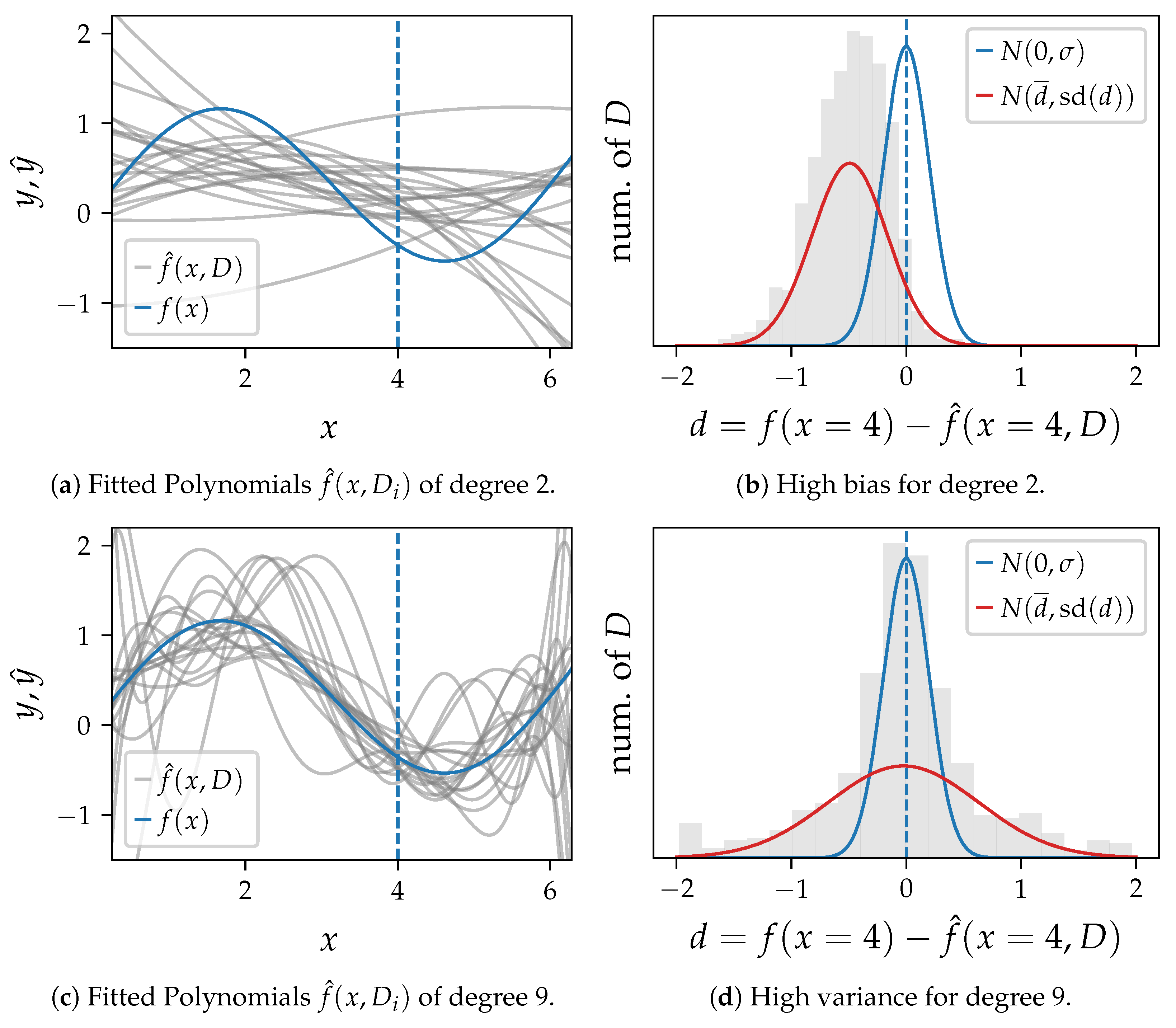
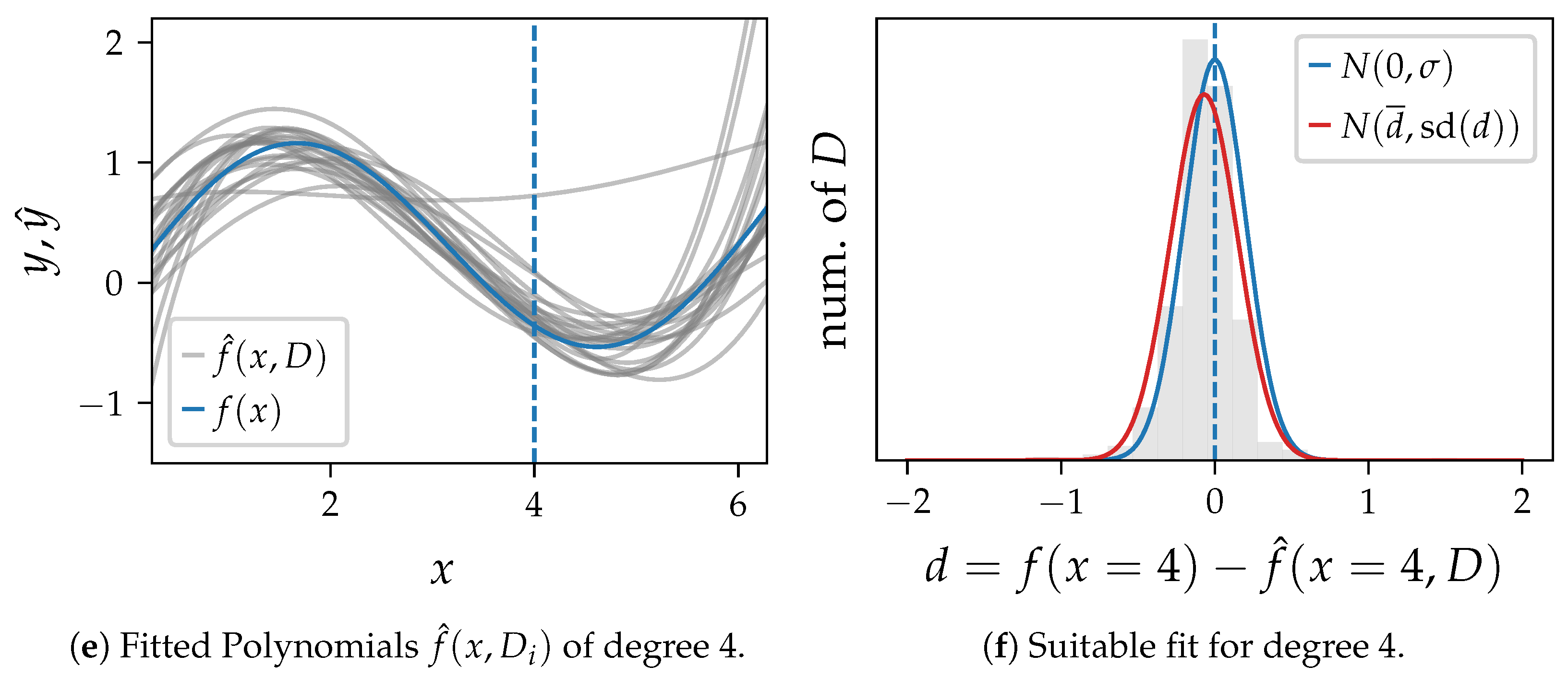
In
Figure 2 we draw 1000 sets of samples
D and learn one polynomial for each training set for three algorithm settings. 20 exemplary polynomials are shown for each setting in
Figure 2a,c,e. The distribution of the difference
d between the ground truth and the model output at
is shown as histogram in
Figure 2b,d,f. These plots also compare the probability density function of
and a normal distribution with the mean and standard deviation (
) of
d as parameters.
Figure 2a,b show polynomials of degree 2, which cannot capture oscillations in the data. The variance is high, and the average error
clearly differs from the ground truth
.
Figure 2c,d show polynomials of degree 9 with the highest variance and the smallest bias in this example. The error of single predictions at that point is often very high, which makes a single model unusable. However, their average fits the ground truth well, as there is nearly no bias.
Figure 2e,f show a polynomial of degree 4, which appears to be an appropriate setting with low bias and variance close to the error variance.
As described before, the bias and variance of an algorithm is directly linked to its model generalization capabilities, as the prediction error on previously unseen data can be decomposed into the square of its bias and its variance [
16]. Therefore, we expect algorithms with models that generalize well to be both low in bias and in variance. Of course, not every algorithm is equally capable to produce high quality results, as shown in SRBench. While the error of models is well measured, it is often unclear whether it is primarily caused by variance or bias. As described in
Section 1, GP-based algorithms exhibit variance even when there is no change in the training data. On the other hand, deterministic algorithms like FFX and AIFeynman produce equal results on equal inputs. Therefore, we expect more similar models and less variance for those algorithms.
3. Experiments
Our analysis builds upon the data sets and models of the SRBench benchmark [
8]. SRBench provides an in-depth analysis of performance and model size of contemporary SR methods on many data sets. The methodology and the results of this benchmark fit perfectly for our purpose, and we can rely on a reviewed setting of algorithms and benchmark data.
SRBench uses the data sets of the PMLB benchmark suite [
9,
18], the Feynman Symbolic Regression Database [
19] and the ODE-Strogatz repository [
20]. In total, it contains both real world data and generated data, where the ground truth and noise distribution is known. In this work, we will use SRBench’s results on the 116 datasets from the Feynman Symbolic Regression Database because the ground truth and data distribution is known, so we can arbitrarily generate new data. We refer to those problems as Feynman problems in the following. The Feynman problems are mostly low-dimensional, nonlinear expression that were taken from physics textbooks [
21].
In SRBench, four different noise levels
are used for Feynman problems. Given a problem-specific ground truth
, training data
y for one problem are generated with
and
, with
meaning no noise. This results in 464 combinations of data sets and noise levels. For each of those 464 problems, ten models were trained in the SRBench benchmark per algorithm. A different training set is sampled for each of the ten models [
8]. All ten models were trained with the same hyperparameters, which is suitable for our study, because bias and variance are only caused by the search procedure and not by differences in hyperparameters.
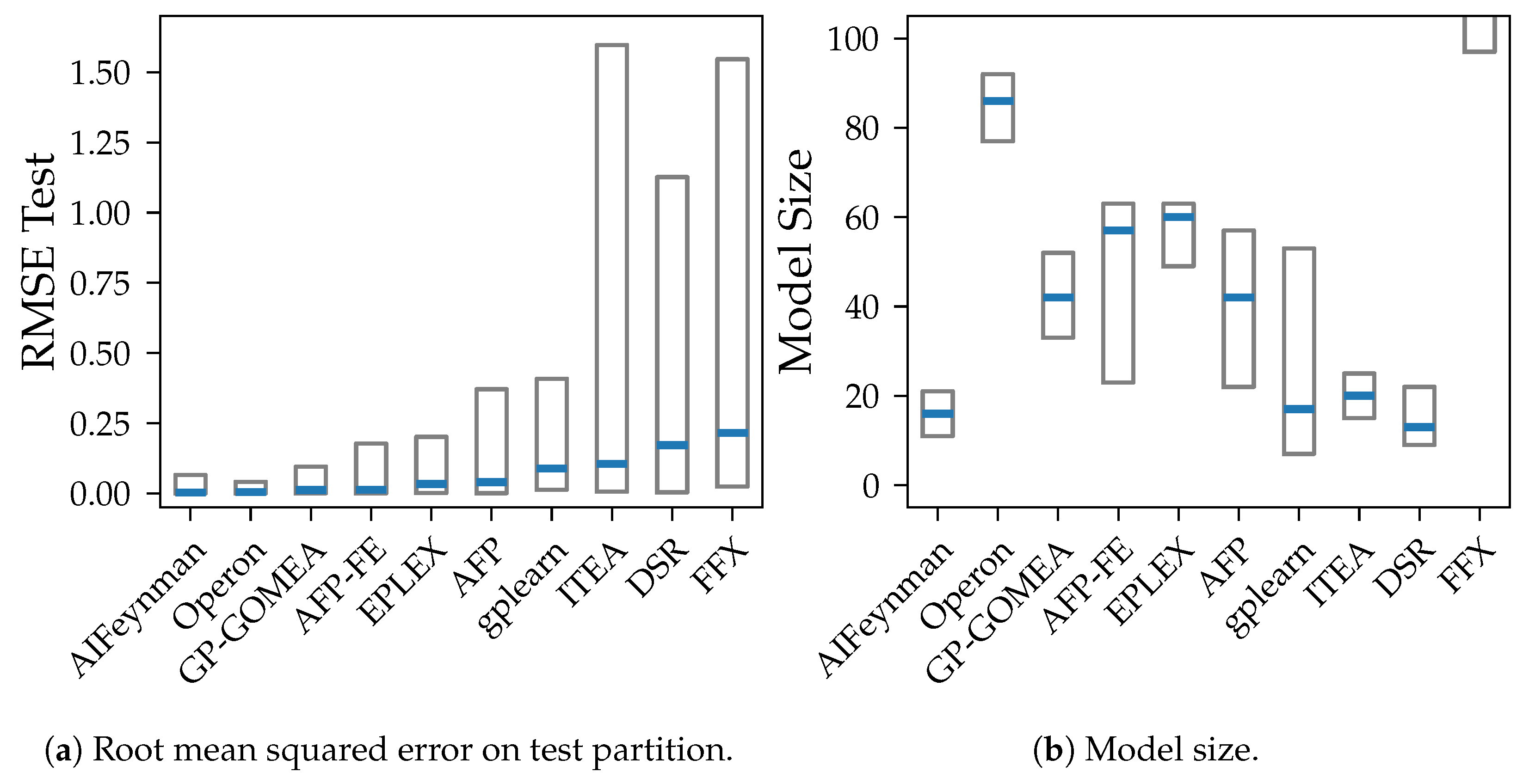
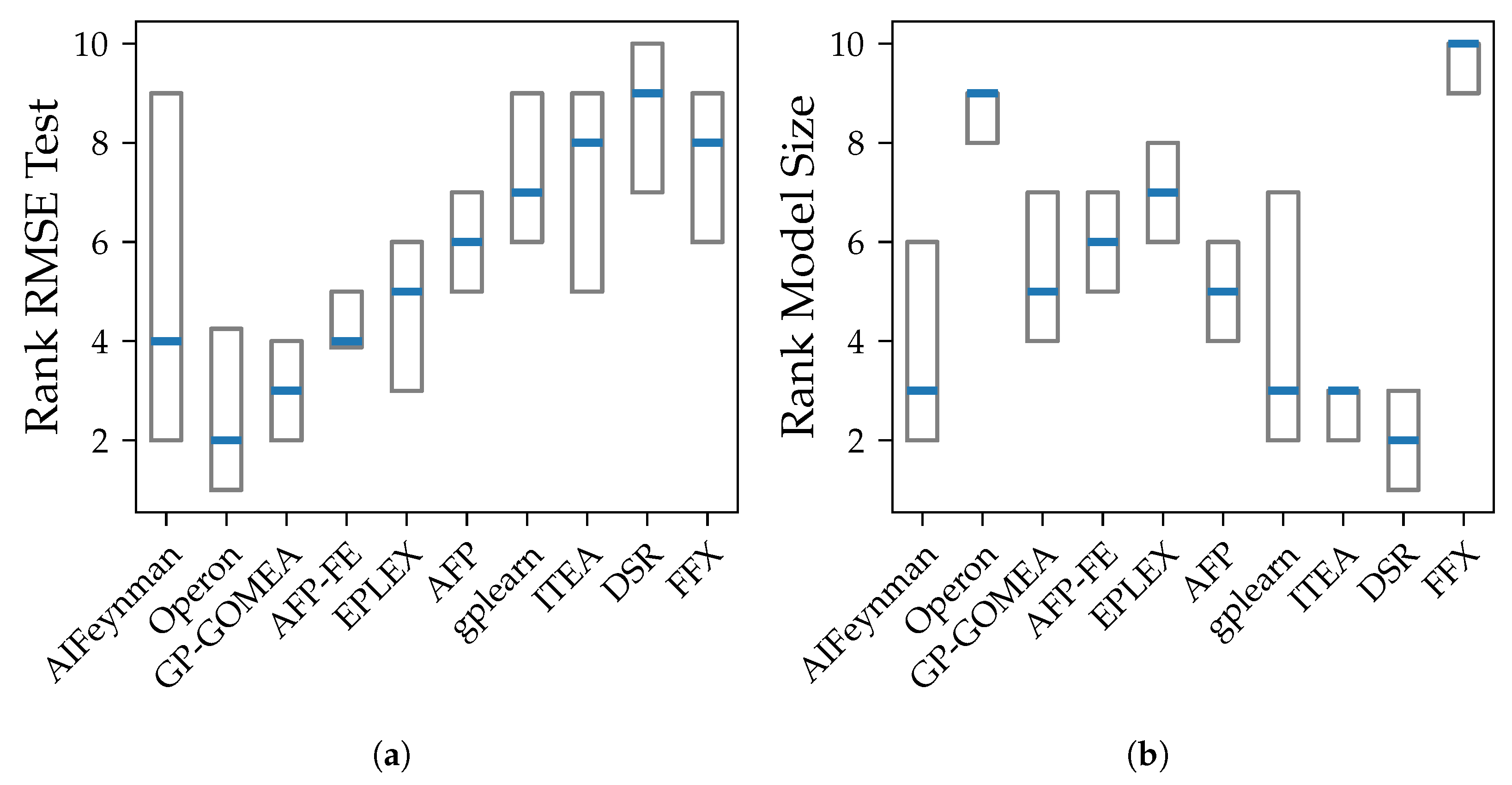
We compare both the accuracy, the model size, bias, and variance. We use the root mean squared error (RMSE) to measure the accuracy, since it is on the same scale as the applied noise and therefore the bias and variance. The model size is the number of symbols in the model as defined in SRBench [
13]. The model size describes the inverse of parsimony and is used as a notion of simplicity of a model.
3.1. Bias/Variance Calculation
To compare the bias and variance between problems and algorithms, we use the expected value of the bias and variance over x. Given are a feature value distribution P, a ground truth , a noise level with a data generating function , and a distribution of training sets with , , . P, , are problem-specific and defined in SRBench.
We reuse the ten models that were trained in SRBench for each problem. Every model was trained on different training set drawn from
D. We generate a new set of data
to estimate the expected value for bias and variance for those ten models. We define the estimators as the average of bias and variance over the ten models for all values in
X, as defined in Equation (
3) for bias and Equation (
4) for variance. To prevent sample-specific outliers, we use the one set of samples
X per problem for all algorithms.
The bias for one value
defined in Equation (
1) can become both negative and positive. However, we are not interested in the sign of the bias but only in its absolute value. To be consistent with the bias/variance decomposition by Hastie et al. [
1], which decomposes the error into the sum of variance, the square of bias and an irreducible error, we also use the square of the bias
from Equation (
1) for the calculation of bias of the overall problem and algorithm in
. We take the square root of the average of the squared bias to be on the same scale as the error of a model.
3.2. SRBench Algorithms and Models
The SR algorithms tested in SRBench provide different approaches to limit the size and/or structure of their produced models to counteract over-fitting and therefore reduce the algorithm’s variance. Therefore, we expect clear differences. E.g., GP implementations such as GP-GOMEA [
22], Operon [
23], AFP [
24] and AFP-FE [
25] adapt their search procedure. GP-GOMEA identifies building blocks that cover essential dependencies. Operon, AFP and AFP-FE use a multi-objective search procedure to incorporate both accuracy and parsimony in their objective function [
23,
24,
25]. EPLEX, AFP and AFP-FE use the
-lexicase selection in their GP procedure [
26], in which instead of one aggregated error value multiple tests are performed over different regions of the training data. ITEA [
27] deliberately restricts the structure of their models. gplearn (
gplearn.readthedocs.io, accessed on 21 November 2024) provides a GP implementation that is close to the very first ideas about GP-based SR by Koza [
5]. DSR [
28] is a non-GP-based algorithm that considers SR as a reinforcement learning problem and uses a neural network, another high-variance method, to produce a distribution of small symbolic models. FFX [
29] and AIFeynman 2.0 [
19] do also not build upon GP but run deterministic search strategies in restricted search spaces. We expect that both determinism and restrictions in the search space result in higher bias and therefore lower variance [
1].
We reuse the string representation of all models from the published SRBench GitHub-repository. The strings are parsed in the same way as in SRBench [
8] with the SymPy [
30] Python framework. However, certain algorithms, such as Operon, lack in precision in their string representation, which printed only three decimal places for real-valued parameters in the model. We re-tuned those parameters for models that didn’t achieve the in SRBench reported accuracy when just evaluating the model using the reported parameter values. We re-tune the parameters with the L-BFGS-B algorithm [
31,
32] as implemented in the SciPy library [
33]. We constrain the optimization so that we only optimize decimal places of the reported parameters, e.g., the optimization of the reported value
is constrained to the interval
. The goal is to change the original model as little as possible and prevent further distortion of the overall results, while still be able to reuse results from SRBench.
We skip algorithms, where we could not reproduce their reported accuracy with the corresponding reported string representation of their models. We excluded Bayesian Symbolic Regression (BSR) [
34], Feature Engineering Automation Tool (FEAT) [
35], Multiple Regression GP (MRGP) [
36] and Semantic Backpropagation GP (SBGP) [
37].
5. Conclusions and Outlook
In this study, we analyze the bias and variance of ten contemporary SR methods. We show, how small differences in training data affect the behavior of the models besides differences in error metrics. We use the models that were trained in the SRBench benchmark [
8], as they provide a well-established setting for a fair algorithm comparison.
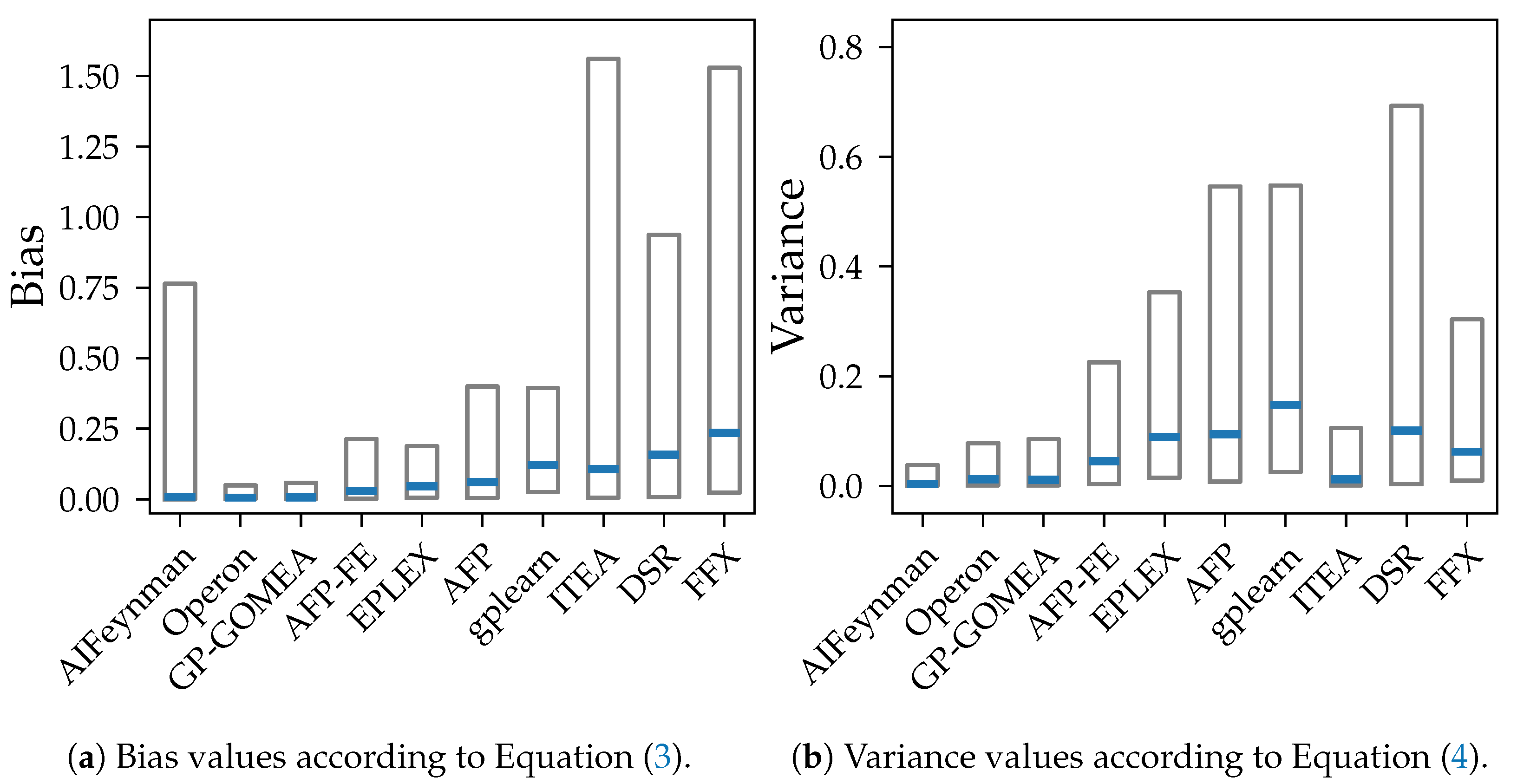
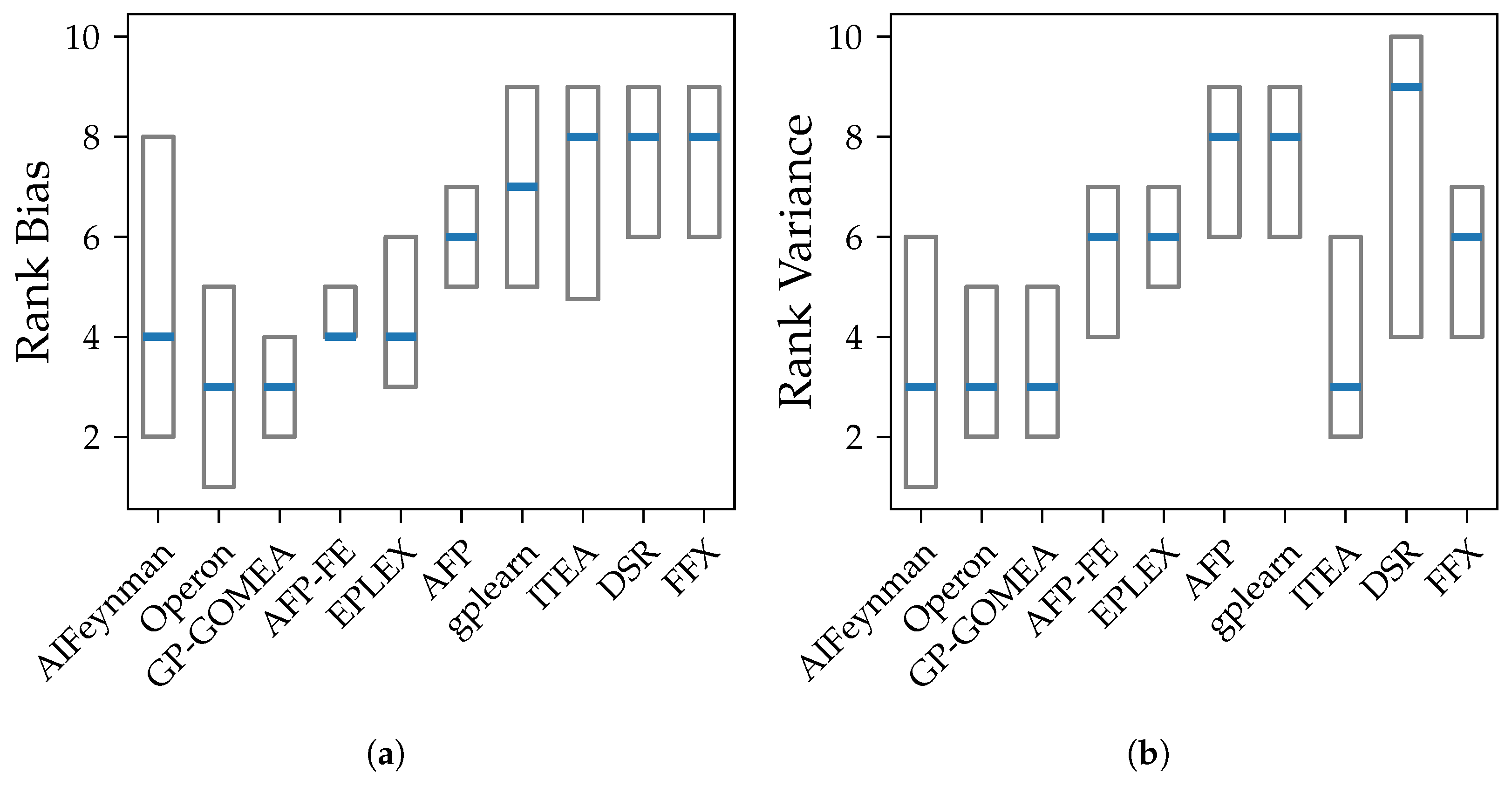
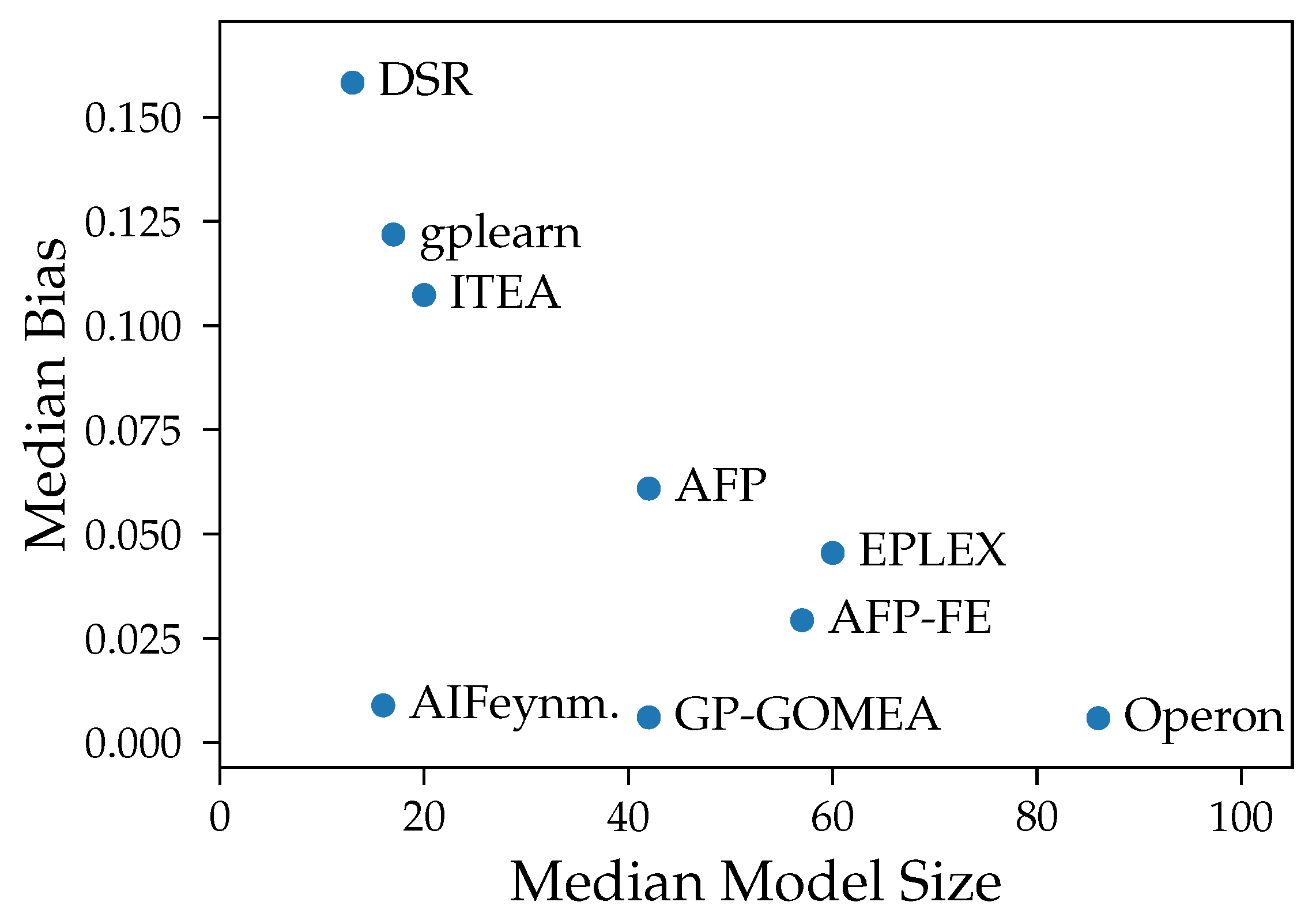
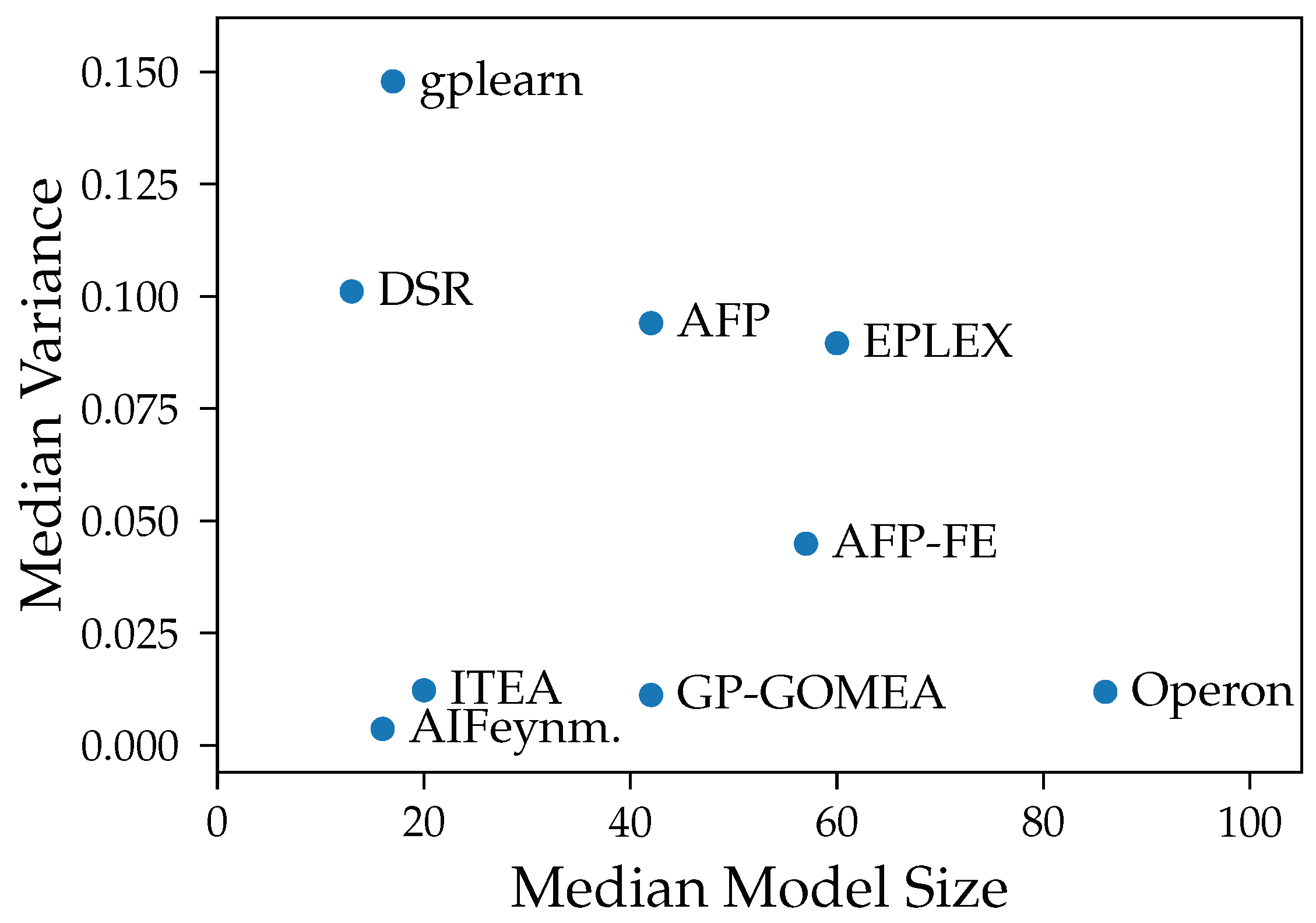
We show that both bias and variance increase with the test error of models over most algorithms. Exceptions are the algorithms AIFeynman, ITEA and FFX, whose error is primarily caused by bias. This is expected as stronger restrictions of the search space should lead to more similar outputs despite changes in training data but also to a consistent, systematic error in all models. Another assumption for the high bias for AIFeynman and FFX are their non-evolutionary heuristic search, which induces bias.
Our experiments prove our expectation that larger models tend to have smaller bias, except from FFX. However, despite the common assumption that larger models are susceptible to high variance, we could not observe a clear connection between those two properties in our analysis. The connection between variance and model size is distorted by the different median accuracy of the methods. Given that high accuracy is achieved both by algorithms with small and large models, also small variance occurs in algorithms with models of any size. E.g., Operon and GP-GOMEA provide the smallest error and variance across all noise levels, however, the size of their models differs clearly. While GP-GOMEA’s average model size ranks between the other algorithms, Operon found the largest models of all GP-based methods. However, this still implies, that even though Operon’s models are large and might look very different between multiple algorithm runs, their behavior on the Feynman datasets are very consistent.
This work is a first step towards the analysis of bias and variance in the symbolic regression domain. Although bias and variance belongs to the basic knowledge in machine learning, recent studies for other machine learning methods challenge the common understanding of this topic. Therefore, we also suggest further research in this direction for symbolic regression. The most obvious extension of our work would be an analysis on real world problems, as this work was restricted to generated benchmark data of problems with limited dimensionality. While the presence of actually unknown noise, an unknown ground truth and usually a too small number of observations in such scenarios make a fair comparison hard, it would give even further insight in the practicality of SR methods. Moreover, our analysis, especially regarding variance, was limited by the high accuracy of multiple algorithms on the given synthetic problems. Further research regarding the relationship between variance and model size would benefit from harder problems, where all algorithms yield a certain level of error. This would allow a more in-depth analysis of variance and highlight more differences between algorithms, especially between GP-GOMEA and Operon. Another SR-specific aspect for further research is the analysis of the symbolic structure of SR models. While this work only focuses on the behavior and output and therefore the semantic of models, another important aspect for practitioners is whether the formulas found in SR are similar from a syntactic perspective.