Abstract
The rapid development of high-throughput technology has generated a large amount of protein–protein interaction (PPI) data, which provide a large amount of data support for constructing dynamic protein–protein interaction networks (PPINs). Constructing dynamic PPINs and applying them to recognize protein complexes has become a hot research topic. Most existing methods for complex recognition cannot fully mine the information of PPINs. To address this problem, we propose a construction method of dynamic weighted protein network by multi-level embedding (DWPNMLE). It can reflect the protein network’s dynamics and the protein network’s higher-order proximity. Firstly, the protein active period is calculated to divide the protein subnetworks at different time points. Then, the connection probability is used for the proteins possessing the same time points to judge whether there is an interaction relationship between them. Then, the corresponding protein subnetworks (multiple adjacency matrices) are constructed. Secondly, the multiple feature matrices are constructed using one-hot coding with the gene ontology (GO) information. Next, the first embedding is performed using variational graph auto-encoders (VGAEs) to aggregate features efficiently, followed by the second embedding using deep attributed network embedding (DANE) to strengthen the node representations learned in the first embedding and to maintain the first-order and higher-order proximity of the original network; finally, we compute the cosine similarity to obtain the final dynamic weighted PPIN. To evaluate the effectiveness of DWPNMLE, we apply four classical protein-complex-recognition algorithms on the DWPNMLE and compare them with two other dynamic protein network construction methods. The experimental results demonstrate that DWPNMLE significantly enhances the accuracy of complex recognition with high robustness, and the algorithms’ efficiency is also within a reasonable range.
1. Introduction
Proteins are complex macromolecules formed from various amino acids linked by peptide bonds following specific patterns. As vital components of cellular structures, proteins are crucial to the biochemical processes of living organisms, forming the foundational substance for the execution of life’s various functions [1]. Proteins participate in almost all biological interactions and play crucial physiological roles. The characteristics of many identified proteins have not been experimentally determined, leaving their functions unknown. Biological functions are not the purview of solitary proteins but rather occur through the complex interplay among myriad proteins. The protein–protein interaction network (PPIN) alludes to the topological structure of the genetic or physical connections among all proteins within an organism, commonly called the protein network. Constructing a dynamic network of these proteins is fundamental to numerous proteomic studies, such as recognizing protein complexes and discovering essential proteins, which rely on a highly reliable protein network. In recent years, an extensive volume of protein–protein interaction (PPI) data [2] have been accumulated, stemming from advancements in high-throughput experimental methodologies. These accessible PPI data contribute to constructing vast and complex protein networks for numerous organisms [3,4]. A considerable portion of false positives and false negatives within the data have led to the generation of largely unreliable protein networks. Thus, these networks’ precise modeling and subsequent analyses remain prominent in contemporary bioinformatics research. Static protein networks can be constructed by employing high-throughput experimental methodologies, especially yeast two hybrid (Y2H) [5] and tandem affinity purification (TAP) [6]. These methods authenticate the presence of interaction edges by ascertaining the interlinks between protein nodes and do not proffer temporal and spatial data corresponding to PPIs and are greatly prone to encompassing false-positive and false-negative results [7,8]. To address the inherent constraints of static PPINs, researchers have advocated novel approaches, utilizing the dynamic expression of proteins at variable time points and integrating multi-source heterogeneous biological data to excavate the complexity of the interaction between proteins in multiple dimensions, encompassing facets such as network topology and functional annotation [9]. Researchers have developed dynamic PPINs on this premise, aiming to precisely illustrate the dynamics underlying protein regulation within biological entities. Li et al. [10] amalgamated gene expression profiles with subcellular localization information to architect a dynamic network. Building on this, Li et al. [11] also proposed a methodology that fuses temporal process gene expression data with subcellular localization data to construct dynamic protein interaction networks. Nevertheless, discrepancies and inconsistencies sourced from multiple data often result in diminished accuracy and reliability of the constructed network.
Currently, graph representation learning has gained wide application within the analysis of PPINs. Such methods take the entire network as input and learn to transform its topological structure into a vector representation (embeddings), which can be applied to tasks such as label prediction and edge prediction [12,13,14]. Meng et al. [15] employed a hierarchical compression strategy to condense the inputted protein interaction network into a multi-layer PPI network, learning different granularity levels of protein embeddings through network embedding methods and constructing a weighted PPI network using the similarity of protein embeddings. Zhang et al. [16] used network embedding to learn the vector representation of gene ontology (GO) terms, mapped GO term vector sequences to protein vector representations using long short-term memory (LSTM) encoding, and predicted whether two proteins interact using a feed-forward neural network.
Within the protein network, each node contains multiple features, such as protein structural domain information, GO annotation information, and gene expression profiles, which significantly impact the performance and function of PPINs [17]. Therefore, we propose a construction method of dynamic weighted protein network through multi-level embedding (DWPNMLE), which makes full use of the topological structure and biological information to study the construction of protein networks and combines variational graph auto-encoders (VGAEs) with deep attributed network embedding (DANE) for dual embedding to build the final dynamic weighted protein interaction network. This method can fully mine the high-order structural information and node attribute information of PPINs. VGAE and DANE are combined because, in recent years, research has proven that attributed network embedding methods are very effective in generating vector representations of nodes in networks [18]. In the transformed vector space, the topological proximity between nodes and the affinity of node attributes is maintained. The essence of the dynamic construction of protein networks is the prediction of interactions between proteins, which can be seen as a label-propagation problem on complex biological networks. VGAE and DANE have shown excellent performance in the feature representation of nodes on graph-structured data, which can mine the network’s high-order structural information as well as maintain the attribute information of the nodes, and have significant application value in the graph-structured data problems of bioinformatics [19,20]. The experimental results show that the DWPNMLE proposed in this paper has advantages regarding F1-measure, robustness, and algorithm efficiency when applied to four different complex identification algorithms, compared with the other two most advanced dynamic protein network construction methods.
2. Dynamic Weighted Protein Network Construction
Figure 1 shows the critical workflow of DWPNMLE, which consists of three steps: adjacency matrix construction, feature matrix construction, and dynamic weighted network generation through multi-level embedding.
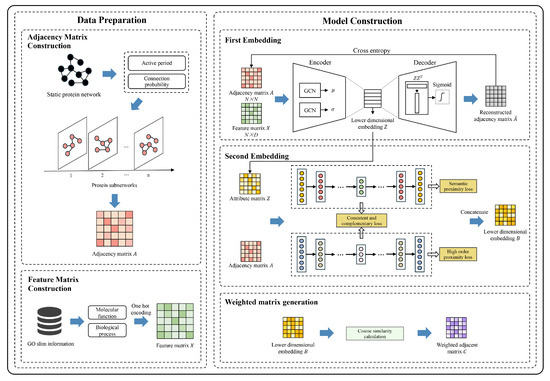
Figure 1.
Process of the DWPNMLE method.
- Adjacency matrix construction. Firstly, the protein activity cycles are computed using a dynamic threshold function. Then, the interactions between different proteins are measured by the connection probability (CP), and if it exceeds the threshold, an edge is added. Finally, the resulting collection of protein subnetworks at different time points is represented by the adjacency matrices .
- Feature matrix construction. After the first step is completed and the adjacency matrices composed of multiple protein subnetworks are obtained, each protein within the subnetworks is annotated using GO Slim information, followed by conducting one-hot encoding. This process constructs multiple feature matrices that represent the GO Slim annotations of the protein subnetworks.
- Dynamic weighted network generation by multi-level embedding. Each adjacency and feature matrix is input into VGAE for the first embedding. After learning the low-dimensional representations of the nodes, a node attribute matrix collection is generated to represent the node attribute information in each protein subnetwork. Then, each attribute and adjacency matrix are input into DNAE for the second embedding, generating multiple low-dimensional embeddings . These embeddings are concatenated in series to construct a weighted PPIN by cosine similarity.
2.1. Construction of the Adjacency Matrix
To facilitate description, the relevant definitions used in the construction process are given as follows.
Definition 1.
Dynamic Protein Network. Define a dynamic protein network, represented as , with representing the temporal snapshot of the protein network at time n. is the protein network at time i, represents the set of proteins expressed at time i, and , represents the set of protein interactions at time i.
Definition 2.
Protein Activity Cycle. For any given protein u, if the average gene expression value of protein u is not lower than the threshold within a given time cycle, then this time cycle is called the activity cycle of protein u.
Definition 3.
Protein Subnetwork. By distinguishing the active time points and activity levels of proteins according to the activity threshold, the activity of all proteins can be calculated to obtain sets of proteins with different time slice sizes. Subnetworks are constructed for the proteins in each set, resulting in multiple protein subnetworks.
2.1.1. Active Period Calculation
Proteins have their active periods in the cell, and gene expression data can reflect the dynamic information of proteins at time points [21,22]. Generally, the expression level of a protein will decrease after the protein has completed its function. Therefore, the protein is active at time points when the related gene expression data are at a high level. Gene expression data always contain unavoidable noise. Even though an active threshold is used for each gene, active proteins with low expression values will likely be filtered out. To address this problem, we calculate the active probability of each protein at different time points based on the 3-sigma [23] criterion. Suppose the gene expression value of protein u at time t is . The mean and standard deviation of gene expression for protein u are and . The activity threshold for protein u is calculated through the following expression:
Based on Equations (1) and (2), the function is defined to reflect the fluctuation of the gene expression curve for protein u,
where . Proteins play different roles and perform multiple functions in biological processes, so their expression value varies across different periods and organizations [24]. Generally, a threshold is used to determine whether a protein is expressed at a time point or condition. A protein is considered to be expressed when its expression value exceeds the given threshold. However, proteins with lower expression values might be filtered out. Therefore, the active probability of a protein is calculated based on the mean and standard deviation of the corresponding gene. The threshold function is defined as follows:
Three standard deviations include about 99% of all observations. Since using a threshold of , where is less than , results in a lower threshold, more gene expression profiles will be retained. The calculation of applies to all possible values of . This paper selects 2.5 as the value of coefficient k [25]. Multiple protein subnetworks with different time points can be divided by calculating the active period of all proteins.
2.1.2. Protein Subnetwork Construction
Let represent the set of n proteins with the same active period. We construct protein subnetworks by analyzing the interactions among proteins within the same active period. The key is to determine whether there is a connection relationship between different proteins within the same time point. To this end, we assess the interactions between different proteins from two perspectives: the number of connection differences and the number of common neighbors, proposing the connection probability (CP). If the CP exceeds a certain threshold, it is assumed that a connection exists between them. Specifically, if there are more common neighbors between two proteins, it indicates a higher interaction probability. The number of connection differences, which is the ratio of the number of directly connected edges between two proteins to the minimum value of the degree of the node, can also indirectly measure the interaction relationship between proteins. The definition of the CP between protein and protein is as follows:
where is the set of neighboring nodes of protein , is the number of edges directly connected between protein and protein , is the degree of protein . The is the sigmoid function. For a given protein, at time point i, if , it is considered that there is an interaction between protein and protein , and an edge is added between them. The threshold is selected based on prior knowledge as 0.9.
A set of protein subnetworks at different time points is obtained at the end of the calculation. The adjacency matrices corresponding to the multiple protein subnetworks within this collection serve as inputs for the subsequent VGAE model. All the adjacency matrices required for multi-level embedding have been constructed. In the following, for the convenience of model description, we use A to replace the adjacency matrix of the protein subnetwork that needs to be input each time, namely . The feature, attribute, and low-dimensional embedding matrices are represented similarly.
2.2. Construction of the Feature Matrix
In bioinformatics, the standardization of terminologies is crucial, and functional annotation based on gene ontology (GO) has become the standard and dominant approach. The main objective of gene ontology design is to provide consistent and standardized annotations of gene and protein functions. They describe the organization of functional information into a directed acyclic graph (DAG) consisting of three primary functional levels: molecular function (MF), biological process (BP), and cellular component (CC). The simplified version, GO Slim, is used instead of the complete GO annotations; in some cases, GO annotations could be improved to avoid overly subdividing functionally similar proteins and better reflect their interactive relations within the protein network. Furthermore, we focus on mainly molecular functions and biological processes, excluding cellular components, due to the interference caused by the annotations related to protein complexes within cellular components during experimental validation. The construction of the protein feature matrix employs one-hot encoding. In brief, each GO Slim annotation is assigned a column, and each protein is assigned a row; if a protein has a particular GO Slim annotation, the corresponding column mark is set to 1; otherwise, it is set to 0. After this processing, multiple feature matrices were generated for each protein subnetwork. Figure 2 illustrates the entire data processing workflow.
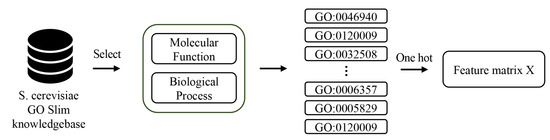
Figure 2.
Construction process of feature matrix.
2.3. Multi-Level Embedding Method Construction
The multi-level embedding method consists of the VGAE model and the DANE model. Firstly, the adjacency matrix A and feature matrix X are input into VGAE for processing to obtain the low-dimensional embedding matrix . Then, Z is input into DANE as an attribute matrix and the adjacency matrix for processing. Finally, multiple low-dimensional embeddings are obtained. The final weighted PPIN is derived by calculating cosine similarity and integrating it with the dynamic PPIN.
2.3.1. Variational Graph Auto-Encoders
VGAE is an unsupervised learning framework based on variational auto-encoders (VAEs) for graph structure data. It utilizes latent variables to learn interpretable latent representations of undirected graphs. The first embedding is performed by inputting the adjacency matrix A and the feature matrix X into an encoder consisting of two layers of graph convolution network (GCN), which are defined as follows:
The first layer generates the low-dimensional representation , and the second layer captures the low-dimensional representation and generates and , which are the mean and variance of the protein node vector, respectively. refers to the symmetrically normalized adjacency matrix, while and refer to the initialization parameter matrix and the update parameter matrix after the GCN layer. and share the first layer weights.
Once the mean and variance are obtained, these parameterized latent variables allow for the definition of a uniquely deterministic multidimensional Gaussian distribution for each protein node. The protein nodes are sampled from this distribution to obtain the embedding representation Z of the protein nodes. This representation captures and quantifies the degree of interactions between proteins and the dynamics of the proteins over time. The matrix the embedding Z is defined as follows:
where represents the embedding vector of node i, and and are the mean and variance of node i. After the mean and variance of all the node vectors are obtained, the node vectors are then formed by resampling; that is, the obeying a standard Gaussian distribution is sampled first, and then z is computed based on the parameters with the following equation:
After obtaining the embedding Z, the distance metric can be calculated using the inner product without altering the vector size. Thus, the reconstructed adjacency matrix is obtained by learning the similarity of each row in Z through the vector inner product [26], as shown in the following equation:
The loss function consists of two parts: the first part is the reconstruction loss, and the other part is the Kullback–Leibler Divergence. Their equations are defined below:
where represents an element from the original data, while represents the corresponding element in the model-reconstructed data. Equation (11) uses the cross-entropy loss function to measure the difference between the model-reconstructed and original adjacency matrix, accurately capturing the dynamic changes and associations between nodes in PPIN. Equation (12) measures the difference between the learned potential spatial distribution and the predefined distribution, which helps better distinguish the dynamic relationships between different nodes.
2.3.2. Deep Attributed Network Embedding
DANE is a deep-learning-based network embedding method that can process node attribute and network structure information at the same time. In contrast, traditional network embedding methods can typically only process one. This allows DANE to reflect the complexity of nodes better and thus achieve superior results in various tasks. Next, a second embedding is performed, where the embedding Z obtained from VGAE is constructed as an attribute matrix, which is used as an input along with the adjacency matrix M. DANE has two branches: the first one consists of a multilayered nonlinear function that captures highly nonlinear network structure by mapping the input M to a low-dimensional space, and the second one maps Z to a low-dimensional space to capture the high nonlinearities in the attributes. Each branch is an autoencoder that captures the highly nonlinear structure. To maintain semantic proximity and higher-order approximation and minimize the reconstruction loss, the loss function is as follows:
where and denote the original attribute matrix input by the encoder and the reconstructed attribute similarity matrix output by the decoder, respectively, and represents all the features associated with node i. Similarly, denotes the original adjacency matrix, and denotes the reconstructed one. If two nodes have similar domain structures, the representations learned by minimizing the reconstruction loss will also be similar. The optimization objective adopted here recovers the protein network with high accuracy and highlights the dynamic weights among proteins in the reconstructed network.
Further, first-order proximity and topology in the attributes can be maintained simultaneously by minimizing the negative log-likelihood. In other words, this loss function helps to ensure that the model captures both the protein node attributes and the interaction patterns in the network structure, thus maintaining these critical features in the embedding vector. The equation is defined as follows:
where denotes that the i protein and the j protein are topologically connected, and the following equation can calculate and :
where and define the predicted probabilities of a link between nodes i and j under the graph structure view M and the attribute information view Z, respectively. is based on the topology representation of node i and the topology representation of node j, while is based on their attribute feature representations and . The embedding of the nodes is reflected in the inner product of the embedding vectors of node j, represented by and . The inner product of two points in an embedding reflects their similarity. When the points are similar, the inner product is close to 1. On the other hand, when the points are dissimilar or unrelated, the inner product is neutral, around 1/2.
It is important to consider consistency and differentiation between nodes to ensure that representations contain both topology and attribute information. Thus, the loss function is defined below:
where denotes the joint probability between the representations of the same node in two different modalities; maximizing makes the representation of the same node in two modalities consistent. denotes the joint probability between the representations of two different nodes in two modalities, and denotes that the i node and j node are not connected topologically. At this point, if left unconstrained, and may be relatively similar, thus leading to the confusion of expressions in different nodes, so they need to be pushed apart, and minimizing can achieve this. In addition, without a direct connection between two nodes, our processing logic underpins distinguishing the functional and spatial locations of different protein nodes, which is one of the prerequisites for constructing an accurate model in dynamic protein network analysis.
The joint optimization objective loss function is obtained by combining the semantic similarity, first-order similarity, higher-order similarity, and consistency.
By minimizing the loss function L, the feature representations learned by all nodes from the network structure and node attributes can be obtained. This approach integrates attributes and structural features while directly mapping the functional relationships and dynamical properties of connected nodes to the embedding of each protein node.
After obtaining the embeddings of multiple protein subnetworks, the protein embeddings can be represented by concatenating the embeddings learned from all protein subnetworks as matrix B. For instance, the final embedding of protein u is represented as
where is the protein subnetwork to which protein u belongs. Next, the weighted PPIN is constructed by calculating the cosine similarity (CS). Given the embedding vectors and for protein u and protein v, the cosine similarity between proteins is defined as
The refined adjustment of the reconstructed weighted adjacency matrix is achieved by calculating the cosine similarity and updating the original PPIN to obtain the final weighted adjacency matrix C. Each element of the matrix C reflects the importance of the existing connections and the potential similarity between nodes.
The resulting weighted matrix C contains not only the structure of the original network but also emphasizes the information embedded in the nodes by the cosine similarity of each pair of connected nodes, which will be next applied to the protein-complex recognition algorithm. The general description of the DWPNMLE is presented in Algorithm 1.
| Algorithm 1 Dynamic Weighted Protein Network Construction (DWPNMLE) |
| Input: PPI data; Gene expression data; Threshold ; GO Slim information Output: Dynamic weighted protein network
|
3. Experiments and Results
Two other dynamic protein network construction methods, ST-APIN [11] and FS-DPIN [27], are selected for comparison to verify the effectiveness of the DWPNMLE method. Taking the protein complex recognition task as an example, a variety of more typical protein complex recognition algorithms, ClusterONE [28], COACH [29], MCL [30], and Core [31], were applied to the two datasets to test the effectiveness of the proposed networks.
3.1. Materials
PPI data are more comprehensive than those of other species and have been widely used in protein-complex recognition experiments. We use two protein interaction datasets, DIP [7] and BioGRID [32]; only physical interactions were retained in both datasets. After removing self-interactions and repeat interactions, the protein and interaction counts were determined and are shown in Table 1. CYC2008 [33], which contains 408 protein complexes, is used as the benchmark protein complex dataset. Gene expression data are obtained from the Gene Expression Omnibus (GEO) GSE3431 [34], and GO annotation information was obtained from the GO Consortium [35].

Table 1.
The Statistics of PPI Datasets.
3.2. Evaluation Metrics
To comprehensively evaluate the superiority of the DWPNMLE method, the following metrics are used to evaluate the performance of different methods:
(1) Overlapping score (OS). The OS evaluates the degree of individual match between the protein complexes recognized by the algorithm and the standard complexes. The recognized protein complexes are represented by set I, while the standard complexes are represented by set S; the OS is defined as follows:
where is a recognized protein complex and ; is a standard protein complex and . The indicates the number of intersections of the recognized protein complex with the standard protein complex . According to most previous research, two protein complexes are matched if the calculation of exceeds the threshold of 0.2, so we also set the threshold to 0.2.
(2) Precision, recall, and F1-measure [36]. Precision represents the ratio of correctly recognized protein complexes to the total number of recognized protein complexes. Recall reflects the coverage of the algorithm in recognizing accurate protein complexes; the higher the recall value, the more real protein complexes the algorithm can recognize. The harmonic mean F1-measure of precision and recall is used to evaluate the overall performance of a protein complex prediction algorithm. These metrics are defined below:
where indicates the number of recognized complexes with at least one match to a known complex, and is the number of known complexes with at least one match to a recognized complex.
3.3. Performance Comparison
In addition to the DWPNMLE method, we have selected two other advanced dynamic protein network construction methods, applied all the construction methods to four classic complex recognition algorithms, and performed comparative experiments on two publicly available datasets, DIP and BioGRID. We also perform robustness analysis and algorithm efficiency analysis.
3.3.1. Comparison with Other Methods
Table 2 and Table 3 show the comparative performance of the different methods regarding precision and recall. The comparison results show that the DWPNMLE in our research outperforms the other two construction methods regarding precision and recall performance metrics on the two datasets where the four complex identification algorithms are applied. On the DIP dataset, the DWPNMLE achieved optimal precision and recall values of 0.7315 and 0.7156 when using the ClusterONE algorithm for complex identification, which are 10% and 11.4% higher than the ST-APIN method and 7.6% and 16.7% higher than the FS-DPIN method. In addition, when combined with the three protein complex identification algorithms, COACH, MCL, and Core, respectively, the Precision values of the DWPNMLE method were 14.3%, 29.5%, and 17.7% higher than those of the ST-APIN method and 8.5%, 14% and 10.1% higher than those of the FS-DPIN method. Meanwhile, the Recall values were improved by 28.3%, 34.8%, and 33.3% over the ST-APIN method and 3.2%, 22.6%, and 12.6% over the FS-DPIN method, respectively. On the BioGRID dataset, when the ClusterONE algorithm was used for complex recognition, DWPNMLE obtained the optimal precision value of 0.6872 and a recall value of 0.6935, which are improved by 11.6% and 23.1%, respectively, over the ST-APIN method and by 4.3% and 15.2% over the FS-DPIN method. In addition, when combined with the three complex recognition algorithms COACH, MCL, and Core, the precision values of the DWPNMLE method are improved by 17.8%, 20.3%, and 23.8% compared to the ST-APIN method and by 14.9%, 6%, and 15.1% compared to the FS-DPIN method. The recall values are improved by 31.2%, 18%, and 41.3% over the ST-APIN method and by 25.3%, 15.8%, and 16.5% over the FS-DPIN method.

Table 2.
Performance comparison of different methods on the DIP dataset.

Table 3.
Performance comparison of different methods on BioGRID dataset.
The results of the comparison of F1-measure values are shown in Figure 3 and Figure 4, which clearly show that the DWPNMLE method has the highest F1-measure value. On the DIP dataset, the DWPNMLE achieves the optimal F1-measure value of 0.7235 when using the ClusterONE algorithm for complex identification, which is 10.7% higher than ST-APIN (0.6535) and 12.2% higher than FS-DPIN (0.6450). In addition, the DWPNMLE also obtains the highest F1-measure values when combines with the three complex recognition algorithms, COACH, MCL, and Core, with 0.6539, 0.5959, and 0.6363, respectively, followed by the FS-DPIN method, with F1-measure values of 0.6184, 0.5026, and 0.5714. For the third-ranked ST-APIN method, the corresponding F1-measure values are 0.5377, 0.4505, and 0.5063. On the BioGRID dataset, the DWPNMLE achieves the optimal F1-measure value of 0.6903 when using the ClusterONE algorithm for complex recognition, which is 16.8% higher than that of the ST-APIN (0.5909) and 9.7% higher than that of the FS-DPIN (0.6294). The DWPNMLE likewise obtained the optimal F1 value. The following best F1 measure values are obtained by the FS-DPIN, with the corresponding F1 values of 0.5230, 0.4605, and 0.5030. The third-best F1-measure values are obtained by the ST-APIN, at 0.5044, 0.4293, and 0.4384, respectively.
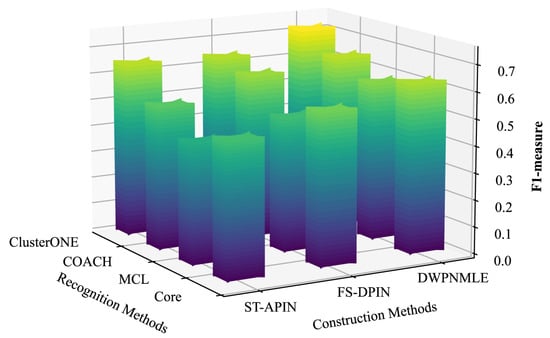
Figure 3.
F1-measure of different methods on the DIP dataset.
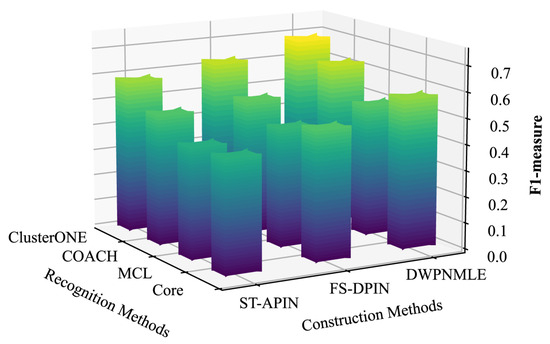
Figure 4.
F1-measure of different methods on the BioGRID dataset.
In summary, our proposed DWPNMLE method outperforms the ST-APIN method and FS-DPIN method in all three evaluation metrics, which fully demonstrates that in effectively fusing biological and topological information to construct dynamically weighted protein networks with multi-level embedding, not only can the rich node representations be deeply mined, but also the higher-order topological patterns of the network can be preserved. Our approach provides an efficient and multidimensional representation learning paradigm for dynamic protein network construction.
3.3.2. Robustness Analysis
PPI data in the real world often include numerous false positives (interactions that are detected but not present) and false negatives (actual interactions but are undetected). These inaccuracies challenge the effectiveness of algorithms meant to recognize protein complexes. To evaluate our network construction method’s reliability, we conducted further analysis by comparing the performance of different complex recognition algorithms when confronted with these incorrect data. Taking the BioGRID dataset as an example, we choose two complex identification algorithms, ClusterONE and Core, and simulate false-positive and false-negative situations in protein networks by randomly adding or removing a certain percentage of edges.
Figure 5 shows that based on the initial protein network, multiple networks with different degrees of false positives are constructed by randomly increasing the proportion of edges, and the performance of the complex recognition algorithms in ClusterONE and MCL on these networks is examined. No matter which algorithm is used for complex recognition, the performance of our method tends to be stable after the proportion of increasing edges reaches 30%. Specifically, the F1-measure values of the combined ClusterONE and MCL algorithms are 0.6038 and 0.4410 when the proportion of added edges reaches 30%, while the corresponding F1-measure values are 0.5901 and 0.4223 when the proportion of added edges reaches 50%. Figure 6 presents the experimental results obtained by randomly deleting the proportion of edges in the network, thus obtaining multiple protein networks with varying degrees of false negatives. The performance of all the recognition algorithms decreases dramatically as the false negatives increase in the network, mainly because the more edges are removed from the network, the more real interactions between the proteins may be disrupted, leading to the loss of complexes. Overall, the F1-measure values of DWPNMLE+ClusterONE and DWPNMLE+MCL are consistently better than those of the other methods, even when the false-negative data increase by up to 50%, fully reflecting the reliability of the protein networks constructed by DWPNMLE.
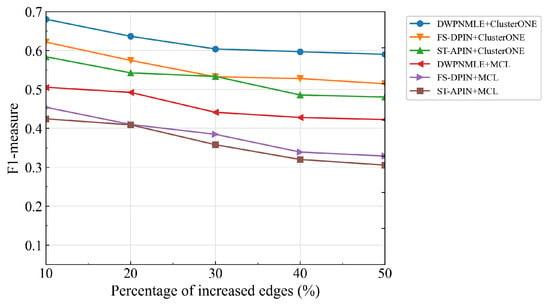
Figure 5.
False positive analysis of different recognition methods.
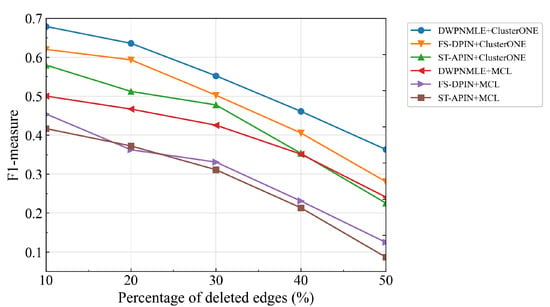
Figure 6.
False negative analysis of different recognition methods.
By simulating these uncertainties, one can assess the robustness and accuracy of the protein network construction method, even when the input data are noisy or incomplete. This provides valuable insights into the practical applicability of the method and its resilience to errors in real experimental data.
3.3.3. Methods Efficiency Analysis
In this section, we separately analyze the efficiency of different methods for complex recognition. Combining the results shown in Figure 7, it can be found that ST-APIN+MCL has the longest running time on both DIP and BioGRID datasets, while FS-DPIN+ClusterONE has the shortest running time. However, DWPNMLE+ClusterONE on both datasets is between the former two, inferior to FS-DPIN+ClusterONE but superior to ST-APIN+MCL.
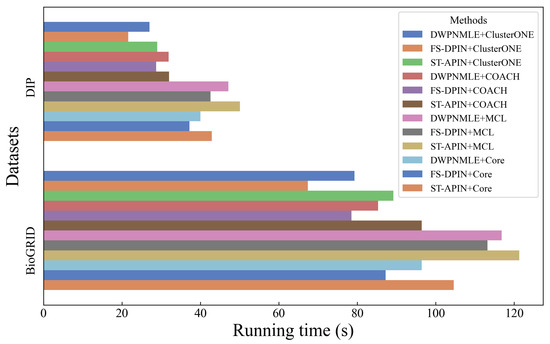
Figure 7.
Efficiency analysis of different methods.
Specifically, on the two datasets, when combining the four complex recognition algorithms of ClusterONE, COACH, MCL, and Core, DWPNMLE has an average efficiency of 9%, 5.9%, 4.9%, and 7.3% over ST-APIN, respectively. On the BioGRID dataset, the advantage of DWPNMLE is even more significant, corresponding to an efficiency improvement over ST-APIN of 11.1%, 11.5%, 3.7%, and 7.8%, respectively. Although DWPNMLE’s overall efficiency is slightly lower than that of FS-DPIN’s, mainly due to the additional computational overhead required to represent multiple protein subnetworks and perform multi-level embeddings, the method sacrifices time efficiency for higher complex identification accuracy.
Overall, DWPNMLE maintains high recognition accuracy while keeping the computational efficiency of the complex recognition task within an acceptable range, showing a good balance between time performance and recognition accuracy.
4. Conclusions
Recognizing biologically significant protein complexes from dynamic PPIN is currently one of the hot research topics in bioinformatics. However, PPI data obtained from biological experiments often contain a large number of false positives and false negatives, which presents significant challenges for protein network construction. In addition, the existing methods cannot fully mine the higher-order information and attribute information of nodes in PPIN, and the shortcomings of the existing methods can be improved by combining graph representation learning methods to fully mine protein network information.
In this paper, we propose a dynamic weighted PPIN construction method using multi-level embedding. The method considers the protein active period, connection probability, and GO information and uses the multi-level embedding model to fully explore the deep information in the network to construct a dynamic weighted protein network. The experimental results indicate that our protein network construction method, DWPNMLE, can effectively improve the accuracy of protein complex recognition while ensuring the method’s efficiency and better robustness compared to the ST-APIN and FS-DPIN. In future work, it will be essential to continue improving the network’s construction using graph representation learning and to develop effective and highly accurate algorithms for protein complex recognition.
Author Contributions
Conception, P.L.; methodology, P.L., S.G. and J.Z.; investigation, P.L., S.G. and C.Z.; data curation, P.L., S.G., C.Z. and M.M.P.; formal analysis, P.L.; writing—original draft preparation, P.L., M.M.P. and S.G.; writing—review and editing, P.L., M.M.P. and J.Z.; visualization, P.L., S.G. and C.Z.; supervision, J.Z.; funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by (1) 2021–2023 National Natural Science Foundation of China under Grant (Youth) No. 52001039; (2) 2022–2025 National Natural Science Foundation of China under Grant No. 52171310.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. DIP data can be found here: https://dip.doe-mbi.ucla.edu/dip/. BioGRID data can be found here: https://thebiogrid.org/. CYC2008 data can be found here: http://wodaklab.org/. Gene expression data can be found here: https://doi.org/10.1126/science.1120499. GO data can be found here: https://geneontology.org/.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Legrain, P.; Wojcik, J.; Gauthier, J.-M. Protein–protein interaction maps: A lead towards cellular functions. Trends Genet. 2001, 17, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The er membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Dooling, L.J.; Tirrell, D.A. Engineering the dynamic properties of protein networks through sequence variation. ACS Cent. Sci. 2016, 2, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.; Bösche, M.; Krause, R.; Grandi, P.; Marzioch, M.; Bauer, A.; Schultz, J.; Rick, J.M.; Michon, A.; Cruciat, C.-M.; et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Xenarios, I.; Salwinski, L.; Duan, X.J.; Higney, P.; Kim, S.-M.; Eisenberg, D. Dip, the database of interacting proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002, 30, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Mrowka, R.; Patzak, A.; Herzel, H. Is there a bias in proteome research? Genome Res. 2001, 11, 1971–1973. [Google Scholar] [CrossRef] [PubMed]
- Cinaglia, P.; Cannataro, M. Network alignment and motif discovery in dynamic networks. Netw. Model. Anal. Health Inform. Bioinform. 2022, 11, 38. [Google Scholar] [CrossRef]
- Li, M.; Ni, P.; Chen, X.; Wang, J.; Wu, F.-X.; Pan, Y. Construction of refined protein interaction network for predicting essential proteins. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 16, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Meng, X.; Zheng, R.; Wu, F.-X.; Li, Y.; Pan, Y.; Wang, J. Identification of protein complexes by using a spatial and temporal active protein interaction network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 17, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.; Zitnik, M.; Wang, B.; Leskovec, J.; Goldenberg, A.; Sharan, R. To embed or not: Network embedding as a paradigm in computational biology. Front. Genet. 2019, 10, 452819. [Google Scholar] [CrossRef] [PubMed]
- Badkas, A.; Landtsheer, S.D.; Sauter, T. Construction and contextualization approaches for protein–protein interaction networks. Comput. Struct. Biotechnol. J. 2022, 20, 3280–3290. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Parvej, M.M.; Zhang, C.; Guo, S.; Zhang, J. Advances in the development of representation learning and its innovations against COVID-19. COVID 2023, 3, 1389–1415. [Google Scholar] [CrossRef]
- Meng, X.; Xiang, J.; Zheng, R.; Wu, F.; Li, M. Dpcmne: Detecting protein complexes from protein–protein interaction networks via multi-level network embedding. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, M.; Qian, Y. protein2vec: Predicting protein–protein interactions based on lstm. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 19, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Zahiri, J.; Emamjomeh, A.; Bagheri, S.; Ivazeh, A.; Mahdevar, G.; Tehrani, H.S.; Mirzaie, M.; Fakheri, B.A.; Mohammad-Noori, M. Protein complex prediction: A survey. Genomics 2020, 112, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Li, K.; Zheng, W.; Liu, X.; Zhang, Y.; Zhao, Z.; He, Z. Protein complexes identification based on go attributed network embedding. BMC Bioinform. 2018, 19, 535. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, J.; Pan, X.; Yan, H.; You, Z.-H. Hiscf: Leveraging higher-order structures for clustering analysis in biological networks. Bioinformatics 2021, 37, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.-W.; Hu, L.; You, Z.-H.; Wang, L.; Su, X.-R. Hingrl: Predicting drug–disease associations with graph representation learning on heterogeneous information networks. Brief. Bioinform. 2022, 23, bbab515. [Google Scholar] [CrossRef] [PubMed]
- Rinner, O.; Mueller, L.N.; Hubálek, M.; Müller, M.; Gstaiger, M.; Aebersold, R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat. Biotechnol. 2007, 25, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Geva-Zatorsky, N.; Eden, E.; Frenkel-Morgenstern, M.; Issaeva, I.; Sigal, A.; Milo, R.; Cohen-Saidon, C.; Liron, Y.; Kam, Z.; et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008, 322, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wang, J.; Liu, B.; Li, M.; Chen, G.; Pan, Y. A comparison of the functional modules identified from time course and static ppi network data. BMC Bioinform. 2011, 12, 339. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Peng, X.; Li, M.; Pan, Y. Construction and application of dynamic protein interaction network based on time course gene expression data. Proteomics 2013, 13, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Wang, J.; Peng, X.; Wu, F.-X. Detecting protein complexes from active protein interaction networks constructed with dynamic gene expression profiles. Proteome Sci. 2013, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, J.; Shuai, S.C.; Zhao, Q.; Shuai, J. Predicting potential interactions between lncrnas and proteins via combined graph auto-encoder methods. Brief. Bioinform. 2023, 24, bbac527. [Google Scholar] [CrossRef]
- Sun, J.; Pan, L.; Li, B.; Wang, H.; Yang, B.; Li, W. A construction method of dynamic protein interaction networks by using relevant features of gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 2790–2801. [Google Scholar] [CrossRef]
- Nepusz, T.; Yu, H.; Paccanaro, A. Detecting overlapping protein complexes in protein–protein interaction networks. Nat. Methods 2012, 9, 471–472. [Google Scholar] [CrossRef]
- Wu, M.; Li, X.; Kwoh, C.-K.; Ng, S.-K. A core-attachment based method to detect protein complexes in ppi networks. BMC Bioinform. 2009, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Vlasblom, J.; Wodak, S.J. Markov clustering versus affinity propagation for the partitioning of protein interaction graphs. BMC Bioinform. 2009, 10, 99. [Google Scholar] [CrossRef]
- Leung, H.C.; Xiang, Q.; Yiu, S.M.; Chin, F.Y. Predicting protein complexes from ppi data: A core-attachment approach. J. Comput. Biol. 2009, 16, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The biogrid interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.; Wong, J.; Turner, B.; Cho, E.; Wodak, S.J. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009, 37, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.P.; Kudlicki, A.; Rowicka, M.; McKnight, S.L. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science 2005, 310, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. Gene ontology annotations and resources. Nucleic Acids Res. 2012, 41, D530–D535. [Google Scholar] [CrossRef] [PubMed]
- Zaki, N.; Singh, H.; Mohamed, E.A. Identifying protein complexes in protein–protein interaction data using graph convolutional network. IEEE Access 2021, 9, 123717–123726. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).