Abstract
Clustering ensemble integrates multiple base clustering results to improve the stability and robustness of the single clustering method. It consists of two principal steps: a generation step, which is about the creation of base clusterings, and a consensus function, which is the integration of all clusterings obtained in the generation step. However, most of the existing base clustering algorithms used in the generation step are shallow clustering algorithms such as k-means. These shallow clustering algorithms do not work well or even fail when dealing with large-scale, high-dimensional unstructured data. The emergence of deep clustering algorithms provides a solution to address this challenge. Deep clustering combines the unsupervised commonality of deep representation learning to address complex high-dimensional data clustering, which has achieved excellent performance in many fields. In light of this, we introduce deep clustering into clustering ensemble and propose an improved selective deep-learning-based clustering ensemble algorithm (ISDCE). ISDCE exploits the deep clustering algorithm with different initialization parameters to generate multiple diverse base clusterings. Next, ISDCE constructs ensemble quality and diversity evaluation metrics of base clusterings to select higher-quality and rich-diversity candidate base clusterings. Finally, a weighted graph partition consensus function is utilized to aggregate the candidate base clusterings to obtain a consensus clustering result. Extensive experimental results on various types of datasets demonstrate that ISDCE performs significantly better than existing clustering ensemble approaches.
1. Introduction
With the explosive growth of 5G, big data have penetrated into every aspect of daily life [1]. These data typically present large scales, high dimensions, and complex structures [2]. How to mine valuable information from these complex data has become an urgent challenge at present [3]. Clustering analysis [4] is an essential technique in many fields of research that involves processing multidimensional data, such as pattern recognition, data retrieval, and bioinformatics. It is normally considered an unsupervised method for grouping data based on similarity [5]. Traditional clustering methods, such as k-means [6], spectral clustering [7], and Gaussian mixture clustering [8], have achieved good performance in various fields. However, most traditional clustering algorithms are only able to exploit shallow features of the data and cannot excavate the interdependence of complex data features in latent space [9]. On the other hand, there is no single clustering algorithm capable of suitably applying to all datasets, so various clustering algorithms are proposed and improved. To address this problem, the concept of clustering ensemble (consensus clustering) [10] is introduced, which is inspired by the success of the supervised classifiers ensemble. Clustering ensemble has been applied in many fields [11] and combines a set of clusterings into a final consensus clustering [12].
Generally, the clustering ensemble method refers to two steps: generation and consensus function. Generation is the first step in generating the set of base clusterings. In clustering ensemble, an appropriate generation process is very important because the final consensus result will be affected by the initial clusterings obtained in this step. The consensus function is the second step, and plays a major role in the clustering ensemble. It is a great challenge to define an appropriate consensus function to improve the results of single clustering algorithms. However, most of the existing base clustering algorithms used in the generation step of clustering ensemble methods are traditional shallow clustering algorithms. It is difficult for these traditional clustering algorithms to exploit deep features of the data and excavate the interdependence of complex data features in latent space [5]. Therefore, the performance of these shallow clusterings after aggregation will also be limited.
In recent years, the emergence and development of deep clustering methods [13] have provided the idea to address complex data clustering, which simultaneously learns cluster assignments and feature representations using deep neural networks. Deep clustering typically learns a mapping from the original data space to a lower-dimensional feature space and iteratively optimizes the clustering objective in that feature space. Existing deep clustering algorithms can be roughly divided into three categories based on different deep representation layers [14]: autoencoder-based (AE-based) deep clustering, variational-autoencoder-based (VAE-based) deep clustering, and generative-adversarial-network-based (GAN-based) deep clustering. We elaborate these deep clustering methods in detail in related work. Compared to traditional shallow clustering methods, existing deep clustering algorithms have achieved excellent performance in clustering complex high-dimensional data [15,16]. However, the existing deep clustering is very sensitive to network parameters and hyperparameters, so the clustering results fluctuate greatly and are not robust enough [17].
To address the aforementioned problem in clustering ensemble and deep clustering, we propose a clustering ensemble method called improved selective deep clustering ensemble (ISDCE). It combines the idea of clustering ensemble with deep clustering and incorporates a selective strategy. ISDCE can be divided into two phases: the deep clustering generation phase and the selective clustering ensemble phase. In the deep clustering generation phase, unsupervised deep autoencoder networks with different initializations are used to train the non-clustering loss, and then k-means are applied to initialize the cluster centroids. The similarity between the deep low-dimensional embeddings extracted by the autoencoder and the clustering centroids is calculated as soft cluster assignment through Student t-distribution. The KL divergence loss of the the soft cluster assignment and auxiliary distribution is used as the clustering loss. Here, the auxiliary distribution is computed by normalizing the square of the soft cluster assignment. Finally, the clustering loss is jointly and iteratively optimized to obtain multiple deep clustering results. In the selective clustering ensemble phase, firstly, different deep base clusterings are evaluated. Considering that both the quality and diversity of base clusterings affect the final ensemble performance, we construct the ensemble quality and diversity evaluation metrics of base clusterings. The base clusterings with higher quality and diversity are selected as ensemble candidates. Meanwhile, for the consensus function, we consider the local diversity of clusters within the same base clusterings and construct an entropy-based criterion to measure the reliability of clusters. A weighted graph partition consensus function is utilized to efficiently aggregate the candidate base clusterings. The final consensus clustering result is obtained by using Tcut [18] for graph partitioning.
The main contributions of our work are summarized as follows:
- This paper studies the limitations of clustering ensemble and deep clustering, which is an interest and important topic. Then, we propose an improved selective deep clustering ensemble (ISDCE) method to mitigate the problem. It incorporates deep clustering into selective clustering ensemble to enhance the robustness and clustering performance.
- ISDCE constructs the ensemble quality and diversity evaluation metrics of base clusterings. It is able to select higher-quality and rich-diversity base clusterings to improve ensemble performance. In addition, ISDCE measure the reliability of clusters and the local diversity of clusters within the same base clusterings to further improve the integration performance.
- Extensive experimental results on various types of datasets confirm that ISDCE performs significantly more robustly and better than existing clustering ensemble approaches.
The remainder of this paper is organized as follows: we review related work on deep clustering and ensemble clustering in detail in Section 2. Then, we elaborate on our proposed ISDCE methodology in Section 3. Section 4 presents the experimental results. Finally, Section 5 discusses the conclusions.
2. Related Work
In this paper, we propose an improved selective deep-learning-based clustering ensemble algorithm that is closely related to three branches of research: clustering ensemble, deep clustering, and selection ensemble. The following will introduce the related work in these three fields.
2.1. Clustering Ensemble
Existing clustering ensemble methods can be divided into three main categories [12], including pair-wise-similarity-based approaches, median-partitioning-based approaches, and graph- and hypergraph-partitioning-based approaches. We elaborate on some of the popular and classical clustering ensemble algorithms in recent years.
PTA [19] considers clustering ensemble based on sparse graph representation and probabilistic trajectory analysis. The problem of link uncertainty is solved using the elite neighbor selection strategy, and a similarity measure based on probabilistic trajectories is constructed using the K-elite neighbor sparse graph. PTA constructs a K-elite neighbor sparse graph to refine the local links using the random walk information, and derives the similarity based on the probabilistic trajectories by capturing global structural information through the random walk trajectories. Finally, two consensus functions, namely probabilistic trajectory accumulation and probabilistic-trajectory-based graph partitioning, are further proposed. ECFG [20] was proposed as a new ensemble clustering approach, termed ensemble clustering using a factor graph. It introduces super-object representation to facilitate the computation of ensemble processes and solves the optimization problem using an efficient solver based on the factor graph technique. WSCE [21] was developed as a new clustering ensemble framework, utilizing some concepts from the community detection domain and graph-based clustering that address the combination of different evaluated individual clustering results in the absence of a thresholding process. It introduces two-kernel spectral clustering to generate graph-based individual clustering results and normalized modularity measures to provide diversity estimates. SECWK [22] was proposed as spectral ensemble clustering to utilize the advantages of co-association matrices in information integration, but with higher operational efficiency. The time and space complexity of spectral ensemble clustering are significantly reduced by identifying the equivalence between spectral ensemble clustering and weighted k-means. It is a promising candidate for big data clustering.
LWEA [23] is considered to be the most popular algorithm among the weighted clustering ensemble methods proposed in recent years. It considers the local diversity of clusters and utilizes uncertainty estimation and local weighted strategies. The uncertainty of each cluster is estimated by considering the cluster labels in the whole set through an entropy criterion. The ensemble-driven clustering validity measure was introduced and a locally weighted co-correlation matrix was proposed as a summary of the ensemble of different clusters. Two new consensus functions are proposed by exploiting the local diversity in the ensembles. The authors in [24] studied that the co-association matrix may be dominated by poor base clusterings in clustering ensemble, resulting in inferior performance. Then, they introduced low-rank tensor approximation (LRTA) to clustering ensemble and exploited the low-rankness of a three-dimensional tensor formed by the coherent-link matrix and the co-association matrix. LRTA formulates the algorithm as a convex constrained optimization problem and solves it efficiently. ECCMS [25] was introduced as a novel effective co-association matrix self-enhancement model for ensemble clustering that improved the traditional co-association matrix. It exploites the high-confidence information to form a sparse high-confidence matrix and denoising error connections simultaneously. Technically, ECCMS is formulated as a symmetric constrained convex optimization problem, which is efficiently solved by an alternating iterative algorithm. The authors in [26] considered that conventional clustering ensemble methods may usually be misguided by unreliable samples due to the lack of labels. Therefore, they integrated the active clustering ensemble method and proposed a self-paced learning framework (SPACE). It evaluates their difficulty in selecting unreliable data and applies easy data to ensemble.
2.2. Deep Clustering
The emergence and development of deep clustering methods provides a technique that exploits the commonalities of unsupervised deep networks and clustering to simultaneously learn feature representations and cluster assignments. Deep clustering typically involves learning a mapping from the original data space to a low-dimensional feature space and iteratively optimizing the clustering objective in that feature space [14].
DEC [27] was one of the earliest deep clustering methods. It was inspired by deep learning for computer vision and extended it to unsupervised data clustering. It has been cited in more than 2000 articles and has attracted much attention in the field. DEC uses a pre-trained stacked denoising autoencoder to learn deep feature representations, and then defines a probability distribution based on the centroid and minimizes its KL divergence to an auxiliary target distribution in order to improve the clustering assignment and the feature representations at the same time. IDEC [28] was an improvement on DEC, and argues that the clustering loss defined by DEC corrupts the feature space and leads to unrepresentative features. Therefore, it adds back the decoder to optimize the reconstruction error and clustering loss. Clustering features suitable for local structure preservation are learned by adding the denoising autoencoder of the decoder. DCEC [29] is the convolutional autoencoders version of IDEC, and takes advantage of convolutional neural networks to better extract the hierarchical embedding features. It is superior to stacked autoencoders by incorporating spatial relationships between pixels in images. DFKM [30] is a novel deep clustering method with adaptive loss function and entropy regularization that employs fuzzy k-means and uses fuzzy information to represent a clear deep clustering structure. To enhance the robustness of the model, DFKM utilizes a robust loss function with adaptive weights. GFDC [31] utilizes a deep convolutional network for the feature generator and a graph convolutional network with a softmax layer to perform a clustering assignment. It constructs a topological graph to express the spatial relationship of features. These AE-based methods have the advantage of being easy to implement and extend, but are sensitive to network initialization parameters and hyperparameters, and they also require a certain limited number of layers in the deep network structure.
GMVAE [32] assumes that observations are generated from a multi-modal prior distribution and constructs an inference model that can be directly optimized using reparameterization techniques. VaDE [33] embeds the probabilistic clustering problem into a variational autoencoder framework. It models the data generation process through a GMM model and a neural network. VaDE is optimized by maximizing an evidence lower bound on the log-likelihood of the data using a stochastic gradient variational Bayesian estimator and a reparameterization technique. The main difference between GMVAE and VaDE is that the assumed sample-generating model is different and GMVAE is somewhat more complex than VaDE, with poorer empirical results. DCVA [34] considers that the potential space of the autoencoder does not pursue the same clustering goals as the k-means or Gaussian mixture model. Therefore, it introduces a variational autoencoder probabilistic approach that represents distances in the latent space in terms of KL divergence and uses probability distributions as inputs instead of using points in the latent space. Finally, the potential space is clustered using a Bayesian Gaussian mixture model. These VAE-based methods have the advantage of being able to generate samples with reasonable theoretical guarantees, but have the disadvantage of high computational complexity.
Witnessing the great success of generative adversarial networks (GANs) in estimating complex data distributions, DAC [35] introduces generative adversarial networks [36] to deep clustering. It matches the aggregated posterior of the latent representation with a Gaussian mixture distribution and optimizes three objectives. ClusterGAN [37] argues that the cluster structure may not be better preserved in the GAN latent space, and utilizes discrete and continuous latent variables and co-trains the GAN with the inverse mapping network clustering loss, where the distance geometry of the projection space mirrors the distance geometry of the variables. These GAN-based methods may lead to pattern collapse and difficulties in the convergence of the algorithm.
2.3. Selection Ensemble
Selection ensemble (or ensemble pruning) [38] is a variant of clustering ensemble that selects an appropriate subset of base clusterings and forms a smaller ensemble that performs better than all of the base clusterings.
Hadjitodorov et al. [39] adopted the ARI measure to evaluate the quality and diversity of the ensemble. They constructed four variants of ARI to measure diversity. They showed that the median diversity selections are usually significantly better than a randomly chosen ensemble. Fern et al. [40] studied the literature for simultaneously considering the diversity and quality of the ensemble. They showed that the combination of quality and diversity for selection ensemble can produce better results using the sum of the normalized mutual information measure. Jia et al. [41] made an improvement on an innovative selective clustering, called a selective spectral clustering ensemble algorithm based on the bagging technique. The base ensembles were generated by employing spectral clustering with random initialization. Then, the bagging technique was applied to ranking and evaluating the component clustering. Based on this ranking, the candidate ensemble was selected for the final solution.
3. Improved Selective Deep Clustering Ensemble
The framework of the ISDCE method is illustrated in Figure 1. As can be seen in Figure 1, similar to the classical clustering ensemble step, ISDCE contains two phases: a deep clustering generation step and selective clustering ensemble step. In the deep clustering generation step, the blue and green network structures in Figure 1 represent the network framework for deep autoencoder clustering. We firstly trained S deep autoencoder clusterings on the original input data, such as blue network structures to in Figure 1. The selective clustering ensemble step consists of two main modules: selective strategy and consensus ensemble. Considering that partial ensemble may outperform full ensemble, we incorporated the selective strategy into the and constructed the ensemble quality and diversity evaluation metrics of base clusterings to select M higher-quality and rich-diversity candidate clusterings (such as green network structures to in Figure 1). In the consensus ensemble step, we considered the local diversity of clusters within the same base clusterings. An entropy-based criterion was utilized to estimate the local uncertainty of different ensemble members. Considering that graph-based consensus functions can be applied to larger-scale data, we constructed a weighted graph partition consensus function to efficiently aggregate the candidate clusterings. The final consensus clustering result was obtained by using Tcut [18] for graph partitioning. The details of each module are described below.
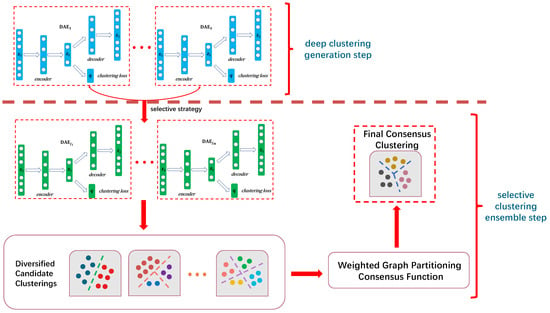
Figure 1.
Illustration of the proposed ISDCE framework.
3.1. Deep Clustering Generation
The framework of deep autoencoder clustering is shown in Figure 2. Deep autoencoder clustering consists of a fully connected autoencoder and a clustering layer, which is connected to the autoencoder embedding layer. In Figure 2, represents the i-th sample of the original input data and indicates the -th reconstructed sample. represents the deep low-dimensional embedding representation of the i-th sample. Given the original input data , deep autoencoder clustering firstly learns a mapping from the original data to a lower-dimensional embedding representation . Then, it maps each embedding point of the input sample to a soft cluster label q using a clustering layer. Therefore, the loss of deep autoencoder clustering contains two parts: non-clustering loss (or reconstruction loss) and clustering loss.
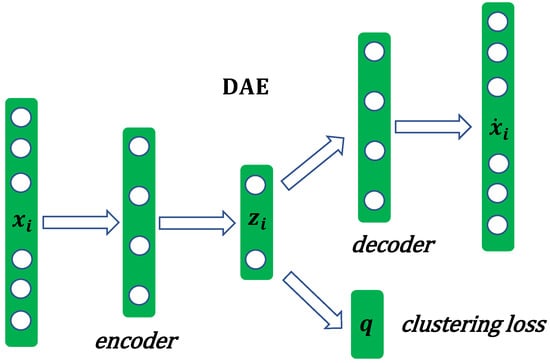
Figure 2.
The structure of deep autoencoder clustering.
The non-clustering loss of deep autoencoder clustering consists of the fully connected autoencoder reconstruction loss, computed as:
Then, Student’s t-distribution is used to measure the similarity between the deep representation and the clustering centroid . The clustering centroid is generated by k-means and used as training weights. The clustering layer maps each embedding point to a soft cluster assignment as follows:
is the degree of freedom, but since it is not possible to cross-validate the validation set in an unsupervised environment, this parameter learning is redundant and, in all later experiments, we set [27]. represents the probability of assigning the i-th sample to the j-th cluster. is the j-th cluster centroid obtained from the initialization of k-means. After that, an auxiliary target distribution is constructed to help iteratively optimize the cluster assignment. We used KL divergence as the clustering loss between the soft assignment Q and the auxiliary distribution P. Here, unlike general autoencoder clustering, our clustering loss was used to scatter the embedded points z and the reconstruction loss. This makes sure that the embedded space preserves the local structure of the data-generating distribution. The clustering loss was computed as follows:
where the auxiliary target distribution P is obtained by normalizing Q squared:
The gradients of for each cluster centroid and for each embedding point were subsequently calculated as:
The gradient of to was passed to the deep network and used for standard backpropagation to compute the parameter gradients. Finally, to obtain the final cluster assignment, the iteration was stopped when it reaches the maximum number of iterations or when the update of the cluster assignment for two consecutive optimization iterations is less than a threshold. In order to clearly show the deep autoencoder clustering computation step, the specific steps are summarized in Algorithm 1.
The whole deep clustering network objective is defined as:
where and are reconstruction loss and clustering loss, respectively, and is a coefficient that controls the degree of distorting embedded space. In subsequent experiments, the gamma value is usually set to 0.1.
| Algorithm 1 Deep Autoencoder Clustering |
| Input: X: Original Input data; k: the number of clusters; S: the number of base deep clustering; : Maximum iteration times; : auxiliary distribution update interval; : stopped threshold; : autoencoder layer parameters. |
| Output: S deep base partitions |
for to S do Step1: Initialize autoencoder network parameters and weights. for to do if then Step2: Calculate all the embedding points according to the parameters of the corresponding dataset. Step3: Initial clustering and clustering centroid were obtained using k-means. Step4: Updating auxiliary target distribution P by Equations (2), (4) and . Step5: Save the cluster assignment pre. Step6: Calculate the new cluster assignments pre. if then Training stopped! end if end if Step7: Update of the autoencoder weights and cluster centroid. end for Step8: Save the rounds-th deep clustering end for return S deep base clustering |
3.2. Selective Clustering Ensemble
3.2.1. Selective Strategy
In the selective strategy, we firstly constructed the ensemble quality and diversity evaluation metrics of ensemble . denotes the r-th base clustering in and indicates the -th cluster of the r-th base clustering. is the number of clusters.
With regard to ensemble quality evaluation, for a given ensemble , we constructed a summational adjusted Rand index (SARI) as a means of evaluating the quality of each base clustering :
Here, the SARI measures the degree of consistency of the overall trend contained in a particular cluster and ensemble . Obviously, when the value of SARI is larger, the quality of is higher. The ARI is the adjusted Rand index, which comprises the common external clustering metrics.
With regard to ensemble diversity evaluation, for a given ensemble , we constructed pair-wise normalized mutual information (PNMI) to evaluate the diversity of the enemble :
NMI is the normalized mutual information, which is an external metric used in clustering to measure the degree of similarity between two clustering results. Here, when the value of PNMI is smaller, the diversity is richer in ensemble .
After defining the ensemble quality and diversity evaluation criteria, a certain number of higher quality and rich diversity candidate ensemble need to be selected from the ensemble . Three selection strategies can be used here:
- (1)
- Quality Strategy (QS). For a given ensemble , the QS adopts to compute all the base clusterings in the ensemble and ranks them in descending order to select the top M () base clusterings with higher SARI values as the candidate ensemble. In general, base clusterings with higher SARI values show more overall trend consistency. Base clusterings with lower SARI values can be considered outliers in the ensemble and may be unfavorable for inclusion in the ensemble.
- (2)
- Diversity Strategy (DS). The DS is a strategy that seeks to maximize ensemble diversity. For a given ensemble , we select M () base clusterings to minimize the PNMI. We can view this objective as a problem of finding weight-maximizing subgraphs, where the edge weight of each vertex is . However, this problem is NP-hard. Therefore, we approximate the solution of the problem using a simple greedy strategy. First, the highest-quality base clusterings are selected to form a new ensemble E using SARI computation, and then a base clustering from the ensemble is gradually selected to be added to the ensemble E so as to minimize the PNMI value. This process is repeated until the number of base clusterings in the ensemble E reaches M.
- (3)
- Balance Strategy (BS). The BS is a combination of the above two strategies. The two metrics SARI and PNMI are combined to form a new metric balance strategy index (BSI) as follows:where is an adjustment factor used to control for quality and diversity. Similarly, this joint metric BSI is solved greedily using a DS-like approach.
3.2.2. Consensus Ensemble
In the consensus ensemble of ISDCE, we considered cluster-wise diversity inside the same base clusterings to enhance the consensus performance [23]. We adopted the local weighting idea to evaluate the reliability of clusters. Given the ensemble , where denotes the m-th base clustering in and indicates the i-th cluster, the uncertainty of cluster for is computed as follows:
where
where is the number of clusters in and is the j-th cluster in . indicates the number of objects in .
Therefore, the uncertainty (or entropy) of for ensemble can be calculated as follows:
where M is the number of base clusterings in .
Then, we constructed a ensemble cluster index (ECI) to compute the reliability of clusters, which is defined as follows:
It is easy to see from the above equation that (0,1] and, the smaller the uncertainty of the cluster, the larger the ECI value. is a coefficient that adjusts for the effects of cluster uncertainty.
Then, based on ECI, we exploited the weighted graph partitioning consensus function. We defined the weighted graph as and computed the link weight between two nodes and as follows:
where X is the original data and is all of the clusters in the ensemble . denotes the sum of clusters in .
The time complexity of ISDCE can be roughly divided into two parts: deep clustering and clustering ensemble. The time complexity of deep clustering section is , where , k, and D are, respectively, the dimension of the embedding layer, number of clusters, and maximum number of neurons in hidden layers. The time complexity of clustering ensemble section is , where is the number of clusters in and l is the average number of links connecting to a node in the graph.
4. Experiments
In this section, to verify the effectiveness of our proposed ISDCE algorithm, we conducted a series of experiments on 7 datasets to compare ISDCE against 11 clustering ensemble algorithms.
4.1. Datasets and Evaluation Measures
We conducted experiments on three widely used UCI datasets (http://archive.ics.uci.edu/ (accessed on 3 November 2023)), two classical image datasets (https://github.com/zhoujielaoyu/2018-NC-DREC (accessed on 3 November 2023)), and two real-world biological datasets (https://github.com/BinPro/CONCOCT/tree/develop (accessed on 3 November 2023)), where a summary of the statistics of these datasets can be found in Table 1.

Table 1.
Statistics of the datasets.
- Cars: Cars dataset is the 1983 ASA Data Exposition dataset, which contains mpg, cylinders, displacement, etc. (8 variables), for 392 different cars.
- Iris: Iris dataset [42] contains 150 samples for 3 classes, where each class refers to a category of iris plant.
- Wine: Wine dataset [43] is the results of a chemical analysis of wines grown in the same region, which determined the quantities of 13 constituents found in each of the three types of wines.
- MNIST: MNIST dataset is a set of well-known image data of 70,000 handwritten digits (10 class labels) with 784 pixels. Mnist5 is the subset of the MNIST [44].
- Strain: Strain dataset is a set of synthetic mock metagenome data [45]. This dataset was constructed to investigate the impact of strain-level variation on clustering.
- Species: Species dataset is also a set of synthetic mock metagenome data [45]. It was designed to resolve species-level variation in a complex community.
To evaluate the quality of the clustering result, we adopted normalized mutual information (NMI) and adjusted Rand index (ARI) as evaluation measures. Greater values of these metrics represent better clustering performance. Detailed introduction can be referred to in the following literature: [46].
4.2. Experimental Settings and Clustering Performance Comparison
In this section, we compare the proposed ISDCE algorithm with 11 comparison algorithms, i.e., WSCE [21], PTAAL, PTACL, PTASL [19], PTGP [19], SECWK [22], LWEA [23], LWGP [23], LRTA [24], ECCMS [25], and SPACE [26]. For the 11 comparison algorithms, the parameters will be set as suggested by their corresponding papers. The total number of base clusterings is 50. The number of base clusterings involved in the ensemble is 20. For each algorithm, the final number of clusters was set to the true number of categories on the dataset. The running time of the algorithm is 20. The NMI, ARI, mean, and variance of the 20 running results were used as the evaluation results.
For our proposed ISDCE algorithm, the initial ensemble size S = 50 and the top M is in the range of [10, ]. The number of clusters k was set to the true number of categories on the dataset. The parameter , which adjusts for the effects of cluster uncertainty, was set uniformly to 0.4 (a range of [0.2, 1] is recommended). Here, we experimented with three selective strategies (quality strategy, diversity strategy, and balance strategy) in the ISDCE, where the adjustment factor in the balance strategy. Regarding the parameter settings for deep base clustering, we used randomly initialized network parameters. The optimizer is uniformly Adam and the activation function is RELU, The corresponding autoencoder network structure for the experimental data is: Cars—[8,8,6], Iris—[4,4,4], Wine—[13,12,10], Mnist5—[784,500,500,2000,10], MNIST—[784,500,500,2000,10], Strain—[200,100,30], Species—[232,100,30].
The clustering performance comparison result is shown in Table 2, Table 3, Table 4 and Table 5. For each dataset, we ran it 20 times and computed average and standard deviation result for evaluation measures. All the bolded results in Table 2, Table 3, Table 4 and Table 5 are statistically (according to pair-wise t-test at significance level) superior to the other methods. ISDCEQS denotes that ISDCE uses quality strategy for ensemble, and ISDCEDS indicates that ISDCE adopts diversity strategy for ensemble. ISDCEBS represents that ISDCE employs balance strategy for ensemble.

Table 2.
Average NMI results of the comparison method with ISDCE on three UCI datasets (The best three scores in each column are highlighted in bold).

Table 3.
Average ARI results of the comparison method with ISDCE on three UCI datasets (The best three scores in each column are highlighted in bold).

Table 4.
Average NMI results of the comparison method with ISDCE on two image and two biological datasets (The best three scores in each column are highlighted in bold.)

Table 5.
Average ARI results of the comparison method with ISDCE on two image and two biological datasets (The best three scores in each column are highlighted in bold).
Table 2 and Table 3 show the clustering performance of three UCI datasets. For each dataset, the highest three scores are highlighted in bold. As can be seen from the NMI results, three strategies of ISDCE achieved the highest score, except for the Cars dataset. SPACE achieved the best NMI on the Cars dataset, but its ARI value is obviously lower than ISDCE in Table 3. This shows that the ISDCE method, especially ISDCEDS, has a better clustering performance and better robustness. In addition to the Cars dataset, three strategies of ISDCE achieved the best ARI scores on the other two datasets. ISDCEDS achieved the best ARI on the Cars dataset, but ISDCEQS and ISDCEBS seem to not be good enough. The reason for why ISDCEDS has a much larger value than other methods may be that, for Cars data, rich ensemble diversity may have its main role in the integration process. ISDCEDS utilizes the rich diversity of base partitions to integrally generate better consensus clustering results. LWEA, LWGP, and ECCMS perform better than ISDCEQS and ISDCEBS, while they perform far worse than ISDCEDS. In comparison experiments on these three small UCI data, we observe that all methods, except for the LRTA and SPACE method, obtain clustering results in less than 1 s. This indicates that these methods can obtain the final ensemble results quickly at a small data scale.
Table 4 and Table 5 show the clustering performance of two image datasets and two biological datasets. “timeout” means that the clustering results were not obtained for a long time. “error” means that, when the eigs function was calculated, dnaupd did not find any eigenvalues that achieved sufficient accuracy using the WSCE method. As can be seen from the NMI results in Table 4, three strategies of ISDCE achieved the best NMI scores on all image and biological datasets. From the ARI results in Table 5, it can be observed that three strategies of ISDCE also obtained the best scores on all image and biological datasets. The overall results from Table 2, Table 3, Table 4 and Table 5 demonstrate the excellent clustering performance of our ISDCE method.
Due to the large number of comparison methods and experimental data involved, this paper does not show the execution time of each algorithm. However, from the experimental results of three image datasets with gradually increasing sample sizes, it is observed that the SECWK algorithm is the fastest, followed by LWGP and our ISDCE. The LRTA method has the slowest running efficiency. In addition, WSCE, LRTA, ECCMS, and SPACE grow exponentially in running efficiency with an increase in data samples, thus failing to obtain the final clustering results on MNIST data for a longer period of time. The execution time of the remaining methods is in the same order of magnitude. In the following, we will conduct a series of experiments to verify the validity of each component of our ISDCE approach.
4.3. ISDCE Component Ablation Experiment
In this section, to validate the ensemble effectiveness of the ISDCE algorithm, we compared the ISDCE with a single deep autoencoder clustering algorithm. To demonstrate the validity of the selection strategy, we ran the ISDCE without a selection strategy (ISDCE_noSS). The experimental results are shown in Table 6 and Table 7 below. From Table 6 and Table 7, it is easy to conclude that combining the ensemble idea and selection strategy in the ISDCE algorithm significantly improves the clustering performance. The clustering performance of ISDCE_noSS is better than the single deep autoencoder clustering, while the performance of ISDCE is almost better than ISDCE_noCC and the clustering results are more stable.

Table 6.
Average NMI results of the ISDCE component ablation experiment.

Table 7.
Average ARI results of the ISDCE component ablation experiment.
4.4. ISDCE t-SNE Visualization
In this section, to verify that ISDCE could achieve good clustering results, we adopted t-SNE to visualize the initial embedding and final embedding clustering results on Iris and Mnist5 datasets. The visualization results are shown in Figure 3, Figure 4, Figure 5 and Figure 6. The horizontal and vertical coordinates of tsne_1, tsne_2 in the figure are the scalar numerical coordinates of the initial embedding and the final embedding reduced to 2D features after t-SNE. The number of clusters visualized is the true number of categories on the dataset. Points of different colors represent samples in different clusters. It can be seen that the final embedding after joint optimization with clustering loss is more cluster-friendly and produces higher-quality clusters. We can easily observe that the final embedding trained by the joint clustering loss forms denser clusters with fewer mixed clusters. For the visualization results of the Iris data, the data are characterized in that only one class in the Iris data is linearly separable, and the other two classes are linearly indivisible. Therefore, visualizing the Iris results with t-SNE results in a situation where there are two clusters that are indistinguishable. But, in fact, from the two clustering evaluation measures, ISDCE indeed achieved good performance on these data.
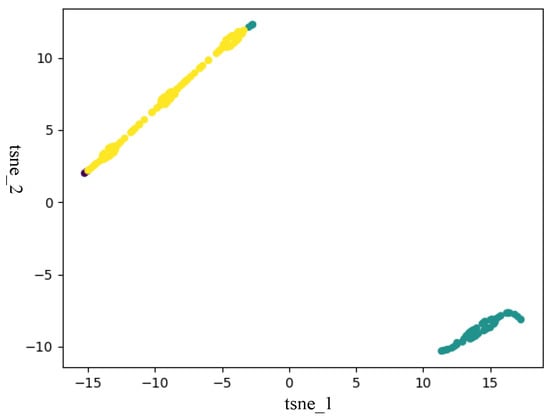
Figure 3.
Initial embedding t-SNE visualization of Iris dataset.
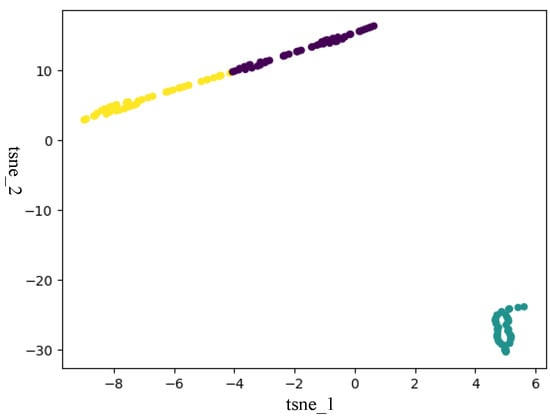
Figure 4.
Final embedding t-SNE visualization of Iris dataset.
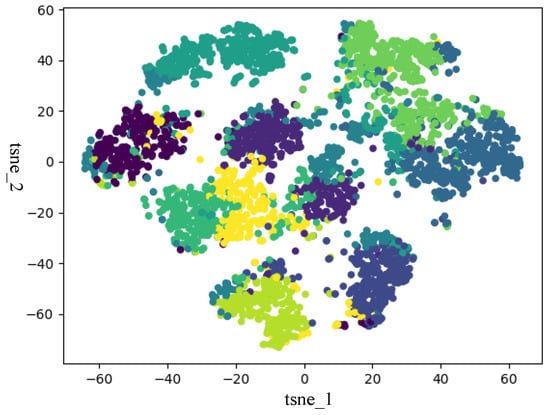
Figure 5.
Initial embedding t-SNE visualization of Mnist5 dataset.
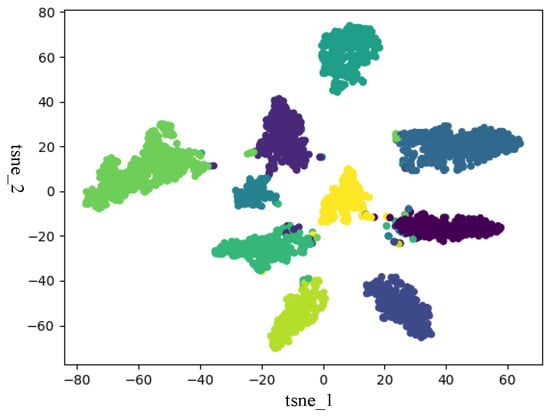
Figure 6.
Final embedding t-SNE visualization of Mnist5 dataset.
5. Conclusions
In this paper, we propose an improved selective deep-learning-based clustering ensemble algorithm (ISDCE). By combining selective strategies and clustering ensemble techniques with deep clustering algorithms, it effectively addresses the challenges faced by existing clustering ensemble and deep clustering algorithms and enables them to achieve more stable and higher-quality clustering results when facing different types of complex data. Comparison experiments with a variety of integrated clustering algorithms on different types of data show that the clustering results produced by the ISDCE algorithm are more robust, with better clustering performance and strong practicality, which can provide basic support for the development of clustering technology. The limitation of ISDCE is the need to ensure data completeness. Combining incomplete data clustering with the proposed method is the main direction of our future work.
Author Contributions
Conceptualization, Y.Q., L.Z. and S.Y.; methodology, Y.Q., L.Z. and S.Y. software, S.Y.; validation, L.Z., S.Y., T.W. and Y.H.; formal analysis, Y.Q. and S.Y.; investigation, Y.Q. and S.Y.; resources, Y.Q. and S.Y.; data curation, L.Z., S.Y. and Y.H.; writing—original draft preparation, Y.Q., L.Z. and S.Y.; writing—review and editing, Y.Q., L.Z., S.Y., T.W. and Y.H.; visualization, T.W. and Y.H.; supervision, Y.Q., S.Y., T.W. and Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. Mnist5 and Mnist10 data can be found at https://github.com/zhoujielaoyu/2018-NC-DREC (accessed on 3 November 2023). Sharon, Strain, and Species data can be found at https://github.com/BinPro/CONCOCT/tree/develop (accessed on 3 November 2023). The UCI data can be found at http://archive.ics.uci.edu/ (accessed on 3 November 2023).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Naeem, M.; Jamal, T.; Diaz-Martinez, J.; Butt, S.A.; Montesano, N.; Tariq, M.I.; De-la Hoz-Franco, E.; De-La-Hoz-Valdiris, E. Trends and future perspective challenges in big data. In Advances in Intelligent Data Analysis and Applications; Springer: Berlin/Heidelberg, Germany, 2022; pp. 309–325. [Google Scholar]
- Shi, Y. Advances in Big Data Analytics. Theory, Algorithms and Practices; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Chamikara, M.A.P.; Bertók, P.; Liu, D.; Camtepe, S.; Khalil, I. Efficient privacy preservation of big data for accurate data mining. Inf. Sci. 2020, 527, 420–443. [Google Scholar] [CrossRef]
- Ezhilmaran, D.; Vinoth Indira, D. A survey on clustering techniques in pattern recognition. In Proceedings of the Advances in Applicable Mathematics—ICAAM2020, Coimbatore, India, 21–22 February 2020; AIP Publishing LLC: Melville, NY, USA, 2020; Volume 2261, p. 030093. [Google Scholar]
- Ghosal, A.; Nandy, A.; Das, A.K.; Goswami, S.; Panday, M. A short review on different clustering techniques and their applications. In Emerging Technology in Modelling and Graphics; Springer: Berlin/Heidelberg, Germany, 2020; pp. 69–83. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Ng, A.; Jordan, M.; Weiss, Y. On spectral clustering: Analysis and an algorithm. Adv. Neural Inf. Process. Syst. 2001, 14, 849–856. [Google Scholar]
- Reynolds, D.A. Gaussian mixture models. In Encyclopedia of Biometrics; Springer: Berlin/Heidelberg, Germany, 2009; Volume 741. [Google Scholar]
- Steinbach, M.; Ertöz, L.; Kumar, V. The challenges of clustering high dimensional data. In New Directions in Statistical Physics: Econophysics, Bioinformatics, and Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2004; pp. 273–309. [Google Scholar]
- Zhang, M. Weighted clustering ensemble: A review. Pattern Recognit. 2021, 124, 108428. [Google Scholar] [CrossRef]
- Niu, H.; Khozouie, N.; Parvin, H.; Alinejad-Rokny, H.; Beheshti, A.; Mahmoudi, M.R. An ensemble of locally reliable cluster solutions. Appl. Sci. 2020, 10, 1891. [Google Scholar] [CrossRef]
- Vega-Pons, S.; Ruiz-Shulcloper, J. A survey of clustering ensemble algorithms. Int. J. Pattern Recognit. Artif. Intell. 2011, 25, 337–372. [Google Scholar] [CrossRef]
- Ren, Y.; Pu, J.; Yang, Z.; Xu, J.; Li, G.; Pu, X.; Yu, P.S.; He, L. Deep clustering: A comprehensive survey. arXiv 2022, arXiv:2210.04142. [Google Scholar]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A survey of clustering with deep learning: From the perspective of network architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- Khan, A.; Hao, J.; Dong, Z.; Li, J. Adaptive Deep Clustering Network for Retinal Blood Vessel and Foveal Avascular Zone Segmentation. Appl. Sci. 2023, 13, 11259. [Google Scholar] [CrossRef]
- Ru, T.; Zhu, Z. Deep Clustering Efficient Learning Network for Motion Recognition Based on Self-Attention Mechanism. Appl. Sci. 2023, 13, 2996. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, H.; Zheng, Z.; Chen, J.; Bu, J.; Wu, J.; Wang, X.; Zhu, W.; Ester, M. A comprehensive survey on deep clustering: Taxonomy, challenges, and future directions. arXiv 2022, arXiv:2206.07579. [Google Scholar]
- Li, Z.; Wu, X.M.; Chang, S.F. Segmentation using superpixels: A bipartite graph partitioning approach. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 789–796. [Google Scholar]
- Huang, D.; Lai, J.H.; Wang, C.D. Robust ensemble clustering using probability trajectories. IEEE Trans. Knowl. Data Eng. 2015, 28, 1312–1326. [Google Scholar] [CrossRef]
- Huang, D.; Lai, J.; Wang, C.D. Ensemble clustering using factor graph. Pattern Recognit. 2016, 50, 131–142. [Google Scholar] [CrossRef]
- Yousefnezhad, M.; Zhang, D. Weighted spectral cluster ensemble. In Proceedings of the 2015 IEEE International Conference on Data Mining, Atlantic City, NJ, USA, 14–17 November 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 549–558. [Google Scholar]
- Liu, H.; Wu, J.; Liu, T.; Tao, D.; Fu, Y. Spectral ensemble clustering via weighted k-means: Theoretical and practical evidence. IEEE Trans. Knowl. Data Eng. 2017, 29, 1129–1143. [Google Scholar] [CrossRef]
- Huang, D.; Wang, C.D.; Lai, J.H. Locally weighted ensemble clustering. IEEE Trans. Cybern. 2017, 48, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, H.; Hou, J.; Zhang, Q. Clustering ensemble meets low-rank tensor approximation. AAAI Conf. Artif. Intell. 2021, 35, 7970–7978. [Google Scholar] [CrossRef]
- Jia, Y.; Tao, S.; Wang, R.; Wang, Y. Ensemble Clustering via Co-association Matrix Self-enhancement. arXiv 2022, arXiv:2205.05937. [Google Scholar] [CrossRef]
- Zhou, P.; Sun, B.; Liu, X.; Du, L.; Li, X. Active Clustering Ensemble with Self-Paced Learning. IEEE Trans. Neural Netw. Learn. Syst. 2023, 1–5. [Google Scholar] [CrossRef]
- Xie, J.; Girshick, R.; Farhadi, A. Unsupervised deep embedding for clustering analysis. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 478–487. [Google Scholar]
- Guo, X.; Gao, L.; Liu, X.; Yin, J. Improved deep embedded clustering with local structure preservation. In Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence. Melbourne, Australia 19–25 August 2017; pp. 1753–1759. [Google Scholar]
- Guo, X.; Liu, X.; Zhu, E.; Yin, J. Deep clustering with convolutional autoencoders. In Proceedings of the International Conference on Neural Information Processing, Guangzhou, China, 14–18 November 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 373–382. [Google Scholar]
- Zhang, R.; Li, X.; Zhang, H.; Nie, F. Deep fuzzy k-means with adaptive loss and entropy regularization. IEEE Trans. Fuzzy Syst. 2019, 28, 2814–2824. [Google Scholar] [CrossRef]
- Chen, J.; Han, J.; Meng, X.; Li, Y.; Li, H. Graph convolutional network combined with semantic feature guidance for deep clustering. Tsinghua Sci. Technol. 2022, 27, 855–868. [Google Scholar] [CrossRef]
- Dilokthanakul, N.; Mediano, P.A.; Garnelo, M.; Lee, M.C.; Salimbeni, H.; Arulkumaran, K.; Shanahan, M. Deep unsupervised clustering with gaussian mixture variational autoencoders. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Jiang, Z.; Zheng, Y.; Tan, H.; Tang, B.; Zhou, H. Variational deep embedding: An unsupervised and generative approach to clustering. arXiv 2017, arXiv:1611.05148. [Google Scholar]
- Lim, K.L.; Jiang, X.; Yi, C. Deep clustering with variational autoencoder. IEEE Signal Process. Lett. 2020, 27, 231–235. [Google Scholar] [CrossRef]
- Harchaoui, W.; Mattei, P.A.; Bouveyron, C. Deep adversarial Gaussian mixture auto-encoder for clustering. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Makhzani, A.; Shlens, J.; Jaitly, N.; Goodfellow, I.; Frey, B. Adversarial autoencoders. arXiv 2015, arXiv:1511.05644. [Google Scholar]
- Mukherjee, S.; Asnani, H.; Lin, E.; Kannan, S. Clustergan: Latent space clustering in generative adversarial networks. AAAI Conf. Artif. Intell. 2019, 33, 4610–4617. [Google Scholar] [CrossRef]
- Golalipour, K.; Akbari, E.; Hamidi, S.S.; Lee, M.; Enayatifar, R. From clustering to clustering ensemble selection: A review. Eng. Appl. Artif. Intell. 2021, 104, 104388. [Google Scholar] [CrossRef]
- Hadjitodorov, S.T.; Kuncheva, L.I.; Todorova, L.P. Moderate diversity for better cluster ensembles. Inf. Fusion 2006, 7, 264–275. [Google Scholar] [CrossRef]
- Fern, X.Z.; Lin, W. Cluster ensemble selection. Stat. Anal. Data Mining ASA Data Sci. J. 2008, 1, 128–141. [Google Scholar] [CrossRef]
- Jia, J.; Xiao, X.; Liu, B.; Jiao, L. Bagging-based spectral clustering ensemble selection. Pattern Recognit. Lett. 2011, 32, 1456–1467. [Google Scholar] [CrossRef]
- Iris; University of CaliforniaIrvine: Los Angeles, CA, USA, 30 June 1988. [CrossRef]
- Wine; University of CaliforniaIrvine: Los Angeles, CA, USA, 30 June 1991. [CrossRef]
- Zhou, J.; Zheng, H.; Pan, L. Ensemble clustering based on dense representation. Neurocomputing 2019, 357, 66–76. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; De Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Ezugwu, A.E.; Ikotun, A.M.; Oyelade, O.O.; Abualigah, L.; Agushaka, J.O.; Eke, C.I.; Akinyelu, A.A. A comprehensive survey of clustering algorithms: State-of-the-art machine learning applications, taxonomy, challenges, and future research prospects. Eng. Appl. Artif. Intell. 2022, 110, 104743. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).