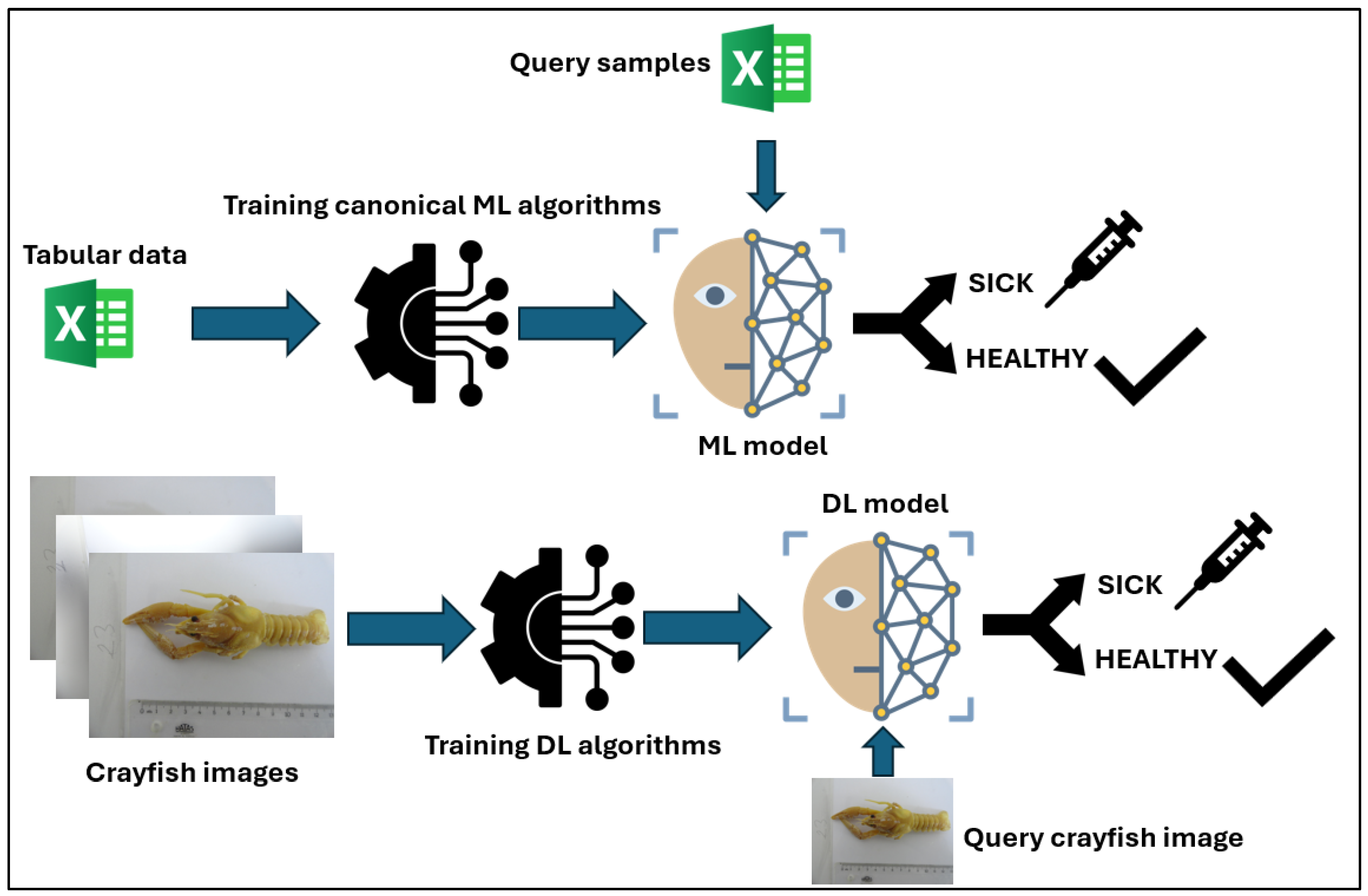
3.1. Evaluation Metrics
In our dataset, the terms true positives (TP) and true negatives (TN) denote the count of cases accurately identified as sick and healthy, respectively. Conversely, false positives (FP) refer to the instances wrongly labeled as sick, while false negatives (FN) pertain to the cases mistakenly classified as healthy.
Accuracy represents the proportion of correct predictions made by a classification model, compared to the overall number of predictions. It is calculated by dividing the count of accurate predictions by the dataset’s total number of predictions. Mathematically, accuracy can be expressed as:
Sensitivity, or recall, measures a classification model’s effectiveness in correctly identifying positive cases. It is calculated as the fraction of true positives out of the total positive instances, which includes both true positives and false negatives. It is particularly important in applications where identifying positive instances is critical, such as in medical diagnosis or fraud detection. Sensitivity can be expressed as:
Specificity assesses a classification model’s accuracy in identifying negative cases. It is determined by the proportion of true negatives relative to the combined count of true negatives and false positives. Mathematically, specificity can be expressed as:
Precision is a metric that gauges the accuracy of a model’s positive predictions in classification tasks. It is expressed as the ratio of true positive predictions to the overall number of positive predictions made:
The F1 score represents a balanced metric that combines precision and recall through their harmonic mean, offering a unified measure that equally weighs both aspects. It can be expressed as:
The Matthews correlation coefficient (MCC) serves as an evaluative metric for the effectiveness of binary classification. It considers every quadrant of the confusion matrix. The MCC is mathematically formulated as follows:
The area under the curve (AUC) is a metric for assessing binary classification models’ performance. It quantifies the area beneath the receiver operating characteristic (ROC) curve—a plot that depicts a binary classifier’s diagnostic capacity as the discrimination threshold changes. The ROC curve plots the true positive rate (TPR) against the false positive rate (FPR) across different thresholds. The AUC reflects the likelihood that the model will assign a higher score to a randomly selected positive instance over a negative one. AUC values range from 0 to 1, where 0.5 signifies no discriminative power—equivalent to random chance—and 1.0 signifies flawless discrimination, with all positive and negative instances correctly identified.
3.3. Experimental Setup
Given the dataset’s modest size and the structured nature of the data, the 10-fold cross-validation technique was employed. This resampling strategy is instrumental in assessing a model’s performance and its ability to generalize. It involves dividing the dataset into 10 equally sized segments, known as folds. The training occurs on 9 folds, while the 10th fold is used for testing. This cycle is conducted 10 times, ensuring each fold is used once as the test set. Such a method is effective in reducing overfitting and yields a more dependable performance metric for the model on new data. Additionally, the ratio of class imbalance was preserved across each fold.
Additionally, min-max normalization was applied to the tabular data to observe whether it would have an effect on the results. Min-max normalization, commonly referred to as feature scaling, is a method that adjusts the values of a feature to fall within a designated range, often between 0 and 1. The transformation is defined as follows:
where
x is an original value, min (
x) is the minimum value of the feature, max (
x) is the maximum value of the feature, and
x′ is the normalized new value. This normalization process ensures that the values of the feature are scaled proportionally within the specified range, preserving the relationships among the original data while enabling more effective comparison and processing by machine learning algorithms.
During the training phase of the deep learning algorithms, both transfer learning and training from scratch were applied. The stopping condition was set to 60 epochs or a training loss of less than 0.01. Stochastic gradient descent (SGD) [
49] was utilized in all deep learning models. ConvNeXt-T was trained with adaptive moment estimation with weight decay regularization (AdamW) [
50] because it is a newer model compared to the others. A batch size of 4 was used in all deep learning architectures.
In the hybrid model, the RF and ResNet50 models, which had the highest accuracy among canonical machine learning and deep learning models, were utilized. When selecting deep learning models, those utilizing SGD for optimization were considered. In the RF-ResNet50 hybrid model, features automatically extracted from crayfish images were utilized to feed the RF algorithm. The features were obtained from the average pooling layer immediately preceding the fully connected layer of the ResNet50 model. By providing these features as input to the RF algorithm, the class of the image was determined. The architecture of the hybrid model is given in
Figure 3.
3.4. Experimental Results
The hyper-parameters for canonical machine learning algorithms without applying min-max normalization are given below. During the fine-tuning process, the accuracy evaluation metric was maximized, and the grid search method was utilized.
For MLP, the number of hidden layers was found as 3. The number of neurons was 50, 10, and 10 for the hidden layers, respectively. Since naive Bayes is a probabilistic classifier, it does not have any hyperparameters. For KNN, the number of the nearest neighbors was found as 17. For SVM, the cost and the gamma parameters were found as 1 × 106 and 1 × 10−8, respectively. For RF, the maximum depth and the number of trees were found as 3 and 100, respectively.
The hyper-parameters for canonical machine learning algorithms with applying min-max normalization are given below. During the fine-tuning process, the accuracy evaluation metric was maximized, and the grid search method was utilized.
For MLP, the number of hidden layers was found as 1. The number of neurons was 100 in the hidden layer. For KNN, the number of the nearest neighbors was found as 25. For SVM, the cost and the gamma parameters were found as 1 × 109 and 0.001, respectively. For RF, the maximum depth and the number of trees were found as 2 and 100, respectively.
The classification results for canonical machine learning algorithms without data normalization and with data normalization are given in
Table 2 and
Table 3, respectively.
According to
Table 2, the best accuracy, sensitivity, precision, F1-score, and MCC performances were obtained with 0.661, 0.404, 0.655, 0.5, and 0.282, respectively, by utilizing the RF model. On the other hand, the best specificity performance was obtained with 0.862 by utilizing the NB model. It can be said that the imbalanced data led to poor performance in detecting sick individuals.
According to
Table 3, the best accuracy, precision, and MCC performances were obtained with 0.643, 0.684, and 0.242, respectively, by utilizing the RF model. The best results for sensitivity and F1-score were obtained with 0.447 and 0.494, respectively, by utilizing the SVM model. Finally, the best specificity performance was obtained with 0.985 by utilizing the NB model. All models, except SVM, experienced a performance loss in accuracy and sensitivity evaluation metrics. Therefore, it can be said that the data normalization had no influence on the performances of the canonical machine learning classifiers for the dataset. On the contrary, the data normalization resulted in a performance increase for detecting sick individuals with SVM, while it caused a performance decrease in identifying healthy individuals.
The classification results for deep learning algorithms and MaxViT vision transformer with TL and FS are given in
Table 4. The results were obtained by utilizing the independent test set.
According to
Table 4, it can be seen that the models with transfer learning outperformed the same models trained from scratch. The best accuracy, precision, F1-score, and MCC performances were achieved with the ResNet50 model. For the specificity evaluation metric, the MaxVit model outperformed the other models. Additionally, the RF-ResNet50 hybrid model obtained the best sensitivity performance with 100 trees having a maximum depth of 3. Among the models evaluated, ResNet50 achieved the minimum number of epochs to complete the training process. The ConvNeXt-T model trained with the AdamW optimizer outperformed the other models in all metrics except for the sensitivity.
AUC scores for deep learning and vision transformer models are given in
Table 5.
According to
Table 5, the highest AUC score of 0.997 was obtained utilizing the ConvNeXt-T AdamW model. With an SGD optimizer, the highest AUC score of 0.987 was achieved by utilizing the AlexNet and ResNet50 models. The RF-ResNet50 hybrid model ranked third in terms of AUC. As can also be seen in the table, the models with transfer learning outperformed the same models trained from scratch.
In the tables presenting the Wilcoxon and McNemar’s test results for statistical significance, bold values indicate a significant difference between the two models at the 5% level. Arrows further indicate which model achieved higher accuracy. If both a left arrow and an upward arrow are present next to a value, it should be understood that the two models have the same accuracy value.
Wilcoxon test results for canonical machine learning algorithms without the data normalization and with the data normalization are given in
Table 6 and
Table 7, respectively.
According to
Table 6, there is a statistical significance between the results of the kNN and SVM, MLP and SVM, and SVM and NB models. The results show that the SVM model achieves statistically significant superiority in classification performance.
According to
Table 7, there is a statistical significance between the results of the kNN and SVM, kNN and NB, MLP and SVM, MLP and NB, SVM and RF, SVM and NB, and RF and NB models. The results show that the SVM model achieves statistically significant superiority in classification performance.
McNemar’s test results for canonical machine learning algorithms without the data normalization and with the data normalization are given in
Table 8 and
Table 9, respectively.
According to
Table 8 and
Table 9, there is no statistical significance among the canonical machine learning models. Data normalization also eliminated the statistically significant differences observed between the models in their unnormalized state.
Wilcoxon test results for deep learning algorithms are given in
Table 10.
According to
Table 10, Resnet50 and ConvNext-T with AdamW models have statistically significant superiority in classification performance. There is a statistical significance between the results of the AlexNet and ResNet50, VGG, EffNetv2, ConvNeXt-T with AdamW; ResNet50 and VGG, ConvNeXt-T; VGG and EffNetv2, ConvNeXt-T with AdamW; EffNetv2 and MaxViT, ConvNeXt-T; ConvNeXt-T and ConvNeXt-T with AdamW models.
McNemar’s test results for deep learning algorithms are given in
Table 11.
According to
Table 11, ConvNext-T with AdamW and the RF-ResNet50 hybrid models have statistically significant superiority in classification performance. There is a statistical significance between the results of the AlexNet and VGG, EffNetv2, ConvNeXt-T, ConvNeXt-T with AdamW, RF-ResNet50 hybrid; ResNet50 and VGG, EffNetv2, ConvNeXt-T, ConvNeXt-T with AdamW, RF-ResNet50 hybrid; VGG and ConvNeXt-T with AdamW, RF-ResNet50 hybrid; EffNetv2 and ConvNeXt-T with AdamW, RF-ResNet50 hybrid; MaxViT and ConvNeXt-T, ConvNeXt-T with AdamW, RF-ResNet50 hybrid; ConvNeXt-T and ConvNeXt-T with AdamW, RF-ResNet50 hybrid; ConvNeXt-T with AdamW and RF-ResNet50 hybrid models.