Ontology Attention Layer for Medical Named Entity Recognition
Abstract
1. Introduction
2. Related Works
2.1. Medical Named Entity Recognition
2.2. Attention Mechanism
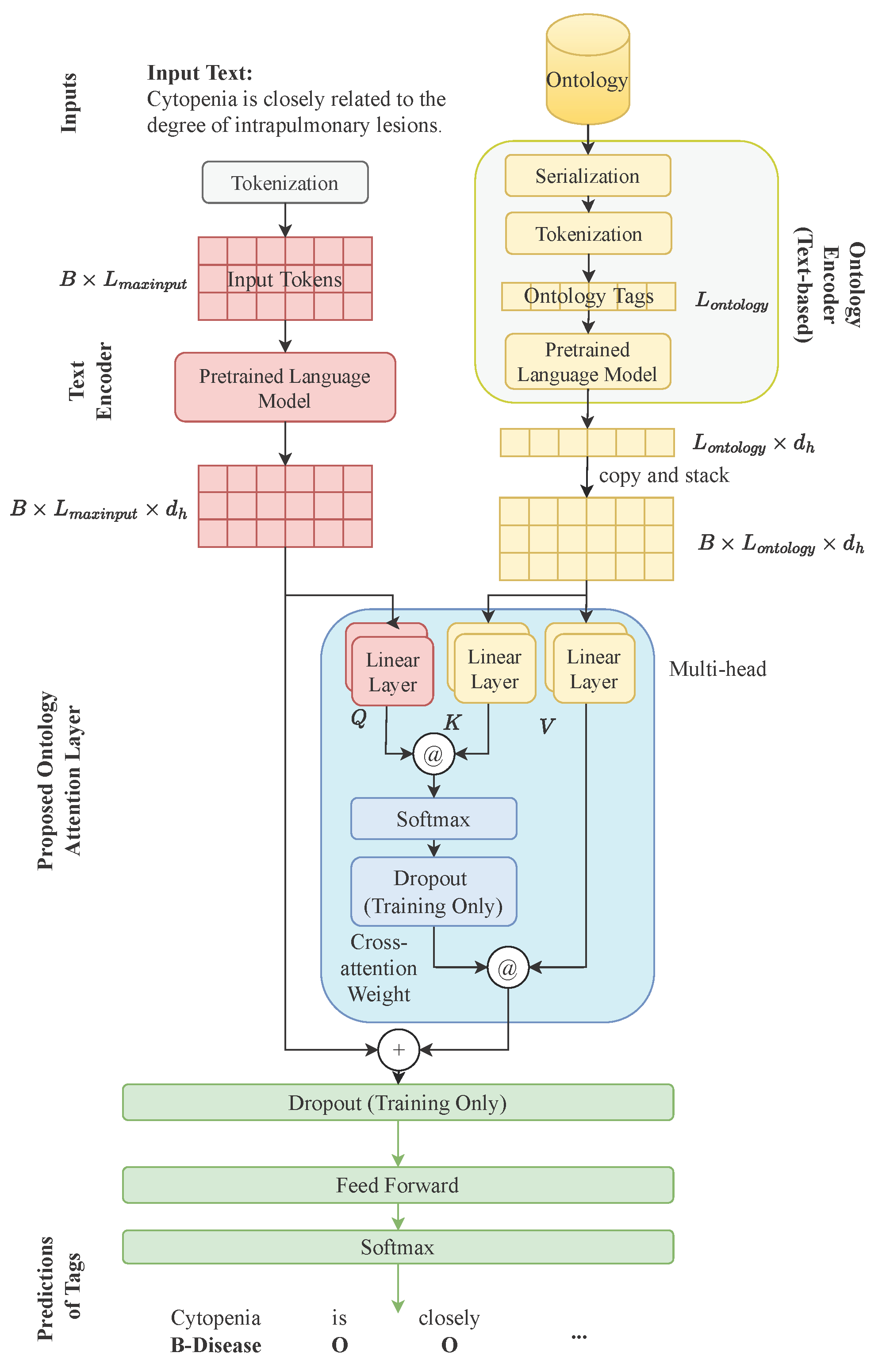
3. Proposed Methods
3.1. Overall Architecture
3.2. Text Encoder
3.3. Ontology Encoder
3.3.1. Serialization and Tokenization
- The relative position of two tokens is able to show the relationship between the corresponding class tags or properties.
- The positions are fixed in every training epoch.
3.3.2. Encoding the Serialized Ontology
3.4. Ontology Attention
3.5. Feed-Forward Layer and Output
4. Experiments
4.1. Dataset
4.2. Labeling Policy
4.3. Construction of the Ontology
4.4. Hyperparameters
5. Experimental Results and Discussion
5.1. Performance over Different Pretrained Language Models as the Encoders
5.2. Discussion on the Granularity of the Ontology
5.3. Ablation Study on Effects of Different Property Types
5.4. Discussion on the Number of Ontology Classes
5.5. Discussion on the Usage of the Ontology Class Name
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Study on Post-Order Traversal and the Combination of Pre-Order and Post-Order Traversal
| Dataset | Pre-Order DFS | Post-Order DFS | Bi-Order DFS |
|---|---|---|---|
| NCBI disease | 66.27% | 65.80% | 64.90% |
| Traversal Method | Serialization Result |
|---|---|
| Pre-order DFS | anatomical location anatomical property anatomy part name disease cause clinical manifestations diagnostic criteria disease name drug contraindication dosage drug component drug name indications hospital department department name departmental functions range of action service object medical equipment equipment name operational requirements symptoms suitable for use microbe biological characteristics microbial name microbiology functions pathogenicity sympton symptom property symptom signs symptom timing |
| Post-order DFS | disease name cause clinical manifestations diagnostic criteria disease symptom signs symptom property symptom timing sympton equipment name symptoms suitable for use operational requirements medical equipment part name anatomy anatomical property anatomical location department name departmental functions service object range of action hospital department microbial name microbiology functions biological characteristics pathogenicity microbe drug name drug component indications contraindication dosage drug |
References
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. In Proceedings of the North American Chapter of the Association for Computational Linguistics, San Diego, CA, USA, 12–17 June 2016. [Google Scholar] [CrossRef]
- Yang, T.; He, Y.; Yang, N. Named Entity Recognition of Medical Text Based on the Deep Neural Network. J. Healthc. Eng. 2022, 2022, 3990563. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hou, L. Named Entity Recognition in Chinese Electronic Medical Records Based on the Model of Bidirectional Long Short-Term Memory with a Conditional Random Field Layer. Stud. Health Technol. Inform. 2019, 264, 1524–1525. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, X.; Li, X.; Ouyang, R.; Wu, F.; Zhang, T.; Li, J.; Wang, G. GPT-NER: Named Entity Recognition via Large Language Models. arXiv 2023, arXiv:2304.10428. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, D.; Cao, T.; Yin, B.; Zhao, T. Named Entity Recognition with Small Strongly Labeled and Large Weakly Labeled Data. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Li, J.; Ding, H.; Shang, J.; McAuley, J.; Feng, Z. Weakly Supervised Named Entity Tagging with Learnable Logical Rules. arXiv 2021, arXiv:2107.02282. [Google Scholar] [CrossRef]
- Li, Y.; Shetty, P.; Liu, L.; Zhang, C.; Song, L. BERTifying the Hidden Markov Model for Multi-Source Weakly Supervised Named Entity Recognition. arXiv 2021, arXiv:2105.12848. [Google Scholar] [CrossRef]
- Aly, R.; Vlachos, A.; McDonald, R. Leveraging Type Descriptions for Zero-shot Named Entity Recognition and Classification. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar]
- Wu, S.; Song, X.; Feng, Z. MECT: Multi-Metadata Embedding based Cross-Transformer for Chinese Named Entity Recognition. arXiv 2021, arXiv:2107.05418. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, Y.; Bach, N.; Wang, T.; Huang, Z.; Huang, F.; Tu, K. Improving Named Entity Recognition by External Context Retrieving and Cooperative Learning. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Wei, X.; Wang, S.; Zhang, D.; Bhatia, P.; Arnold, A. Knowledge Enhanced Pretrained Language Models: A Compreshensive Survey. arXiv 2021, arXiv:2110.08455. [Google Scholar] [CrossRef]
- Zhou, B.; Cai, X.; Zhang, Y.; Yuan, X. An End-to-End Progressive Multi-Task Learning Framework for Medical Named Entity Recognition and Normalization. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Ji, Z.; Xia, T.; Han, M.; Xiao, J. A Neural Transition-based Joint Model for Disease Named Entity Recognition and Normalization. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Shindo, H.; Matsumoto, Y.; Watanabe, T. Nested Named Entity Recognition via Explicitly Excluding the Influence of the Best Path. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Li, F.; Wang, Z.; Hui, S.C.; Liao, L.; Song, D.; Xu, J.; He, G.; Jia, M. Modularized Interaction Network for Named Entity Recognition. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Zhu, H.; Liu, T.; Yu, N.; Sun, L. Discontinuous Named Entity Recognition as Maximal Clique Discovery. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Bangkok, Thailand, 22–27 May 2021. [Google Scholar] [CrossRef]
- Li, F.; Lin, Z.; Zhang, M.; Ji, D. A Span-Based Model for Joint Overlapped and Discontinuous Named Entity Recognition. arXiv 2021, arXiv:2106.14373. [Google Scholar] [CrossRef]
- Luong, T.; Pham, H.; Manning, C.D. Effective Approaches to Attention-based Neural Machine Translation. arXiv 2015, arXiv:1508.04025. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, D.; Dyer, C.; He, X.; Smola, A.; Hovy, E.H. Hierarchical Attention Networks for Document Classification. In Proceedings of the North American Chapter of the Association for Computational Linguistics, San Diego, CA, USA, 12–17 June 2016. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, Z.L.; Brandt, J.; Shen, X.; Sclaroff, S. Top-Down Neural Attention by Excitation Backprop. Int. J. Comput. Vis. 2016, 126, 1084–1102. [Google Scholar] [CrossRef]
- Gehring, J.; Auli, M.; Grangier, D.; Yarats, D.; Dauphin, Y. Convolutional Sequence to Sequence Learning. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.M.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Proceedings of the NIPS, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar] [CrossRef]
- Eldele, E.; Chen, Z.; Liu, C.; Wu, M.; Kwoh, C.; Li, X.; Guan, C. An Attention-Based Deep Learning Approach for Sleep Stage Classification With Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, H.; Wang, H.; Rao, Y.; Liu, L.; Huan, J. DELTA: DEep Learning Transfer using Feature Map with Attention for Convolutional Networks. arXiv 2019, arXiv:1901.09229. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019. [Google Scholar] [CrossRef]
- Cui, Y.; Che, W.; Liu, T.; Qin, B.; Wang, S.; Hu, G. Revisiting Pre-Trained Models for Chinese Natural Language Processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: Findings, Online, 16–20 November 2020; pp. 657–668. [Google Scholar]
- Cui, Y.; Che, W.; Liu, T.; Qin, B.; Yang, Z.; Wang, S.; Hu, G. Pre-Training With Whole Word Masking for Chinese BERT. IEEE/ACM Trans. Audio Speech Lang. Process. 2019, 29, 3504–3514. [Google Scholar] [CrossRef]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Glorot, X.; Bordes, A.; Weston, J.; Bengio, Y. A semantic matching energy function for learning with multi-relational data. Mach. Learn. 2013, 94, 233–259. [Google Scholar]
- Dettmers, T.; Minervini, P.; Stenetorp, P.; Riedel, S. Convolutional 2D Knowledge Graph Embeddings. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence and Thirtieth Innovative Applications of Artificial Intelligence Conference and Eighth AAAI Symposium on Educational Advances in Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Vashishth, S.; Sanyal, S.; Nitin, V.; Talukdar, P. Composition-based Multi-Relational Graph Convolutional Networks. In Proceedings of the International Conference on Learning Representations, Online, 26–30 April 2020. [Google Scholar]
- Zhang, N.; Chen, M.; Bi, Z.; Liang, X.; Li, L.; Shang, X.; Yin, K.; Tan, C.; Xu, J.; Huang, F.; et al. CBLUE: A Chinese Biomedical Language Understanding Evaluation Benchmark. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics, Dublin, Ireland, 22–27 May 2022. [Google Scholar] [CrossRef]
- Doğan, R.I.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]





| Ontology Class | Entity-Type Property | Attribute-Type Property | Relationship-Type Property |
|---|---|---|---|
| 疾病 disease | 疾病名称 disease name 疾病病因 cause | 临床表现 clinical manifestations 诊断标准 diagnostic criteria | 诊断 diagnosis 治疗 treat 预防 prophylaxis |
| 临床表现 symptom | 症状 symptom signs | 表现的性质 symptom property 表现的时序 symptom timing | 表现与疾病的关系诊断和治疗 diagnosis and treatment of the relationship between manifestations and disease |
| 医疗设备 medical equipment | 设备名称 equipment name 设备功能 equipment function | 适合使用的症状 symptoms suitable for use 操作要求 operational requirements | 设备使用 device use 设备操作 device operation |
| 身体 anatomical location | 部位名称 part name 解刨结构 anatomy | 部位性质 anatomical property | 部位与疾病的临床表现 location and clinical manifestations of the disease |
| 医院部门 hospital department | 部门名称 department name 部门职能 departmental functions | 服务对象 service object 作用范围 range of action | 部门与疾病治疗的关系 the relationship between the department and the treatment of diseases |
| 微生物 microbe | 微生物名称 microbial name 微生物工作职能 microbiology functions | 生物学特征 biological characteristics 致病性 pathogenicity | 微生物与药物之间的关系 the relationship between microorganisms and drugs |
| 药物 drug | 药物名称 drug name 药物成分 drug component | 适应症 indications 禁忌症 contraindication 剂量 dosage | 药物治疗疗程与疾病的关系 the relationship between the course of drug therapy and the disease |
| Hyperparameter | Value |
|---|---|
| Training Batch Size | 16 |
| Evaluation Batch Size | 64 |
| Learning Rate | 5 × 10−5 |
| Epochs | 10 |
| ID | Text | Plain Base Model | w/ Proposed Method | w/ Self-Attention |
|---|---|---|---|---|
| 11 | As constitutional DNA was not available, a putative hereditary predisposition to T-PLL will require further investigation. | (None) | T- | T- |
| 18 | Neither the content nor the activity of Na+/K+ ATPase and sarcoplasmic reticulum Ca2+- ATPase are affected by DMPK absence. | (None) | (None) | DMPK absence |
| 81 | Saamis (2%) and Mordvinians (1.8%) had significantly lower frequencies of the Tyr allele. | Saamis | (None) | (None) |
| 94 | Numerous cytogenetic and allelotype studies have reported frequent loss of heterozygosity on chromosomal arm 10q in sporadic prostate cancer. | prostate cancer | sporadic prostate cancer | prostate cancer |
| 138 | We conclude that paternal transmission of congenital DM is rare and preferentially occurs with onset of DM past 30 years in the father. | DM, DM | congenital DM, DM | DM, DM |
| 155 | Mutations in the SMAD4/DPC4 tumor suppressor gene, a key signal transducer in most TGFbeta-related pathways, are involved in 50 % of pancreatic cancers. | pancreatic cancers | tumor, pancreatic cancers | tumor, pancreatic cancers |
| Group | Contained Property Types |
|---|---|
| Coarse-grained | class name only without properties |
| Medium-grained | entity-type properties |
| Fine-grained | entity-type properties, attribute-type properties |
| Ontology Class | Entity-Type Property | Attribute-Type Property | Relationship-Type Property |
|---|---|---|---|
| 医疗程序 medical process | 医务人员 personnel 检验样本 sample | 医疗程序的执行 execution of medical process 医学检验项目的目的 the purpose of the test | 医疗程序的操作步骤 the process of performing medical procedure 医疗程序的选择 choice of medical procedure |
| 医疗检验项目 test items | 医疗设备 equipment 检测设备 | 操作技术 operational technique 医学检验项目的指标 indicator | 检验流程 inspection process 质量控制 quality control |
| Settings | Coarse-Grained | Medium-Grained | Fine-Grained |
|---|---|---|---|
| w/ BERT & original ontology (7 classes) | 56.27% | 56.41% | 56.21% |
| w/ BERT & extended ontology (9 classes) | 56.35% | 56.20% | 56.21% |
| w/ BERT-WWM & original ontology (7 classes) | 58.73% | 58.91% | 58.60% |
| w/ BERT-WWM & extended ontology (9 classes) | 58.78% | 58.71% | 58.75% |
| w/ RoBERTa & original ontology (7 classes) | 59.27% | 59.23% | 59.00% |
| w/ RoBERTa & extended ontology (9 classes) | 59.18% | 59.15% | 59.15% |
| Settings | Medium-Grained | Fine-Grained |
|---|---|---|
| w/ BERT and class name | 56.41% | 56.21% |
| w/ BERT, w/o class name | 56.14% | 56.57% |
| w/ BERT-WWM and class name | 58.91% | 58.60% |
| w/ BERT-WWM, w/o class name | 58.53% | 58.92% |
| w/ RoBERTa and class name | 59.23% | 59.00% |
| w/ RoBERTa, w/o class name | 59.38% | 59.19% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zha, Y.; Ke, Y.; Hu, X.; Xiong, C. Ontology Attention Layer for Medical Named Entity Recognition. Appl. Sci. 2024, 14, 421. https://doi.org/10.3390/app14010421
Zha Y, Ke Y, Hu X, Xiong C. Ontology Attention Layer for Medical Named Entity Recognition. Applied Sciences. 2024; 14(1):421. https://doi.org/10.3390/app14010421
Chicago/Turabian StyleZha, Yue, Yuanzhi Ke, Xiao Hu, and Caiquan Xiong. 2024. "Ontology Attention Layer for Medical Named Entity Recognition" Applied Sciences 14, no. 1: 421. https://doi.org/10.3390/app14010421
APA StyleZha, Y., Ke, Y., Hu, X., & Xiong, C. (2024). Ontology Attention Layer for Medical Named Entity Recognition. Applied Sciences, 14(1), 421. https://doi.org/10.3390/app14010421






