On the Importance of Diversity When Training Deep Learning Segmentation Models with Error-Prone Pseudo-Labels
Abstract
1. Introduction
2. Related Work
3. Methods
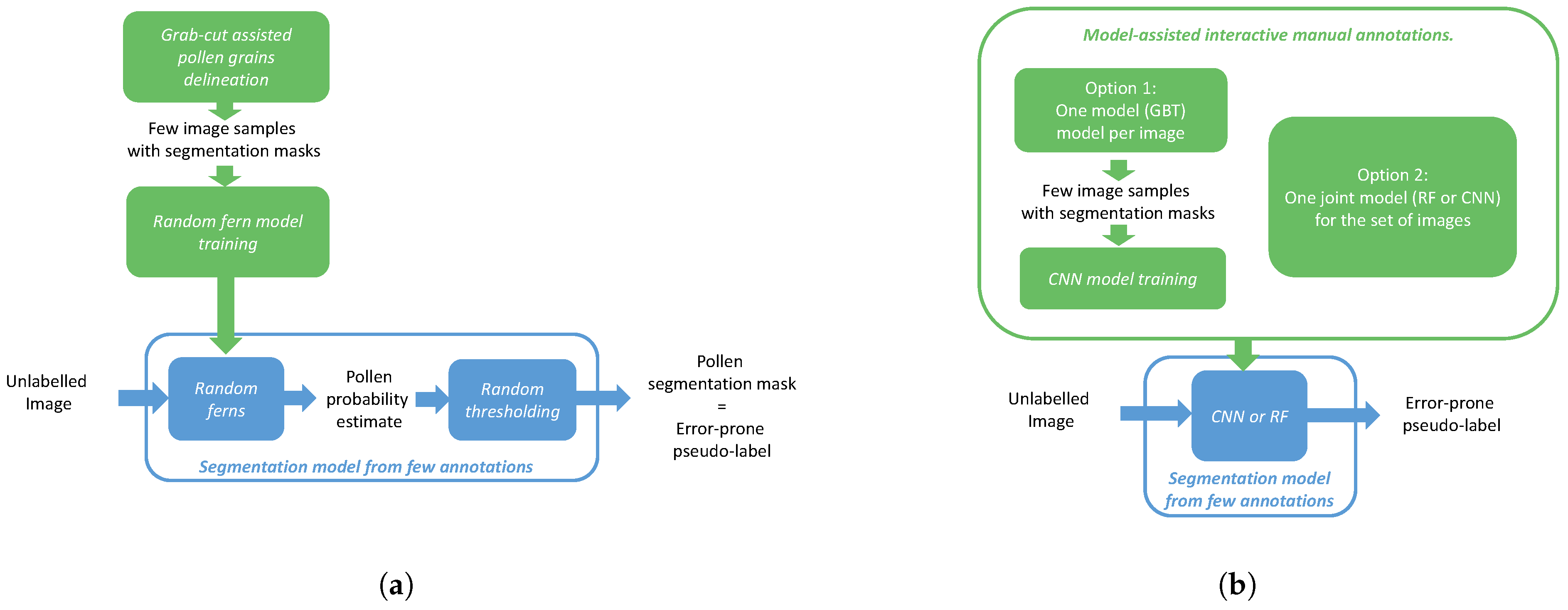
3.1. (Semi-)Manual Annotation
- The manual delineation of the foreground object contours. This solution is suited to smooth and regular object shapes, and is adopted in the following to delineate the pollen grains in our first experimental scenario.
- Semi-manual methods, relying on interactive user annotations to refine and increment the training set that is used to train a machine learning model. Those learning-based methods are suited in cases where the foreground and background are visually dissimilar but are separated by an irregular border, requiring time for fully manual delineation.
3.2. Training a Pseudo-Label Generator
3.3. Training a CNN from Pseudo-Labels
4. Experimental Validation
4.1. Datasets
4.1.1. Pollen Grain Microscopy Images
4.1.2. RGB Canopy Images
4.2. Methods and Terminology
4.2.1. Pollen Grain Microscopy
4.2.2. Outdoor RGB Canopy Images
4.3. Metrics and Implementation Details
4.3.1. Evaluation Metrics
4.3.2. CNN Network
4.3.3. Loss
4.3.4. Training
4.4. Results and Discussion
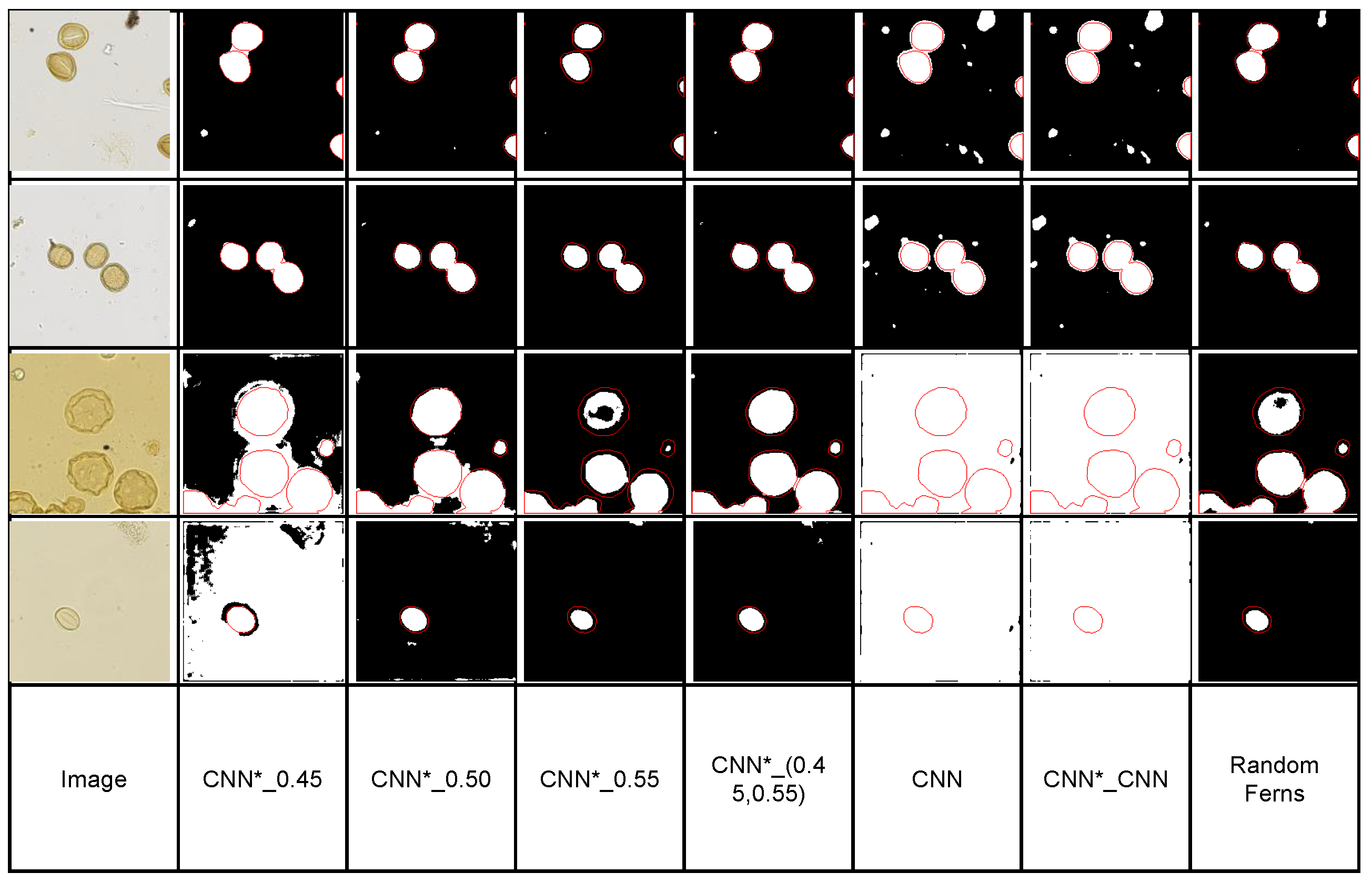
4.4.1. Pollen Grain Microscopy
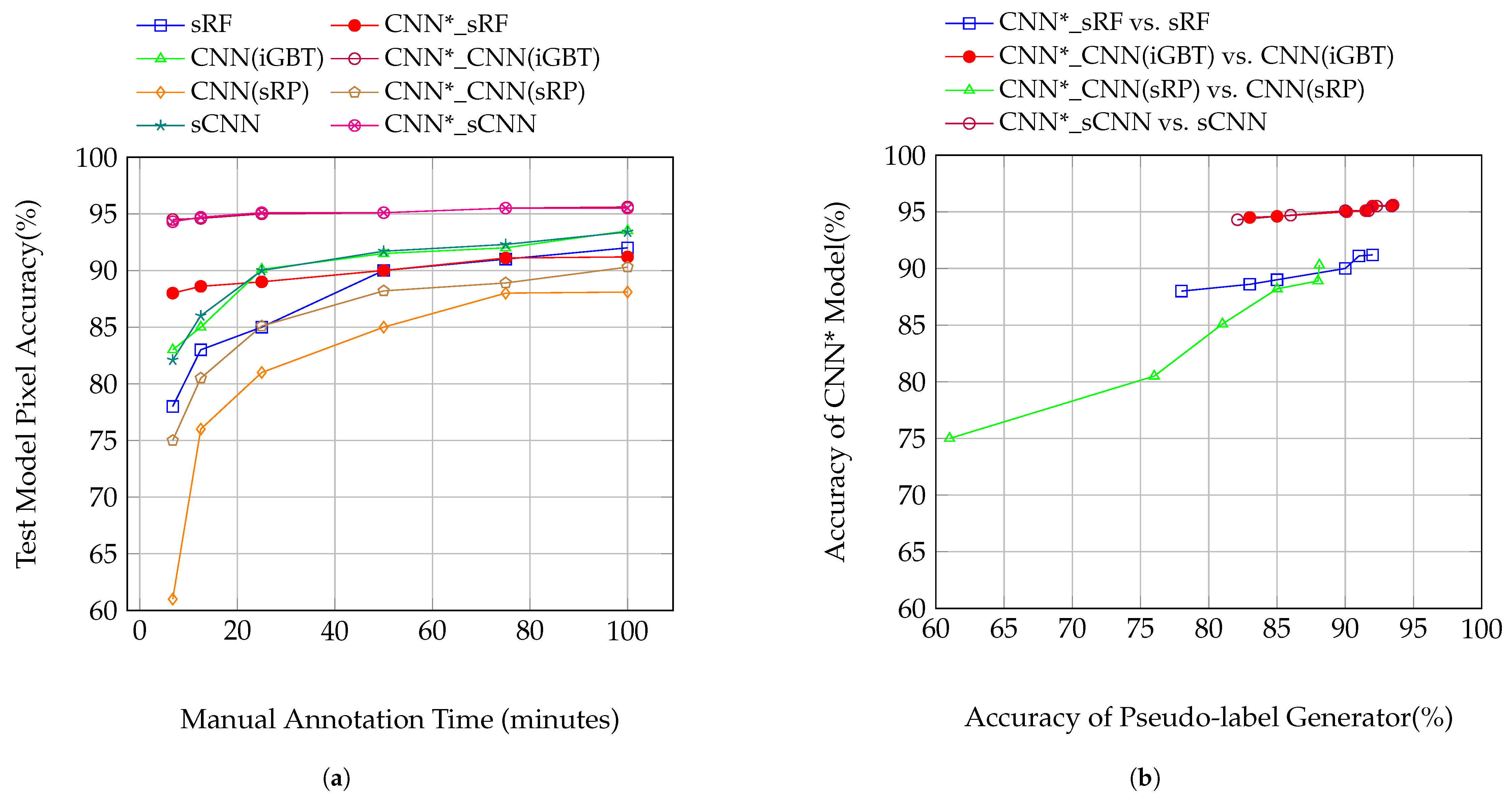
4.4.2. Outdoor RGB Canopy Images
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Apley, D.W.; Zhu, J. Visualizing the effects of predictor variables in black box supervised learning models. J. R. Stat. Soc. Ser. B Stat. Methodol. 2020, 82, 1059–1086. [Google Scholar] [CrossRef]
- Castelli, M.; Vanneschi, L.; Largo, Á.R. Supervised learning: Classification. Encycl. Bioinform. Comput. Biol. 2018, 1, 342–349. [Google Scholar]
- Sarmadi, H.; Entezami, A. Application of supervised learning to validation of damage detection. Arch. Appl. Mech. 2021, 91, 393–410. [Google Scholar] [CrossRef]
- Zhou, Z.H. Semi-supervised learning. In Machine Learning; Springer: Singapore, 2021; pp. 315–341. [Google Scholar]
- Ouali, Y.; Hudelot, C.; Tami, M. An overview of deep semi-supervised learning. arXiv 2020, arXiv:2006.05278. [Google Scholar]
- Zheng, M.; You, S.; Huang, L.; Wang, F.; Qian, C.; Xu, C. SimMatch: Semi-supervised Learning with Similarity Matching. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 14471–14481. [Google Scholar]
- Sayez, N.; De Vleeschouwer, C. Accelerating the creation of instance segmentation training sets through bounding box annotation. In Proceedings of the 2022 26th International Conference on Pattern Recognition (ICPR), Montreal, QC, Canada, 21–25 August 2022; pp. 252–258. [Google Scholar]
- Mendel, R.; De Souza, L.A.; Rauber, D.; Papa, J.P.; Palm, C. Semi-supervised segmentation based on error-correcting supervision. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part XXIX 16. pp. 141–157. [Google Scholar]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Arazo, E.; Ortego, D.; Albert, P.; O’Connor, N.E.; McGuinness, K. Pseudo-labeling and confirmation bias in deep semi-supervised learning. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Yang, L.; Zhuo, W.; Qi, L.; Shi, Y.; Gao, Y. St++: Make self-training work better for semi-supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 4268–4277. [Google Scholar]
- Zhu, Y.; Zhang, Z.; Wu, C.; Zhang, Z.; He, T.; Zhang, H.; Manmatha, R.; Li, M.; Smola, A.J. Improving semantic segmentation via efficient self-training. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 46, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Smith, A. Image segmentation scale parameter optimization and land cover classification using the Random Forest algorithm. J. Spat. Sci. 2010, 55, 69–79. [Google Scholar] [CrossRef]
- Chen, H.; Wu, L.; Chen, J.; Lu, W.; Ding, J. A comparative study of automated legal text classification using random forests and deep learning. Inf. Process. Manag. 2022, 59, 102798. [Google Scholar] [CrossRef]
- Fröhlich, B.; Rodner, E.; Denzler, J. Semantic segmentation with millions of features: Integrating multiple cues in a combined random forest approach. In Proceedings of the Asian Conference on Computer Vision, Daejeon, Republic of Korea, 5–9 November 2012; pp. 218–231. [Google Scholar]
- Wei, P.; Hänsch, R. Random Ferns for Semantic Segmentation of PolSAR Images. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5218212. [Google Scholar] [CrossRef]
- Mo, Y.; Wu, Y.; Yang, X.; Liu, F.; Liao, Y. Review the state-of-the-art technologies of semantic segmentation based on deep learning. Neurocomputing 2022, 493, 626–646. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, B.; Geng, L.; Zhang, F.; Liu, Y. Segmentation of lung nodules using improved 3D-UNet neural network. Symmetry 2020, 12, 1787. [Google Scholar] [CrossRef]
- Rizve, M.N.; Duarte, K.; Rawat, Y.S.; Shah, M. In defense of pseudo-labeling: An uncertainty-aware pseudo-label selection framework for semi-supervised learning. arXiv 2021, arXiv:2101.06329. [Google Scholar]
- Zhang, B.; Wang, Y.; Hou, W.; Wu, H.; Wang, J.; Okumura, M.; Shinozaki, T. Flexmatch: Boosting semi-supervised learning with curriculum pseudo labeling. Adv. Neural Inf. Process. Syst. 2021, 34, 18408–18419. [Google Scholar]
- Wei, C.; Shen, K.; Chen, Y.; Ma, T. Theoretical analysis of self-training with deep networks on unlabeled data. arXiv 2020, arXiv:2010.03622. [Google Scholar]
- Liu, J.; Yao, J.; Bagheri, M.; Sandfort, V.; Summers, R.M. A semi-supervised CNN learning method with pseudo-class labels for atherosclerotic vascular calcification detection. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 780–783. [Google Scholar]
- Liu, B.; Yu, X.; Zhang, P.; Tan, X.; Yu, A.; Xue, Z. A semi-supervised convolutional neural network for hyperspectral image classification. Remote Sens. Lett. 2017, 8, 839–848. [Google Scholar] [CrossRef]
- Pintelas, E.; Livieris, I.E.; Pintelas, P. A grey-box ensemble model exploiting black-box accuracy and white-box intrinsic interpretability. Algorithms 2020, 13, 17. [Google Scholar] [CrossRef]
- Chakravarthy, A.D.; Abeyrathna, D.; Subramaniam, M.; Chundi, P.; Gadhamshetty, V. Semantic image segmentation using scant pixel annotations. Mach. Learn. Knowl. Extr. 2022, 4, 621–640. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, Z.; Zhang, H.; Li, C.L.; Bian, X.; Huang, J.B.; Pfister, T. Pseudoseg: Designing pseudo labels for semantic segmentation. arXiv 2020, arXiv:2010.09713. [Google Scholar]
- Xu, H.; Liu, L.; Bian, Q.; Yang, Z. Semi-supervised semantic segmentation with prototype-based consistency regularization. Adv. Neural Inf. Process. Syst. 2022, 35, 26007–26020. [Google Scholar]
- Zhang, B.; Zhang, Y.; Li, Y.; Wan, Y.; Guo, H.; Zheng, Z.; Yang, K. Semi-supervised deep learning via transformation consistency regularization for remote sensing image semantic segmentation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 16, 5782–5796. [Google Scholar] [CrossRef]
- Vu, T.H.; Jain, H.; Bucher, M.; Cord, M.; Pérez, P. Advent: Adversarial entropy minimization for domain adaptation in semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2517–2526. [Google Scholar]
- Vesal, S.; Gu, M.; Kosti, R.; Maier, A.; Ravikumar, N. Adapt everywhere: Unsupervised adaptation of point-clouds and entropy minimization for multi-modal cardiac image segmentation. IEEE Trans. Med Imaging 2021, 40, 1838–1851. [Google Scholar] [CrossRef]
- Zeng, G.; Peng, H.; Li, A.; Liu, Z.; Liu, C.; Yu, P.S.; He, L. Unsupervised Skin Lesion Segmentation via Structural Entropy Minimization on Multi-Scale Superpixel Graphs. arXiv 2023, arXiv:2309.01899. [Google Scholar]
- Cioppa, A.; Deliege, A.; Istasse, M.; De Vleeschouwer, C.; Van Droogenbroeck, M. ARTHuS: Adaptive real-time human segmentation in sports through online distillation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Baldeon Calisto, M. Teacher-student semi-supervised approach for medical image segmentation. In MICCAI Challenge on Fast and Low-Resource Semi-supervised Abdominal Organ Segmentation; Springer: Cham, Switzerland, 2022; pp. 152–162. [Google Scholar]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive machine learning for (bio) image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Smith, A.G.; Han, E.; Petersen, J.; Olsen, N.A.F.; Giese, C.; Athmann, M.; Dresbøll, D.B.; Thorup-Kristensen, K. RootPainter: Deep learning segmentation of biological images with corrective annotation. New Phytol. 2022, 236, 774–791. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Zhang, H.; Keerthi, S.S.; Mahajan, D.; Dhillon, I.S.; Hsieh, C.J. Gradient boosted decision trees for high dimensional sparse output. In Proceedings of the 34th International Conference on Machine Learning, PMLR, Sydney, NSW, Australia, 6–11 August 2017; pp. 3182–3190. [Google Scholar]
- Schonlau, M.; Zou, R.Y. The random forest algorithm for statistical learning. Stata J. 2020, 20, 3–29. [Google Scholar] [CrossRef]
- Browet, A.; De Vleeschouwer, C.; Jacques, L.; Mathiah, N.; Saykali, B.; Migeotte, I. Cell segmentation with random ferns and graph-cuts. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 4145–4149. [Google Scholar]
- Parisot, P.; De Vleeschouwer, C. Scene-specific classifier for effective and efficient team sport players detection from a single calibrated camera. Comput. Vis. Image Underst. 2017, 159, 74–88. [Google Scholar] [CrossRef]
- Bay, Y.Y.; Yearick, K.A. Machine Learning vs. Deep Learning: The Generalization Problem. arXiv 2024, arXiv:2403.01621. [Google Scholar]
- Neyshabur, B.; Bhojanapalli, S.; McAllester, D.; Srebro, N. Exploring generalization in deep learning. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Kawaguchi, K.; Kaelbling, L.P.; Bengio, Y. Generalization in Deep Learning. In Mathematical Aspects of Deep Learning; Cambridge University Press: Cambridge, UK, 2022. [Google Scholar] [CrossRef]
- Léger, J.; Leyssens, L.; Kerckhofs, G.; De Vleeschouwer, C. Ensemble learning and test-time augmentation for the segmentation of mineralized cartilage versus bone in high-resolution microCT images. Comput. Biol. Med. 2022, 148, 105932. [Google Scholar] [CrossRef] [PubMed]
- Gal, Y. Uncertainty in Deep Learning. 2016. Available online: http://106.54.215.74/2019/20190729-liuzy.pdf (accessed on 8 June 2024).
- Smith, L.; Gal, Y. Understanding measures of uncertainty for adversarial example detection. arXiv 2018, arXiv:1803.08533. [Google Scholar]
- Rubens, U.; Hoyoux, R.; Vanosmael, L.; Ouras, M.; Tasset, M.; Hamilton, C.; Longuespée, R.; Marée, R. Cytomine: Toward an open and collaborative software platform for digital pathology bridged to molecular investigations. PROTEOMICS-Appl. 2019, 13, 1800057. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Gao, P.; Jiang, L.; Chen, M. Grab cut image segmentation based on image region. In Proceedings of the 2018 IEEE 3rd international conference on image, vision and computing (ICIVC), Chongqing, China, 27–29 June 2018; pp. 311–315. [Google Scholar]
- Ozuysal, M.; Calonder, M.; Lepetit, V.; Fua, P. Fast keypoint recognition using random ferns. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 32, 448–461. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Joos, V.; Jacquemart, A.L.; Buyens, C.; De Vleeschouwer, C. Using Pure Pollen Species When Training a CNN To Segment Pollen Mixtures. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, New Orleans, LA, USA, 18–24 June 2022; pp. 1695–1704. [Google Scholar]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Zhang, C.; Bengio, S.; Hardt, M.; Recht, B.; Vinyals, O. Understanding deep learning (still) requires rethinking generalization. Commun. ACM 2021, 64, 107–115. [Google Scholar] [CrossRef]


| (Semi-)Manual Annotation | Pseudo-Label Generator | Test Model |
|---|---|---|
| sRF [34] | X | sRF |
| sRF | CNN*_sRF | |
| sRP | CNN(sRP) | CNN*_CNN(sRP) |
| X | CNN(sRP) | |
| iGBT | CNN(iGBT) | CNN*_CNN(iGBT) |
| X | CNN(iGBT) | |
| sCNN [35] | sCNN | CNN*_sCNN |
| X | sCNN |
| Foreground IoU (%) | ||||||
|---|---|---|---|---|---|---|
| Method | 1 Images | 5 Images | 10 Images | 15 Images | 25 Images | 50 Images |
| CNN | 37.4 | 58.9 | 69.9 | 72.4 | 80.4 | 81.6 |
| Random Ferns | 74.7 | 74.0 | 77.3 | 80.0 | 79.6 | 79.2 |
| CNN*_0.45 | 72.2 | 76.0 | 76.0 | 73.7 | 74.7 | 74.3 |
| CNN*_0.50 | 80.0 | 81.3 | 82.2 | 82.4 | 82.6 | 82.7 |
| CNN*_0.55 | 75.0 | 73.3 | 76.2 | 80.3 | 79.7 | 78.2 |
| CNN*_(0.45,0.55) | 84.5 | 83.3 | 84.3 | 84.8 | 85.0 | 85.1 |
| CNN*_CNN | 45.7 | 65.9 | 73.8 | 75.7 | 82.8 | 82.9 |
| Pixel Accuracy (%) | ||||||
|---|---|---|---|---|---|---|
| Method | 1 Images | 5 Images | 10 Images | 15 Images | 25 Images | 50 Images |
| CNN | 75.2 | 86.2 | 94.6 | 95.1 | 97.3 | 97.4 |
| Random Ferns | 97.0 | 96.9 | 97.3 | 97.6 | 97.5 | 97.5 |
| CNN*_0.45 | 90.1 | 94.4 | 96.0 | 96.2 | 95.5 | 95.7 |
| CNN*_0.50 | 97.0 | 97.9 | 98.0 | 97.4 | 97.7 | 97.8 |
| CNN*_0.55 | 97.6 | 97.4 | 97.6 | 98.1 | 98.0 | 97.9 |
| CNN*_(0.45,0.55) | 98.4 | 98.1 | 98.2 | 98.2 | 98.3 | 98.2 |
| CNN*_CNN | 73.5 | 83.3 | 91.9 | 92.7 | 98.2 | 98.1 |
| (Semi-)Manual Annotation | Pseudo-Label Generator | Test Model |
|---|---|---|
| Manual Delineation | X | CNN |
| X | Random Ferns | |
| Random Ferns | _ | |
| Random Ferns (,) | CNN*_(,) | |
| CNN | CNN*_CNN |
| Foreground IoU (%) | ||||||
|---|---|---|---|---|---|---|
| Annotation Time | 6.75 | 12.5 | 25 | 50 | 75 | 100 |
| sRF | 63.0 | 69.5 | 72.3 | 81.7 | 83.3 | 84.2 |
| CNN*_sRF | 79.6 | 80.2 | 81.8 | 82.8 | 83.5 | 83.7 |
| CNN(iGBT) | 69.5 | 72.4 | 81.8 | 83.5 | 84.3 | 86.9 |
| CNN*_CNN(iGBT) | 88.7 | 89.2 | 89.6 | 89.9 | 90.7 | 90.8 |
| CNN(sRP) | 42.9 | 67.2 | 74.2 | 78.7 | 79.9 | 82.2 |
| CNN*_CNN(sRP) | 61.3 | 68.1 | 78.6 | 80.2 | 81.8 | 83.5 |
| sCNN | 69.3 | 72.6 | 81.9 | 83.3 | 84.3 | 86.9 |
| CNN*_sCNN | 89.0 | 89.4 | 89.6 | 89.9 | 90.8 | 91.0 |
| Pixel Accuracy (%) | ||||||
|---|---|---|---|---|---|---|
| Annotation Time | 6.75 | 12.5 | 25 | 50 | 75 | 100 |
| sRF | 78.3 | 83.2 | 85.1 | 89.7 | 91.0 | 91.8 |
| CNN*_sRF | 88.1 | 88.6 | 89.1 | 90.0 | 91.1 | 91.2 |
| CNN(iGBT) | 83.1 | 85.2 | 90.1 | 91.5 | 92.0 | 93.5 |
| CNN*_CNN(iGBT) | 94.5 | 94.6 | 95.0 | 95.1 | 95.5 | 95.6 |
| CNN(sRP) | 61.1 | 75.9 | 81.3 | 85.2 | 87.8 | 88.1 |
| CNN*_CNN(sRP) | 75.2 | 80.5 | 85.1 | 88.2 | 88.9 | 90.3 |
| sCNN | 82.1 | 85.9 | 90.0 | 91.7 | 92.3 | 93.4 |
| CNN*_sCNN | 94.3 | 94.7 | 95.1 | 95.1 | 95.5 | 95.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, N.; Rongione, C.; Jacquemart, A.-L.; Draye, X.; Vleeschouwer, C.D. On the Importance of Diversity When Training Deep Learning Segmentation Models with Error-Prone Pseudo-Labels. Appl. Sci. 2024, 14, 5156. https://doi.org/10.3390/app14125156
Yang N, Rongione C, Jacquemart A-L, Draye X, Vleeschouwer CD. On the Importance of Diversity When Training Deep Learning Segmentation Models with Error-Prone Pseudo-Labels. Applied Sciences. 2024; 14(12):5156. https://doi.org/10.3390/app14125156
Chicago/Turabian StyleYang, Nana, Charles Rongione, Anne-Laure Jacquemart, Xavier Draye, and Christophe De Vleeschouwer. 2024. "On the Importance of Diversity When Training Deep Learning Segmentation Models with Error-Prone Pseudo-Labels" Applied Sciences 14, no. 12: 5156. https://doi.org/10.3390/app14125156
APA StyleYang, N., Rongione, C., Jacquemart, A.-L., Draye, X., & Vleeschouwer, C. D. (2024). On the Importance of Diversity When Training Deep Learning Segmentation Models with Error-Prone Pseudo-Labels. Applied Sciences, 14(12), 5156. https://doi.org/10.3390/app14125156






