Abstract
Lung nodule classification is crucial for the diagnosis and treatment of lung diseases. However, selecting appropriate metrics to evaluate classifier performance is challenging, due to the prevalence of negative samples over positive ones, resulting in imbalanced datasets. This imbalance often necessitates the augmentation of positive samples to train powerful models effectively. Furthermore, specific medical tasks require tailored augmentation methods, the effectiveness of which merits further exploration based on task objectives. This study conducted a detailed analysis of commonly used metrics in lung nodule detection, examining their characteristics and selecting suitable metrics based on this analysis and our experimental findings. The selected metrics were then applied to assessing different combinations of image augmentation techniques for nodule classification. Ultimately, the most effective metric was identified, leading to the determination of the most advantageous augmentation method combinations.
1. Introduction
Lung cancer remains the leading cause of cancer-related mortality globally, with estimates suggesting over 2.2 million new cases and 1.8 million deaths annually [1]. Patients with non-small cell lung cancer at an operable stage have higher survival rates than those presenting with metastatic disease, with five-year survival of 71–77% for stage IA and 58% for stage IB, a statistic reflecting its significant impact on public health [2]. Despite advancements in imaging and diagnostic technologies, the accuracy of lung cancer diagnosis can be compromised by several factors, leading to diagnostic errors. Studies have shown that misdiagnosis rates can be influenced by the quality of imaging technology, the interpretative skills of radiologists, and the inherent difficulties in distinguishing lung cancer from other respiratory conditions in early stages. Enhancing diagnostic accuracy through improved imaging techniques and AI-supported diagnostic tools presents a promising avenue to mitigate these issues, aiming to increase early detection rates and reduce misdiagnoses, thereby improving overall survival rates and reducing the burden of lung cancer. Early detection of lung nodules is crucial for early diagnosis of various diseases; however, the extensive requirements for manual examination by highly skilled specialists pose significant challenges, particularly given the prevalence of negative instances [3]. A single Computed Tomography (CT) scan can encompass over 100 slices, underscoring the labor-intensive and time-consuming nature of manual review processes. This complexity highlights the pressing need for more efficient and precise diagnostic methods in pulmonary oncology. To overcome the obstacles associated with medical image segmentation and classification, a variety of advanced applications have been developed to assist physicians in the diagnostic process. Unlike traditional techniques that require manual feature design, deep learning models excel in autonomously extracting features through training. Key methodologies in deep learning for feature extraction include Autoencoders, Deep Belief Networks, Deep Boltzmann Machines, and CNN. Computer-Aided Diagnosis (CAD) systems, particularly those targeting the detection of pulmonary nodules, have become increasingly significant, due to their role in the early detection of lung cancer—a leading cause of cancer-related deaths. Effective early detection through CAD models is pivotal, as it considerably improves patient prognosis by identifying potentially treatable lung cancer at an early stage. However, the challenges of analyzing lung nodules and their surrounding tissues are amplified by the limited size and variability of the existing datasets, which often compromise the generalization capabilities of deep learning models, leading to unreliable predictions and limiting practical application. The complexity is increased by CT images being 3D grayscale, where lesions typically exhibit low contrast against normal tissues, making diverse feature extraction particularly challenging. Additionally, the small sizes of lesion areas relative to normal tissues often results in the neglect of high-level feature maps by deep learning models. The prevalence of false nodules in datasets further exacerbates the imbalance, potentially causing models to overfit specific samples rather than generalize effectively. Extensive research has focused on augmentation techniques to address overfitting in CAD applications, enhancing classifiers’ generalization and robustness by expanding training datasets. Yet, no single solution fits all; specific augmentation methods need to be tailored to the unique requirements of different models and datasets.
In this study, a 3D Convolutional Neural Network (CNN) was employed to classify pulmonary nodules, assessing candidates for indications of disease. Quality metrics are essential for evaluating classifier performance, providing a means to measure and enhance the effectiveness of classification models. While numerous metrics are available for model assessment, the challenge lies in selecting those that align best with the distinct characteristics of various models and datasets. Such careful selection ensures that performance evaluations are accurate and contextually relevant, which is crucial for advancing medical imaging and improving diagnostic tools.
Our research included a rigorous comparison of prevalent metrics for lung nodule classification, supplemented by a comprehensive investigation of augmentation techniques, categorized into geometric and photometric methods, applied during the training of classification models. Our findings highlight geometric augmentation methods as particularly beneficial in enhancing model performance. Among various metrics, the -score was identified as most suitable, due to its responsiveness to the specific challenges encountered in lung nodule detection. This paper makes two key contributions: First, this study selected the most suitable metrics for nodule classification in medical screening tasks through theoretical analysis and experimentation. Second, the selected metrics guided us in determining the best combinations of augmentation techniques for this classification task. The remainder of the paper is organized as follows: Section 2 discusses related work. Section 3 describes the methodology, outlining the problem construction, comparing metrics, and introducing the classifier architecture. Section 4 details the experiments conducted with refined design, compares the results of several metrics, selects the best metrics, and identifies the top five combinations of augmentation techniques. Section 5 concludes the paper.
2. Related Work
2.1. Medical Image Processing
Medical image processing plays a crucial role in various biomedical applications, including cancer diagnosis and histopathological image segmentation. With the advancement of deep learning techniques, supervised and unsupervised learning methods have been widely explored in the field of medical image processing. Chen et al. [4] reviewed the recent advances and clinical applications of deep learning in medical image analysis, highlighting the success of deep learning models in disease detection and diagnosis. Zhao et al. [5] proposed a deeply supervised active learning framework for biomedical image segmentation, combining active learning and semi-supervised learning strategies to enhance the segmentation performance. Kim et al. [6] introduced an unsupervised deformable image registration method using a cycle-consistent CNN for 3D volume registration, highlighting the importance of deep neural networks in medical image analysis. Tseng et al. [7] proposed a semi-supervised CNN architecture, DNetUnet, for medical image segmentation, demonstrating superior performance compared to existing models. In the realm of medical image segmentation, Lerousseau et al. [8] presented a weakly supervised framework for histopathological tumor segmentation, leveraging standard clinical annotations for whole slide imaging segmentation. Li et al. [9] introduced a semi-supervised approach for medical image segmentation, emphasizing the utilization of limited labeled data and a large quantity of unlabeled images. These studies showcase the growing interest in developing deep learning-based frameworks for medical image segmentation with limited supervision. Overall, the literature review indicates a shift towards leveraging supervised and unsupervised learning techniques, such as CNNs and active learning, in medical image processing for tasks like image registration, segmentation, and disease classification. These advancements in deep learning methods have shown promising results in improving the accuracy and efficiency of medical image analysis, paving the way for enhanced healthcare applications in the future.
2.2. Classification in Medical Image Processing
Deep learning techniques have been widely utilized in medical image classification in the context of pulmonary medical images [10]. The application of deep learning in medical image processing has been particularly significant during the COVID-19 pandemic, with research focusing on utilizing deep learning for processing medical images related to the coronavirus outbreak [11]. Deep learning models have shown promise in medical science, particularly in the field of medical image processing for the diagnosis and prognosis of life-threatening ailments such as lung cancer [12]. Furthermore, the integration of deep learning tools with Internet of Health Things (IoHT) devices has been proposed, to develop a computational intelligence framework for online medical image recognition, including lung nodule images for malignant classification [13]. Deep learning approaches, specifically convolutional neural networks, have been compared for the classification of lung biopsy images, highlighting the automation of biopsy image analysis for cancer diagnosis [14]. A machine learning approach has been employed to diagnose lung and colon cancer, using a deep learning-based classification framework, achieving high accuracy in identifying cancer tissues [15]. Radiomics, which involves extracting features from medical images for automated classification, has been explored in the context of lung cancer screening, emphasizing the challenge of building a cross-validated model based on radiomic signatures [16]. Distant domain transfer learning has been proposed as a method to effectively handle distribution shifts in medical imaging tasks, achieving high classification accuracy compared to existing algorithms [17]. Machine learning and image processing techniques have been utilized for accurate lung cancer classification and prediction, incorporating noise reduction, feature extraction, and analysis of damaged regions in lung images. Moreover, a novel self-supervised pre-training pipeline, Multi-task Self-supervised Continual Learning (MUSCLE), has been developed for multiple medical imaging tasks using X-ray images from various body parts, including lungs [18]. This approach addresses challenges such as data heterogeneity, overfitting, and catastrophic forgetting in multi-task learning scenarios. Overall, the literature highlights the significant role of deep learning and machine learning techniques in medical lung image processing for classification, diagnosis, and prognosis of lung-related ailments.
2.3. Metrics for the Assessment of Models’ Performance
In classification tasks, evaluating performance and interpreting confusion matrices correctly is critical, yet there remains no universal agreement on an optimal metric to be universally adopted in machine learning. Traditionally, accuracy and scores derived from confusion matrices have been popular choices for evaluating binary classifications. However, these metrics often present over-optimistically inflated results, particularly in scenarios involving imbalanced datasets. Zachary C. et al. [19] explored the dynamics between the optimal score and the decision-making threshold that achieves it, revealing insightful correlations that aid in refining performance measurements. Concurrently, Davide et al. [20] conducted comparative analyses between the MCC, score, and accuracy. Their findings suggested that the MCC provides a more reliable assessment of model performance, offering distinct advantages over the score and accuracy, especially in binary classification evaluations. Furthermore, Amalia et al. [21] undertook a systematic review of how class imbalance affects various classification performance metrics. They concluded that while Geometric Mean or Bookmaker Informedness serve well under conditions where classification successes are the primary concern, the MCC emerges as the superior metric when both classification successes and errors need equal consideration. These studies collectively highlight the need for a more discerning selection of performance metrics in binary classification tasks, particularly in light of the challenges posed by dataset imbalances. Such insights are invaluable for developing more accurate and equitable machine learning models.
2.4. Augmentation Methods to Reduce Overfitting
The efficacy of deep learning models is heavily reliant on extensive datasets for training. However, in medical image processing, annotating large datasets is particularly costly and time-consuming, often resulting in a limited number of available samples for training. To address this challenge, data augmentation has become a pivotal strategy in training deep learning models to ensure they generalize effectively to new, unseen data. Phillip et al. [22] have categorized data augmentation techniques for deep learning into three primary groups: basic, deformable, and advanced deep learning augmentation techniques. Fang et al. [23] implemented various augmentation strategies, such as the addition of salt-pepper noise, Gaussian noise, random rotation, and the use of a median filter. These techniques were applied to enhance lung CT scans, enlarging the training dataset by a factor of one hundred through median intensity projections. Similarly, Christina et al. [24] utilized automated low-level segmented PET data to train sophisticated deep learning algorithms for analyzing corresponding CT data. To expand their dataset, they applied rotation and scaling to both CT images and the masks derived from the PET scans, and they also introduced zero-mean Gaussian noise to the CT images. These examples underscore the crucial role of data augmentation in overcoming the inherent limitations posed by small datasets in medical imaging, thus enhancing the performance and applicability of deep learning models in clinical settings.
3. Methodology
3.1. Preliminaries
In this study, a classifier was provided with a list of suspected nodules and was tasked with determining whether each suspect was a nodule or a non-nodule. For this purpose, the model was trained using a supervised method, where it received a large number of samples that guided it on making the final decision. The primary objective of this work was to train the model to become an efficient and accurate classifier, capable of correctly categorizing each input. The suspects were represented by 3D arrays X, and the classifier’s job was to transform these arrays into logits z through a function f, where , and j was the number of classes. These logits were then converted into probabilities for each class through a softmax operation defined as
for each class k.
To enhance the model’s ability to generalize to new, unseen data, this work implemented a series of augmentation techniques, both individually and in combination. These techniques were categorized into geometric and photometric augmentations. Geometric augmentations included rotations, translations, scaling, and flipping of images. These manipulations simulated the variations that might occur due to differences in patient positioning and the orientation of organs in diverse clinical settings, thus preparing the model to handle such discrepancies effectively. On the other hand, photometric augmentations altered the visual attributes of the images, such as adjusting brightness, contrast, and adding noise. These adjustments were crucial for training the model to process images that might vary in visual quality due to differences in imaging equipment or settings across different healthcare facilities. The effectiveness of the augmentation techniques and the overall model performance were quantitatively assessed, using a suite of evaluation metrics. These included Accuracy, Score, Score, Score, and the MCC.
3.2. Convolutional Neural Network for Classification
CNNs are pivotal in image classification systems, particularly following medical image segmentation tasks like those performed by U-Net on CT scans. As illustrated in Figure 1 [25], CNNs are structured with an array of layers including input, multiple convolutional layers, pooling layers, fully connected layers, and an output layer. In medical imaging, such as lung nodule detection, convolutional layers apply filters to detect relevant features like edges and textures, while pooling layers reduce the dimensionality of feature maps, decreasing computational load and preventing overfitting. The fully connected layers integrate these features into high-level representations crucial for classification tasks, such as distinguishing between benign and malignant nodules.
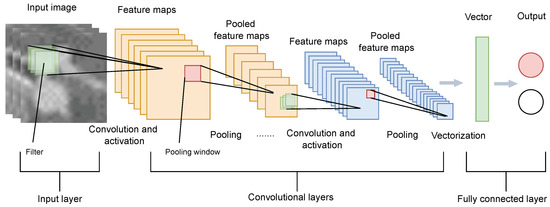
Figure 1.
Network for classification.
The training of CNNs necessitates large volumes of labeled data, a significant challenge in medical imaging due to the scarcity and expense of obtaining precise annotations. Data augmentation methods are often utilized to expand training datasets and boost the model’s generalization capabilities. Extensive validation and testing ensure the CNN’s robust performance across varied clinical settings, enhancing diagnostic accuracy and patient outcomes through more precise disease detection. In this work, augmentation methods were leveraged to improve the generalization capability and balance the positive and negative samples during training.
Cross-entropy loss is employed in training to force the model to learn correct prediction of nodule class. The loss is defined by
In this study, a categorical classification approach was used, where C represented the number of classes, and N was the batch size, with denoting the logit corresponding to class c for each input in the batch. This method is particularly apt for training models in problems involving C distinct classes. Unlike the mean square error loss (MSE), which emphasizes minimizing the numerical differences between the output logits and specific target values, our focus shifts towards ensuring that the logit for the correct class is the highest among all classes. This approach aligns more closely with the objectives of classification tasks where the accuracy of class prediction is more critical than the exact numerical output of logits. However, we acknowledge the potential benefits of exploring alternative loss functions, such as weighted cross-entropy loss. This variant could indeed provide a more nuanced approach by assigning different weights to the classes, thereby addressing the imbalance between positive and negative samples more directly. Regarding regularization techniques, L1 and L2 regularization can be leveraged to mitigate the risk of overfitting. L1 regularization can promote model sparsity and feature selection, and L2 can improve the generalization of the model on unseen data. While these advanced techniques were not implemented in the current study, they represent valuable areas for future research that could enhance the model’s sensitivity to under-represented classes. This work employs Rectified Linear Unit (ReLU) as the activation function, which is defined as
It is characterized by its ability to introduce nonlinearity while maintaining computational efficiency, as it simply thresholds negative values to zero. This attribute not only speeds up computation but also facilitates sparsity within network activations, with only a subset of neurons activating at any given time. ReLU is particularly valuable for its role in mitigating the vanishing gradient problem commonly seen with saturating activation functions like sigmoid or tanh, as its gradient remains constant. Comparatively, when ReLU is used in models for lung nodule detection, it often outperforms other traditional activation functions like sigmoid or tanh, which can suffer from rapid saturation and severe vanishing gradients in deep networks.
3.3. Performance Metrics for Classification
Four metrics were leveraged and compared in this work.
3.3.1. Accuracy
In medical image classification, particularly when identifying lung nodules indicative of potential malignancies, accurate detection is paramount. True Positives (TPs) represent correctly identified nodules, reflecting the ultimate goal of such diagnostics—to ensure no potential threat goes unnoticed. Conversely, False Positives (FPs) occur when non-nodules are mistakenly classified as nodules, and False Negatives (FNs) are actual nodules that are overlooked, misclassified as benign. True Negatives (TNs) correctly identify non-nodules. The importance of distinguishing between these categories is underscored by the imperative to detect real nodules accurately, which can suggest the presence of cancer:
While accuracy calculates the proportion of true results (both TPs and TNs) among the total number of cases examined, it may not always be the best metric, especially in scenarios where the dataset is imbalanced. This imbalance is typical in medical imaging, where positive cases (actual nodules) are significantly outnumbered by negatives. Relying solely on accuracy can lead to misleadingly high-performance metrics, suggesting effective model performance when, in reality, the model may be inadequately identifying critical positive cases. Therefore, in medical diagnostics, where the cost of missing a True Positive is very high, other metrics such as sensitivity (recall) or the F-score, which balance precision and recall, are often more informative and indicative of a model’s practical utility in clinical settings.
3.3.2. Recall and Precision
Recall, or sensitivity, is a critical metric in medical diagnostics, quantifying the proportion of actual positives correctly identified by a classification model. This metric, also known as the True Positive Rate, is essential where the cost of missing a positive diagnosis, such as a disease, could be grave. A model with high recall ensures comprehensive identification of positive cases, which is vital in medical scenarios where overlooking a condition can result in severe health consequences or even fatality. High recall demonstrates that a model is adept at detecting positive cases, a crucial capability in medical testing environments where failing to diagnose a condition timely can be detrimental.
Precision, on the other hand, measures the accuracy of positive predictions made by a model, calculated as the ratio of TPs to the total predicted positives (sum of True and FPs). High precision indicates a high probability that a positive classification by the model is correct, which is especially important in medical contexts where FPs can lead to undue stress, unnecessary invasive procedures, or expensive treatments. The challenge often lies in balancing precision with recall, a dynamic known as the precision–recall trade-off. Enhancing recall may increase FPs, reducing precision, while optimizing precision could cause the model to miss some TPs, thus lowering recall. This balance is particularly critical in healthcare, where the consequences of FNs (missed diagnoses) and FPs (unnecessary treatments) must be carefully weighed. Decision makers must therefore judiciously adjust the model to ensure an optimal balance, reflecting the specific demands and potential repercussions inherent in their diagnostic applications.
3.3.3. F-Score
To evaluate a model’s performance, it needs a metric that combines precision and recall. F-score is a generally accepted way, though it is far away from perfect. It is calculated from the precision and recall of the test, where the precision is the number of True Positive results divided by the number of all positive results, including those not identified correctly, and the recall is the number of True Positive results divided by the number of all samples that should have been identified as positive. Precision is also known as a positive predictive value, and recall is also known as sensitivity in diagnostic binary classification. Among F-score methods, the most popular one is the score, which is the harmonic mean of precision and recall:
Expanding upon the basic score, the allows for varying the emphasis on precision versus recall, accommodating the specific needs of different testing environments. Here, is a parameter that sets the weight of recall relative to precision. A value greater than 1 gives more weight to recall, making it more crucial in scenarios where missing a positive instance carries greater consequences than FPs. Conversely, a less than 1 emphasizes precision, suitable for cases where FPs are more costly or detrimental than FNs. This flexibility makes the score adaptable to diverse applications, providing a tailored metric that can reflect the unique cost dynamics associated with particular diagnostic tests or other classification tasks. The highest value of an score is 1.0, indicating perfect precision and recall, while the lowest is 0, occurring when either the precision or the recall is zero, signifying a complete failure in either identifying TPs or excluding FPs effectively [26]:
3.3.4. Matthews Correlation Coefficient (MCC)
The MCC, named after biochemist Brian W. Matthews, is a robust statistical rate used to evaluate the quality of binary classifications in machine learning. This coefficient effectively incorporates TPs, TNs, FPs, and FNs into its calculation, making it a balanced metric suitable for datasets with imbalanced classes. The MCC is essentially a correlation coefficient between the observed and predicted binary classifications, yielding a value between −1 and +1. A coefficient of +1 indicates perfect prediction accuracy, 0 suggests a performance equivalent to random guessing, and −1 denotes complete disagreement between prediction and observation. It is important to note that values of the MCC that do not reach the extremes of its range (neither −1, 0, nor +1) still provide useful insights into the predictive accuracy and are not indicative of mere random guessing, contrary to some interpretations; rather, they reflect the degree of correlation based on the dataset’s specific characteristics [27]. While there is no perfect way of describing the confusion matrix of True and FPs and Negatives by a single number, the MCC is generally regarded as being one of the best such measures [28]. Other measures, such as the proportion of correct predictions (also termed accuracy), are not useful when the two classes are of very different sizes. For example, assigning every object to the larger set achieves a high proportion of correct predictions, but is not generally a useful classification. Some scientists claim the MCC to be the most informative single score to establish the quality of a binary classifier prediction in a confusion matrix context [29]. The MCC can be calculated directly from the confusion matrix using the following formula:
Selecting the most appropriate metrics for our lung nodule classification project presented significant challenges, due to the inherent advantages and disadvantages of each metric. Extensive research was conducted, including experiments with various augmentation techniques, to determine which metrics were most effective for this specific application.
3.4. Augmentation Methods in Lung Nodule Detection
Insufficient training samples can lead to premature overfitting in models, where they merely memorize labels rather than achieving true generalization. To enhance generalization capabilities, it is crucial to challenge the models to learn general features rather than allowing easy memorization. Increasing the number of training samples for medical image classification, however, is often prohibitively expensive and time-consuming. As a result, augmentation techniques have become a vital strategy to expand training datasets by applying synthetic modifications to individual samples. When implemented effectively, augmentation can significantly enlarge the dataset size beyond the model’s capacity to memorize, compelling it to rely more on generalization. Nevertheless, not all augmentation techniques are equally effective, making their research a focal point in medical image classification, especially with the rise of deep neural networks. Two primary categories of augmentation techniques are geometric and photometric. Geometric augmentation manipulates the spatial attributes of images, such as rotating, flipping, or scaling, to mimic the variability in patient positioning and organ orientation that occurs naturally. On the other hand, photometric augmentation alters image appearance attributes, like brightness, contrast, and noise levels, addressing variations due to different imaging equipment or settings. The efficacy of these techniques in preventing overfitting and promoting robustness makes them indispensable in training deep learning models for medical imaging tasks. Their ongoing evaluation and optimization continue to be critical for advancing the field and improving diagnostic accuracy in clinical settings.
3.4.1. Flipping the Images
Flipping images is a straightforward yet effective geometric augmentation technique widely used in medical image processing. In medical imaging, especially in the context of nodules, the anatomical structure does not inherently possess a ‘correct’ orientation, allowing flipped images to remain clinically valid and representative of potential real-world variations a model might encounter. For instance, flipping a lung CT scan horizontally does not alter the significance of any nodules present, as these biological features do not exhibit directionality bias that affects diagnosis. This method enhances the model’s ability to generalize from the training data by effectively increasing the dataset size with minimal computational overhead. It prepares the model for diverse orientations of input data, thereby reducing the model’s sensitivity to image orientation. This is particularly crucial in scenarios where images may be captured or processed in varying alignments, due to different scanning protocols or operator preferences.
3.4.2. Shifting the Image Around
Image shifting involves moving the image a certain number of pixels in one or more directions. This technique is beneficial because medical images, such as CT scans or MRI, often feature a region of interest, like a tumor or lesion, that may not be perfectly centered. By training the model on images where the nodules have been shifted slightly, it becomes robust to variations in nodule placement, which commonly occur due to patient movement or differences in scanning procedures. Shifting can be implemented in small increments to ensure that the integrity of the nodule’s context within the surrounding anatomical structure is maintained. The model trained with shifted images can better handle cases where nodules appear near the edges of the image, which are typical scenarios in clinical settings, thereby enhancing the model’s diagnostic accuracy.
3.4.3. Scaling the Images
Scaling alters the size of the image or its elements, either enlarging or reducing them while maintaining the same aspect ratio. In medical imaging, scaling is critical as it compensates for the variation in the physical size of anatomical structures among different patients. For example, a nodule in a CT scan might appear larger in a smaller patient compared to a larger patient. By incorporating images scaled at different ratios, the model learns to identify structures of interest across a range of sizes, which is essential for robust diagnostic applications. This technique not only broadens the diversity of the dataset but also mimics potential real-world scenarios where equipment settings or patient physiology result in size variations of anatomical features. Scaling ensures that the model’s performance is consistent regardless of such differences.
3.4.4. Rotating the Images
Rotation, particularly around the head–foot axis in medical scans, is relevant, due to the asymmetrical nature of human anatomy along this axis. This rotation reflects plausible variations during patient positioning in the scanner. Unlike rotations in other planes, which might distort the image due to the anisotropic (non-cubic) nature of voxels in medical imaging, rotating around the head–foot axis preserves the clinical features and their spatial relationships in the scan. This type of rotation helps the model to be invariant to minor misalignment or rotations in the scanned images, accommodating common discrepancies that arise from different scanning protocols. Training with images rotated around this axis enhances the model’s ability to correctly interpret images regardless of slight orientation differences, thereby improving diagnostic accuracy and reliability.
3.4.5. Injecting Noise into the Images
Introducing noise to medical images is a double-edged sword; it simulates realistic scenarios where scans might be affected by various factors like sensor quality or environmental interference, potentially lowering image quality. Noise injection, such as adding Gaussian or salt-and-pepper noise, tests the model’s resilience against real-world, imperfect data conditions. However, excessive noise can obscure critical diagnostic features, complicating the task of the model in identifying subtle cues crucial for accurate diagnoses. This method must be carefully calibrated, to ensure that it does not overwhelm the meaningful content of the images. Despite these risks, noise injection remains a valuable technique for enhancing the robustness of medical imaging models, preparing them for diverse and less-than-ideal conditions they might encounter in clinical use.
3.4.6. Modifying the Intensity of Pixels or Voxels
Adjusting the intensity values of pixels or voxels affects the image’s overall appearance, in terms of brightness and contrast. Techniques such as gamma correction adjust the luminance, potentially highlighting hidden details in lighter or darker regions that were not initially evident. Linear contrast adjustments and histogram equalization alter the dynamic range and distribution of intensities, which can help in better differentiating between regions of interest and the background or between different tissues. These adjustments mimic the variations in image quality that might result from different imaging hardware or settings, ensuring the model is adaptable and performs well across the range of equipment found in various medical facilities. Tailoring pixel intensity also aids in training models to focus on relevant features rather than being misled by artifacts or anomalies in brightness or contrast that are not clinically relevant.
3.5. Blurring the Images
Blurring the images is another effective method of image augmentation, particularly useful in medical imaging where fine details can vary significantly, due to differences in scan quality or machine calibration. By applying a blur filter, such as a Gaussian blur, the images simulate the effects of lower resolution or focus that might occur in real-world clinical environments. This type of augmentation is crucial because it trains the model to perform well even when images are not perfectly sharp, mirroring conditions that might be encountered in clinical settings with varying equipment qualities. Moreover, blurring helps to reduce model overfitting to high-frequency noise that is not clinically relevant, thus promoting a focus on more significant, larger patterns within the data. This generalization ensures that the model remains robust and consistent in its diagnostic capabilities, even when presented with images that are less than ideal in terms of clarity and detail resolution.
Geometric techniques are indispensable when images from diverse patient populations are used, where variations in anatomy and scan orientations are common. Photometric techniques are vital when deploying models across various hospitals or clinics with different imaging equipment and settings, ensuring consistent performance regardless of technological disparities. Both augmentation types are often used in tandem, to prepare robust models capable of handling the range of variations seen in clinical practice. Their combined use ensures comprehensive training, covering both anatomical variations and imaging conditions, which is essential for deploying AI in diverse medical environments. In conclusion, both geometric and photometric augmentation techniques offer distinct and complementary benefits. Their judicious application can significantly enhance the effectiveness and generalizability of deep learning models in medical image processing, ultimately leading to better diagnostic tools and improved patient outcomes.
4. Experiments
4.1. Dataset
The LUNA16 dataset [30], which served as the foundation for training our classification model, consists of 888 computed tomography (CT) scans that have been meticulously annotated by four experienced radiologists via a detailed two-phase process, in which the radiologists evaluated and annotated CT scans to identify and confirm the presence of lung nodules. In the first phase, each of the CT scans in the dataset was independently reviewed by the radiologists, to detect potential lung nodules. The second phase involved a review process where the findings from the initial phase were verified. This dataset is primarily designed for studies focusing on the reduction of FPs in lung nodule detection. It includes a comprehensive collection of 495,958 nodule candidates, generated through the application of three sophisticated detection algorithms, out of which 1215 have been confirmed as positive samples. This equates to a positive-to-negative ratio of approximately 0.0025, indicating a pronounced imbalance that presents substantial challenges for effective model training. To address the reviewer’s query, each nodule candidate within the dataset is annotated with additional attributes that provide deeper insights into their clinical characteristics. The nodules vary in size, typically ranging from 3 mm to 30 mm in diameter, and they are described with shapes that may be spherical, lobulated, spiculated, or irregular, reflecting their diverse physical manifestations in CT scans. Furthermore, the dataset categorizes these nodules into benign or malignant, based on their histopathological evaluation, where available from follow-up procedures or prior diagnostic data. However, it is important to note that not all nodules in the dataset have confirmed malignancy status, due to the limitations in available clinical follow-up. Moreover, the LUNA16 dataset includes information regarding the nature of the nodules, distinguishing between primary lung cancers and metastatic lesions. This distinction is crucial for the model, as it aids in understanding the different patterns associated with primary and secondary lung cancer manifestations in imaging studies. By integrating these detailed attributes into our analysis, the dataset not only facilitated the training of more accurate models for nodule detection but also enhanced the models’ ability to generalize across a more diverse set of pathological features, thereby improving diagnostic precision in clinical settings.
The LUNA16 dataset, which forms the basis of our experimental analysis, primarily includes nodules that are both benign and malignant in nature. However, it is important to clarify that while the dataset provides a robust platform for detecting and classifying lung nodules, it does not specifically differentiate all nodule types by the exact disease or condition they represent.
4.2. Model Architecture
This study adopted the neural network architecture outlined in [25], characterized by a simple design aimed at optimizing performance. The architecture is segmented into three primary sections: a batch normalization tail, a robust four-block backbone, and a decision-making head consisting of a linear layer topped with a softmax function for classification.
Each block within the backbone is meticulously structured to enhance feature extraction and includes two layers of 3D convolution, each followed by ReLU activation to introduce non-linearity and enhance model learning capabilities. Following the convolutions, a 3D MaxPooling layer is applied, serving to progressively reduce the dimensionality of the input arrays while simultaneously increasing the number of feature channels. This systematic reduction and feature enhancement after each block are crucial for efficiently processing the volumetric data typical in medical imaging applications.
4.3. Experiment Design
The training regimen for the model was meticulously crafted, involving numerous epochs at a carefully determined learning rate and batch size, selected to optimize the learning curve and prevent overfitting. The specific hyperparameters included:
- Epochs: 20, to ensure adequate exposure to the training data without overfitting.
- Batch Size: 16, balancing the computational load and the granularity of the gradient updates.
- GPU: RTX 3060
- Learning Rate: 0.001, starting conservatively, to ensure comprehensive convergence over training epochs.
- Optimizer: SGD, chosen for its adaptive learning rate capabilities, helping to fine-tune the model’s weights effectively.
- Loss: Cross-entropy loss.
- Activation function: ReLU.
- Convolution kernal: .
- Pooling layer: Max pooling .
- Loss: Cross-entropy.
- Ratio of training/test: 0.8/0.2.
This study systematically explored the impact of various augmentation strategies on the performance of medical image classification. Initially, we trained the model using the original dataset without any preprocessing. The results were initially promising, as after 20 epochs the model achieved an accuracy rate exceeding 99.8%. However, a deeper analysis revealed a significant issue: while the negative samples were classified with 100% accuracy, the classification of positive samples was entirely incorrect. In other words, the trained model defaulted to classifying all samples as negative, to minimize training loss. This behavior made sense from the model’s perspective, due to the overwhelming imbalance in the dataset, where there was approximately one positive sample for every 400 samples, leading to inevitable overfitting. Augmenting the data could eliminate the imbalance in the dataset, compelling the model to learn the underlying patterns rather than merely memorizing the results. This approach sought to address the skewed distribution of classes and encourage a more robust learning process.
Both geometric and photometric augmentation techniques were implemented, separately combining them to assess their respective contributions to enhancing classification accuracy. The specifics of these combinations are detailed in Table 1 and Table 2. Figure 2 and Figure 3 illustrate a positive instance and the augmented versions with different combinations of augmentation techniques. Additionally, the establishment of a baseline by training the model without any augmentation methods allowed us to measure the incremental benefit provided by each augmentation approach. This comparative analysis helped us to not only gauge the effectiveness of individual methods but also to evaluate their synergistic effects when combined. To rigorously assess the performance of each augmentation method, the calculations of various metrics were carried out. These metrics served as critical indicators of classification success, enabling us to identify the most effective techniques for improving model accuracy in medical image processing. By comparing these metrics across different augmentation scenarios, this work aimed to determine the most suitable strategies for significantly enhancing model performance. This approach ensures that one not only adopts the most effective augmentation methods but also that one tailors metric selection to best reflect the nuanced improvements these methods bring to complex medical imaging tasks.

Table 1.
Combinations of geometric augmentations.

Table 2.
Combinations of photometric augmentations.
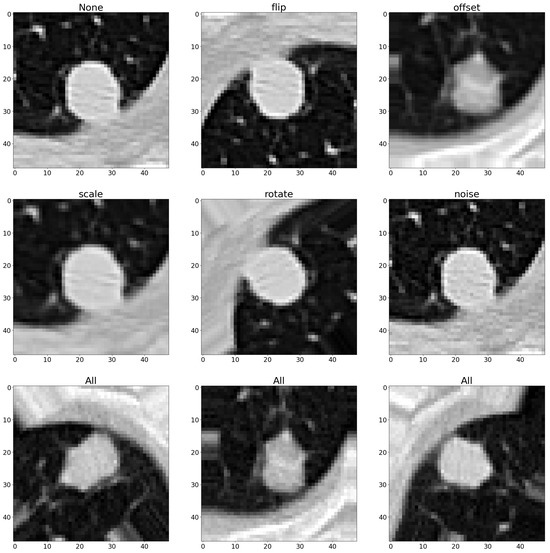
Figure 2.
Augmented samples with geometric techniques.
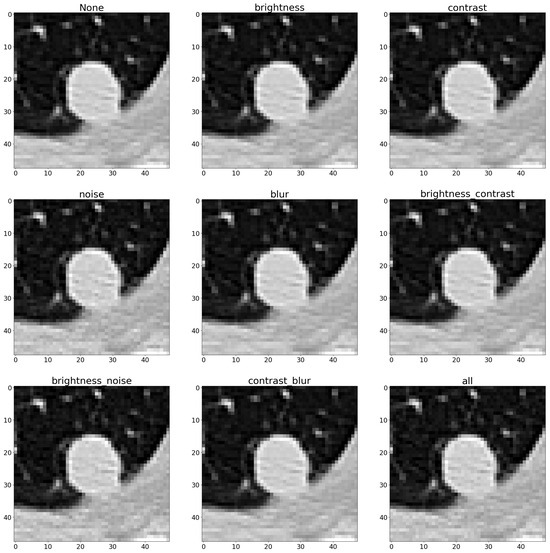
Figure 3.
Augmented samples with photometric techniques.
4.4. Experiment Outcomes
The performance of the model using the original dataset is depicted in Figure 4. In this figure, the training curve is represented by a yellow line, while the validation curve is shown in black. It was observed that the baseline model consistently predicted all samples as negative, a result attributable to the significant imbalance in the dataset, characterized by a negative-to-positive ratio of approximately 400:1. Consequently, while the accuracy across all samples and specifically for negative samples was notably high, the accuracy for positive samples was zero. This indicates that the model failed to identify any positive samples.
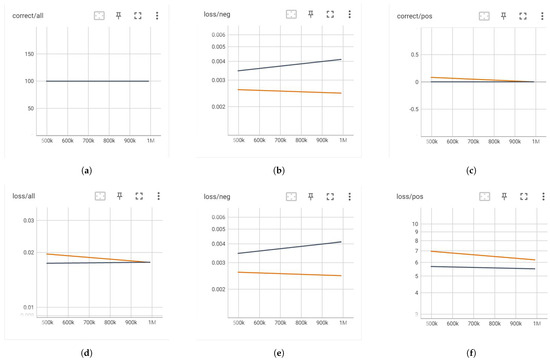
Figure 4.
Training dynamics without image augmentation. (a) Correct/all. (b) Correct/neg. (c) Correct/pos. (d) Loss/all. (e) Loss/neg. (f) Loss/pos.
Figure 5 illustrates the validation curves corresponding to various augmentation combinations applied during training. Specifically, the yellow line reflects the rotation effects, the purple line represents a combination of noise, rotation, scaling, and shifting, and the red line depicts the impact of all geometric augmentations combined. These results demonstrate that different augmentation combinations exert distinct influences on model performance, a topic that will be explored in greater depth in the subsequent section.
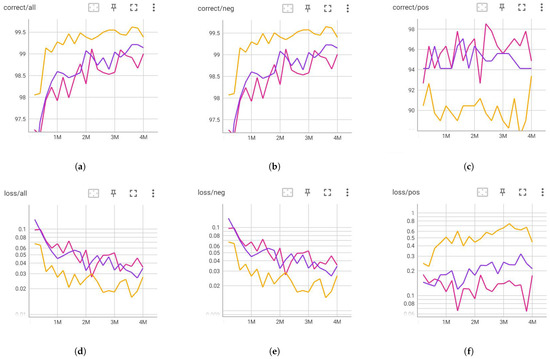
Figure 5.
Training dynamics with image augmentation: (a) Correct/all. (b) Correct/neg. (c) Correct/pos. (d) Loss/all. (e) Loss/neg. (f) Loss/pos.
5. Results and Discussion
Table 3 presents a comprehensive overview of the experimental outcomes, arrayed according to the augmentation combinations detailed in Table 1 and Table 2. The results reveal significant disparities across various augmentation strategies, indicating that the choice of metric profoundly influences classifier ranking. Metrics such as Accuracy, Score, and the MCC demonstrated favorable outcomes with noise injection, which notably enhanced the True Negative Rate, suggesting improved performance under these metrics. Despite these gains, a critical observation is the low True Positive Rate, which indicates a substantial number of real nodules were mistakenly classified as non-nodules. This misclassification poses a severe risk in medical contexts where the early detection of nodules is crucial for timely and effective cancer diagnosis and treatment. Note that the metrics for experiment index 47, which did not utilize any augmentation techniques, indicate that the model tended to incorrectly classify all samples as non-nodules, a bias influenced by the overwhelming majority of non-nodule samples in the unbalanced dataset. To provide a clearer understanding, the results were further dissected based on individual metrics, with a specific focus on highlighting the top five performers for each metric in subsequent analyses.

Table 3.
Metrics for combinations of augmentations.
5.1. Metric Analysis and Selection
Table 4 showcases the top five classifiers in terms of accuracy, which exhibited performances that were narrowly clustered together. Despite this proximity in accuracy, the recall remained alarmingly low, with the highest detection rate of nodules being only 58.68%. Such a low recall rate is inadequate for medical screening purposes, where detecting as many nodules as possible is critical. Moreover, this low recall did not come with a compensatory increase in precision; the precision was also subpar, indicating that only about 60% of the nodules identified by these classifiers were actually genuine. Consequently, this analysis suggests that accuracy alone is an insufficient metric for assessing classifier performance in this medical context.

Table 4.
Nodule prediction rankings based on accuracy.
Table 5 displays the scores in descending order, yet the recall values fluctuated significantly, ranging from 0.5074 to 0.7348. Although this shows some variability in performance, the recall rates remained lower than desirable for a screening project. This leads to the conclusion that while the score considers both recall and precision, it does not reliably identify the best classifiers for this task, as it fails to consistently prioritize the high recall necessary for effective medical screening.

Table 5.
Nodule prediction rankings based on score.
Table 6, which ranks classifiers by their scores, shows a significant improvement in recall compared to the values presented in Table 5, reaching levels that are more acceptable for a medical screening project. However, it is important to note that there was a substantial decrease in precision; a considerable portion of the predicted positive samples were actually negatives. Notably, as the score decreased, recall diminished smoothly, suggesting that the score is a useful indicator in this context, as it effectively balances the trade-off between recall and precision, prioritizing recall slightly more due to its higher weight in the calculation.

Table 6.
Nodule prediction rankings based on score.
Table 7 identifies the most effective model based on recall, which demonstrates a trade-off, sacrificing precision to enhance recall. This model successfully classified over 96% of the nodules, albeit with low precision, indicating that only about one-sixth of the classified nodules were actual positives. From the perspective of a screening task, this trade-off was justified, as it prioritized the detection of potential nodules, which is crucial in medical diagnostics. Consequently, in this study we chose the score as our performance metric. The score places a stronger emphasis on recall, making it well-suited for evaluating the effectiveness of different augmentation methods in enhancing model sensitivity.

Table 7.
Nodule prediction rankings based on score.
Table 8 illustrates that the MCC strives for a balance between recall and precision. This characteristic of the MCC may prove advantageous in scenarios where the cost of FPs is high, making it a potentially valuable metric for diverse tasks that require careful error management.

Table 8.
Nodule prediction rankings based on MCC score.
5.2. Augmentation Method Analysis
Table 9 highlights the performance of the top five augmentation combinations, revealing a distinct preference for geometric augmentations. This preference correlates with the characteristics of the LUNA dataset, where CT scans are aligned in such a way that photometric variations provide minimal additional information. Geometric augmentations effectively mimic the variations in human organs and tissues, significantly enhancing the model’s capability to process CT scans. Notably, the best-performing augmentation combination excluded rotation, while the next four highest-ranked combinations all included rotation, suggesting its individual contribution to model performance. This intriguing observation warrants further investigation in future research, to better understand the role of rotation and other geometric transformations in enhancing diagnostic accuracy. In Figure 6, it is evident that the best augmentation combinations outperformed other strategies, in terms of the score, and that the highest scores consistently correlated with a high recall rate. Because the score aims to balance recall and precision, its specific ability to do so effectively makes it more suitable than other metrics for evaluating performance in this context.

Table 9.
The rank of augmentation combinations based on score.
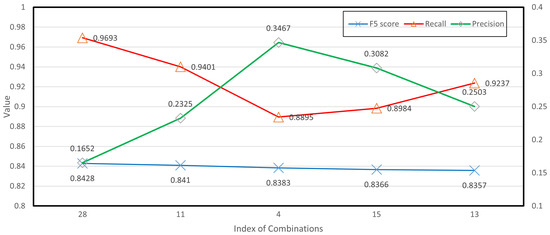
Figure 6.
Top 5 augmentation combinations.
The results indicate a clear preference for geometric augmentations in enhancing model performance. Among these, the most effective combination included flip, shift, scale, and noise, followed by other primarily geometric strategies. This outcome can be attributed to several key factors inherent to the nature of medical imaging and the specific challenges posed by lung nodule detection. Firstly, the superior performance of the combination involving flip, shift, scale, and noise can be linked to their collective ability to mimic real-world variations in lung scans. Flipping the images and shifting the images simulate changes in patient positioning and orientation relative to the imaging equipment. This is crucial because slight variations in how a patient is positioned can significantly alter the appearance of nodules. Scaling addresses the variation in nodule size, which can be affected by the distance of the nodules from the imaging device, among other factors. Noise introduction, while often considered a photometric augmentation, in this context helps model robustness by simulating slight imperfections in imaging technology, such as electronic noise or sensor artifacts. The second-best combination, involving shift and rotate, underscores the importance of rotational and translational invariance in medical imaging analysis. Rotation mimics the axial changes that can occur if a patient is not perfectly aligned in the scanner, while shifts account for lateral movements. These types of augmentations are particularly useful for enhancing the model’s ability to generalize from the training data to new, unseen images that may not be perfectly aligned or scaled. The variations in augmentation performance also suggest that while rotation alone is useful—evidenced by its third-place ranking—its combination with other techniques, particularly noise, further refines the model’s accuracy. This is likely because noise introduces a level of difficulty that forces the model to learn more robust features rather than overfitting to noise-free images in the training set. Lastly, the combination of scale and rotate points to the significance of handling both size and orientation variability together. Each augmentation method does not work in isolation but rather complements others to cover a broader range of real-world variations in lung nodule appearance.
These results highlight the critical role of data augmentation in preparing robust models capable of high accuracy and generalization in medical imaging tasks, particularly for applications as sensitive and variable as lung nodule classification. The gap in performance among the different combinations provides valuable insights into the complex interplay of factors that affect image recognition and classification in a clinical context.
5.3. Comparison with Other Research and Future Research
Recent advancements in lung nodule classification have predominantly centered on deep learning techniques, encompassing ensemble models, transfer learning, attention mechanisms, and capsule networks. In our study, simple CNNs were constructed as the baseline, to concentrate on exploring various metrics and data augmentation strategies. Leading entries on the LUNA16 leaderboard have achieved recall above 90% at 1/8 FPs per scan. In our research, the top performance of combination obtained more than 96% at 1/5 FPs, far outperforming the State-of-the-Art (SOTA) model. Our research also delved into the practical challenges of implementing these models in real-world clinical settings, where inconsistencies in imaging protocols and equipment variability can adversely affect performance. The insights from this study aim to inform ongoing developments in diagnostic technologies, ensuring they are sufficiently robust for use across varied healthcare scenarios, thereby improving patient outcomes through the early and precise identification of critical conditions such as lung nodules. Our future research aims to expand investigation into data augmentation techniques and their synergistic effects, while also deepening our understanding of the interplay between different performance metrics within the framework of SOTA architectures and broader application contexts. This ongoing research is anticipated to enhance the proficiency of deep learning models in medical imaging, thereby revolutionizing diagnostic methodologies and improving the effectiveness of medical treatments.
6. Conclusions
This study meticulously selected and compared metrics, to identify the most suitable ones for lung nodule detection, emphasizing the need for metrics that accurately reflect the true performance of models, especially in unbalanced datasets. This research determined that the score, which gives greater weight to recall, is the most effective metric for evaluating medical image classification models. The appropriate metrics led to the identification of the most potent combinations of augmentations for nodule classification. The results of these experiments were rigorously documented and analyzed, with comprehensive tables comparing the effects of different augmentations. This analysis not only underscored the benefits of specific techniques but also revealed the synergistic impact of combining them, significantly boosting the model’s precision in detecting lung nodules.
Author Contributions
Conceptualization, D.L. and J.B.; methodology, D.L. and I.Y.; software, Y.W., I.Y., and D.L.; writing—original draft preparation, D.L.; writing—review and editing, D.L. and J.B.; visualization, D.L.; supervision, J.B.; project administration, J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study has been supported by MOTIE funding program “Advanced Graduate Education for Management of Convergence Technology”.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MCC | Matthew’s Correlation Coefficients |
| CT | Computed Tomography |
| CNN | Convolutional Neural Network |
| CAD | Computer-Aided Diagnosis |
| FPR | False Positive Rate |
| TPR | True Positive Rate |
| SOTA | State-of-the-Art |
| TPs | True Positives |
| TNs | True Negatives |
| FPs | False Positives |
| FNs | False Negatives |
References
- Leiter, A.; Veluswamy, R.R.; Wisnivesky, J.P. The global burden of lung cancer: Current status and future trends. Nat. Rev. Clin. Oncol. 2023, 20, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Del Ciello, A.; Franchi, P.; Contegiacomo, A.; Cicchetti, G.; Bonomo, L.; Larici, A.R. Missed lung cancer: When, where, and why? Diagn. Interv. Radiol. 2017, 23, 118. [Google Scholar] [CrossRef]
- Du, J.; Guan, K.; Zhou, Y.; Li, Y.; Wang, T. Parameter-free similarity-aware attention module for medical image classification and segmentation. IEEE Trans. Emerg. Top. Comput. Intell. 2022, 7, 845–857. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Zhao, Z.; Zeng, Z.; Xu, K.; Chen, C.; Guan, C. Dsal: Deeply supervised active learning from strong and weak labelers for biomedical image segmentation. IEEE J. Biomed. Health Inform. 2021, 25, 3744–3751. [Google Scholar] [CrossRef]
- Kim, B.; Kim, J.; Lee, J.G.; Kim, D.H.; Park, S.H.; Ye, J.C. Unsupervised deformable image registration using cycle-consistent CNN. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 166–174. [Google Scholar]
- Tseng, K.K.; Zhang, R.; Chen, C.M.; Hassan, M.M. DNetUnet: A semi-supervised CNN of medical image segmentation for super-computing AI service. J. Supercomput. 2021, 77, 3594–3615. [Google Scholar] [CrossRef]
- Lerousseau, M.; Vakalopoulou, M.; Classe, M.; Adam, J.; Battistella, E.; Carré, A.; Estienne, T.; Henry, T.; Deutsch, E.; Paragios, N. Weakly supervised multiple instance learning histopathological tumor segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part V 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 470–479. [Google Scholar]
- Li, Y.; Chen, J.; Xie, X.; Ma, K.; Zheng, Y. Self-loop uncertainty: A novel pseudo-label for semi-supervised medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part I 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 614–623. [Google Scholar]
- Ma, J.; Song, Y.; Tian, X.; Hua, Y.; Zhang, R.; Wu, J. Survey on deep learning for pulmonary medical imaging. Front. Med. 2020, 14, 450–469. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Maddikunta, P.K.R.; Pham, Q.V.; Gadekallu, T.R.; Chowdhary, C.L.; Alazab, M.; Piran, M.J. Deep learning and medical image processing for coronavirus pandemic: A survey. Sustain. Cities Soc. 2021, 65, 102589. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, C.; Kumar, I.; Vijayakumar, V.; Singh, K.U.; Kumar, A. The state of the art of deep learning models in medical science and their challenges. Multimed. Syst. 2021, 27, 599–613. [Google Scholar] [CrossRef]
- Dourado, C.M.; da Silva, S.P.P.; da Nobrega, R.V.M.; Reboucas Filho, P.P.; Muhammad, K.; de Albuquerque, V.H.C. An open IoHT-based deep learning framework for online medical image recognition. IEEE J. Sel. Areas Commun. 2020, 39, 541–548. [Google Scholar] [CrossRef]
- Zhao, Z.; Alzubaidi, L.; Zhang, J.; Duan, Y.; Gu, Y. A comparison review of transfer learning and self-supervised learning: Definitions, applications, advantages and limitations. Expert Syst. Appl. 2023, 242, 122807. [Google Scholar] [CrossRef]
- Masud, M.; Sikder, N.; Nahid, A.A.; Bairagi, A.K.; AlZain, M.A. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors 2021, 21, 748. [Google Scholar] [CrossRef]
- Binczyk, F.; Prazuch, W.; Bozek, P.; Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res. 2021, 10, 1186–1199. [Google Scholar] [CrossRef]
- Niu, S.; Liu, M.; Liu, Y.; Wang, J.; Song, H. Distant domain transfer learning for medical imaging. IEEE J. Biomed. Health Inform. 2021, 25, 3784–3793. [Google Scholar] [CrossRef]
- Liao, W.; Xiong, H.; Wang, Q.; Mo, Y.; Li, X.; Liu, Y.; Chen, Z.; Huang, S.; Dou, D. Muscle: Multi-task self-supervised continual learning to pre-train deep models for x-ray images of multiple body parts. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 151–161. [Google Scholar]
- Lipton, Z.C.; Elkan, C.; Narayanaswamy, B. Thresholding classifiers to maximize F1 score. arXiv 2014, arXiv:1402.1892. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Luque, A.; Carrasco, A.; Martín, A.; de Las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Fang, T. A novel computer-aided lung cancer detection method based on transfer learning from GoogLeNet and median intensity projections. In Proceedings of the 2018 IEEE International Conference on Computer and Communication Engineering Technology (CCET), Beijing, China, 18–20 August 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 286–290. [Google Scholar]
- Gsaxner, C.; Roth, P.M.; Wallner, J.; Egger, J. Exploit fully automatic low-level segmented PET data for training high-level deep learning algorithms for the corresponding CT data. PLoS ONE 2019, 14, e0212550. [Google Scholar] [CrossRef]
- Stevens, E.; Antiga, L.; Viehmann, T. Deep Learning with PyTorch; Manning Publications: Shelter Island, NY, USA, 2020. [Google Scholar]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef]
- Way, T.W.; Sahiner, B.; Chan, H.P.; Hadjiiski, L.; Cascade, P.N.; Chughtai, A.; Bogot, N.; Kazerooni, E. Computer-aided diagnosis of pulmonary nodules on CT scans: Improvement of classification performance with nodule surface features. Med. Phys. 2009, 36, 3086–3098. [Google Scholar] [CrossRef]
- Liu, Y.; Balagurunathan, Y.; Atwater, T.; Antic, S.; Li, Q.; Walker, R.C.; Smith, G.T.; Massion, P.P.; Schabath, M.B.; Gillies, R.J. Radiological image traits predictive of cancer status in pulmonary nodules. Clin. Cancer Res. 2017, 23, 1442–1449. [Google Scholar] [CrossRef]
- Jacobs, C.; Van Rikxoort, E.M.; Twellmann, T.; Scholten, E.T.; De Jong, P.A.; Kuhnigk, J.M.; Oudkerk, M.; De Koning, H.J.; Prokop, M.; Schaefer-Prokop, C.; et al. Automatic detection of subsolid pulmonary nodules in thoracic computed tomography images. Med. Image Anal. 2014, 18, 374–384. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.; Van Den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).