Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data
Abstract
1. Introduction
2. Materials and Methods
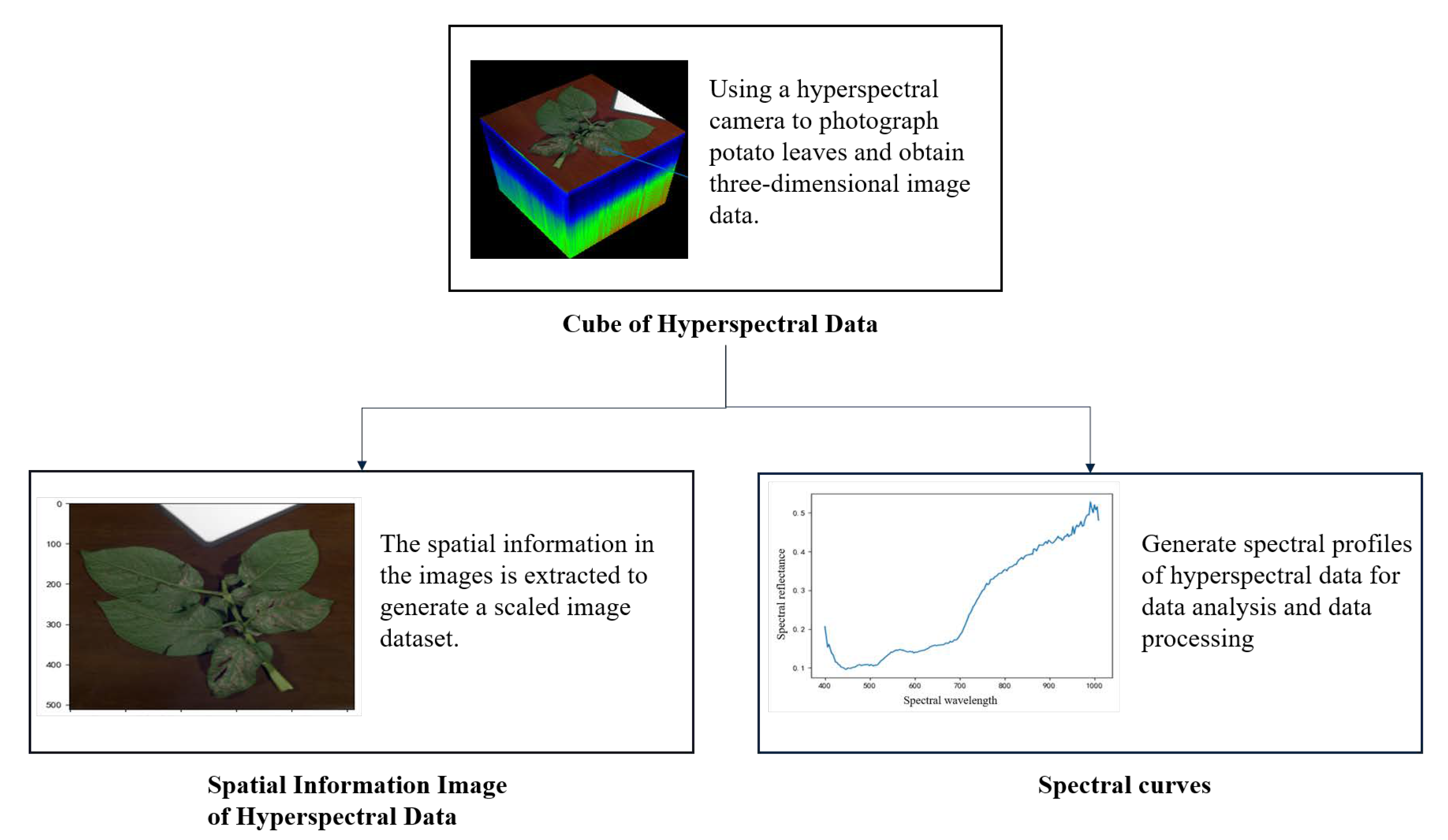
2.1. Data Acquisition and Preprocessing
2.2. Methods
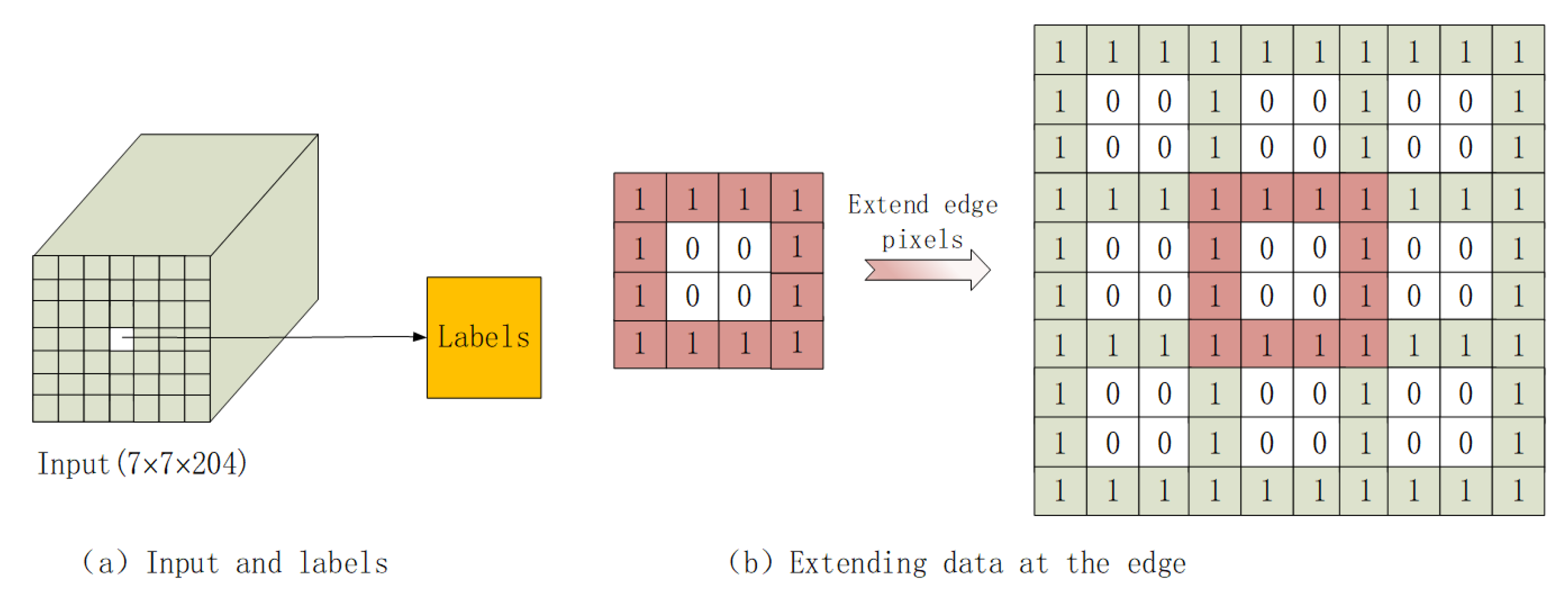
2.2.1. Method of Label Category Selection
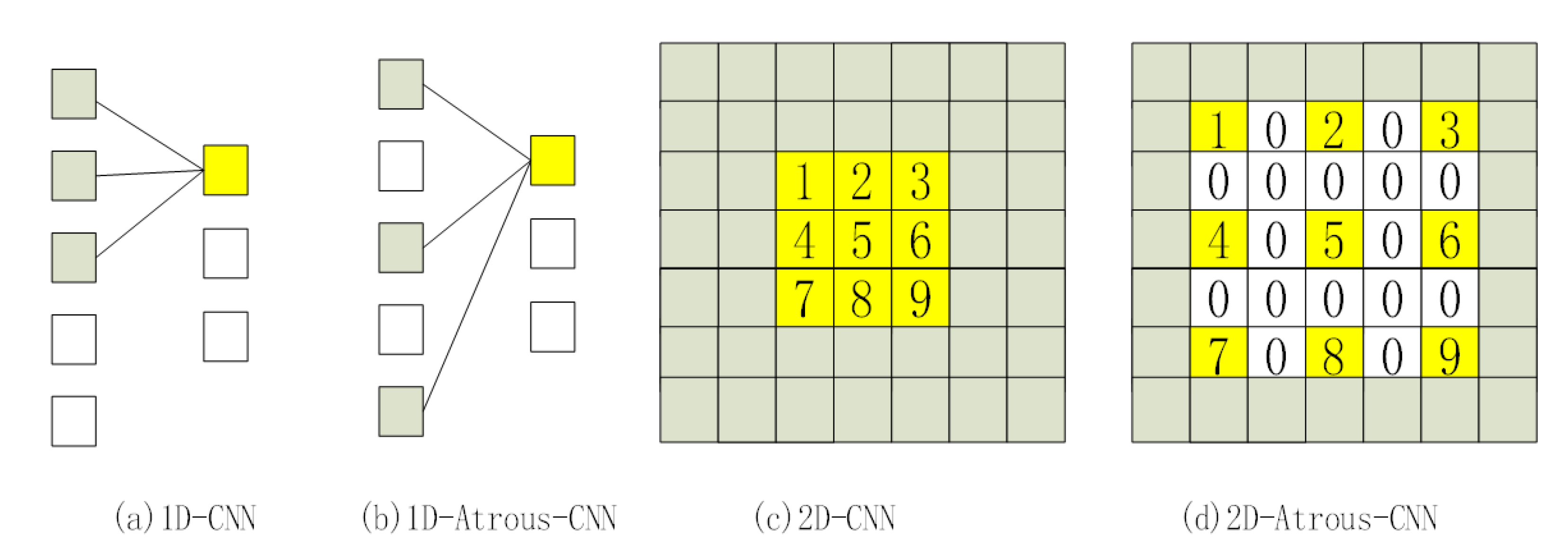
2.2.2. Atrous-CNN
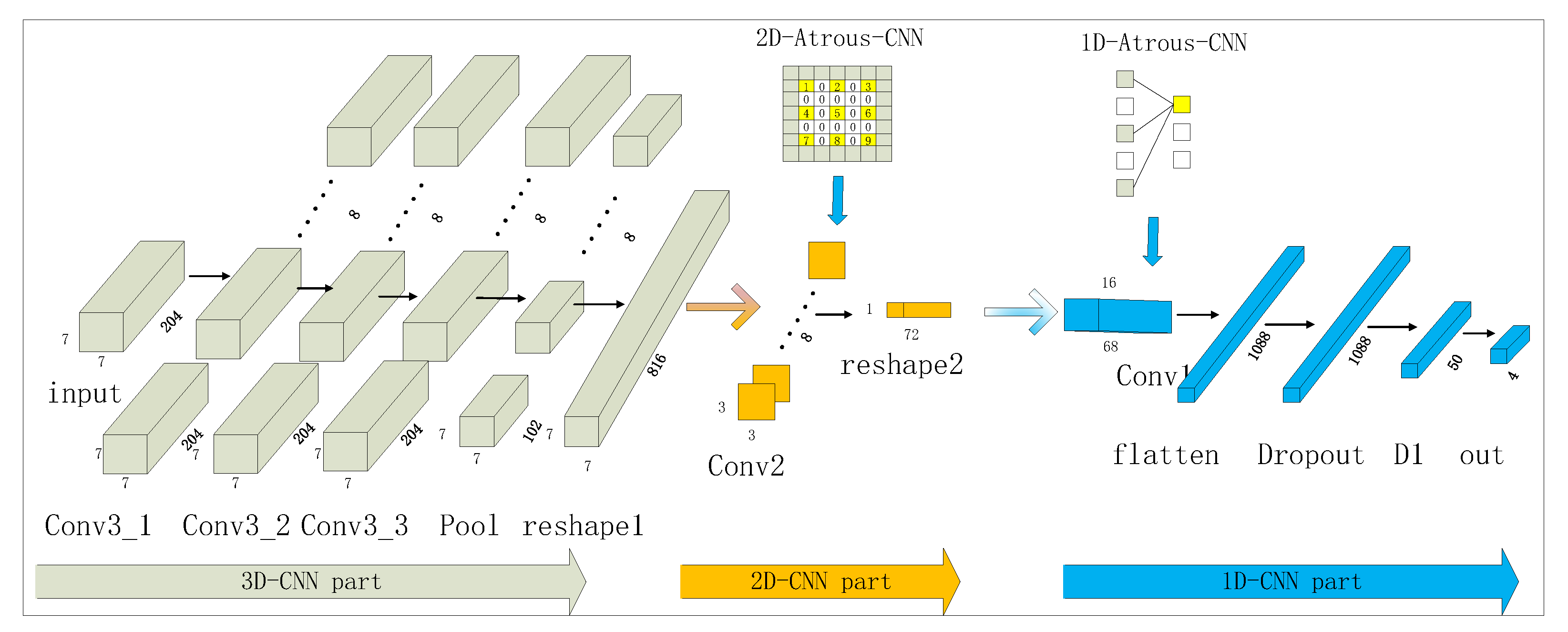
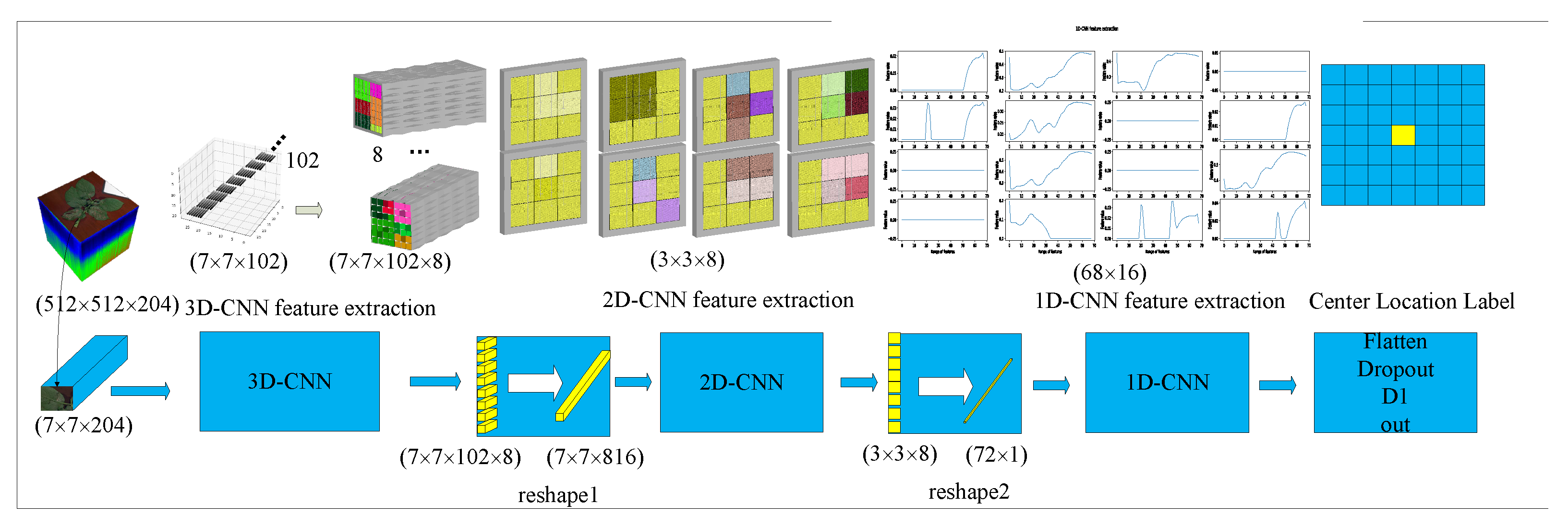
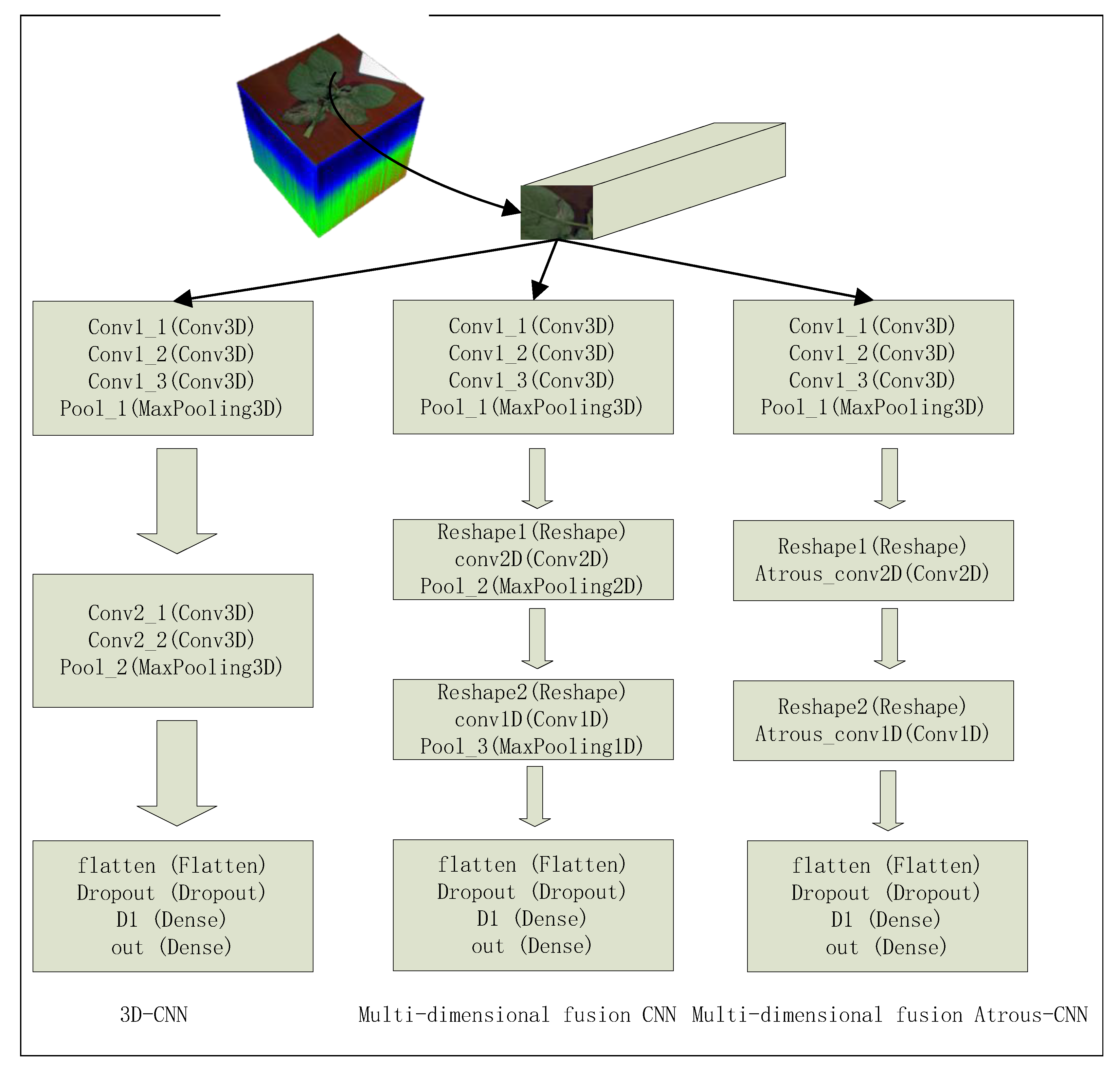
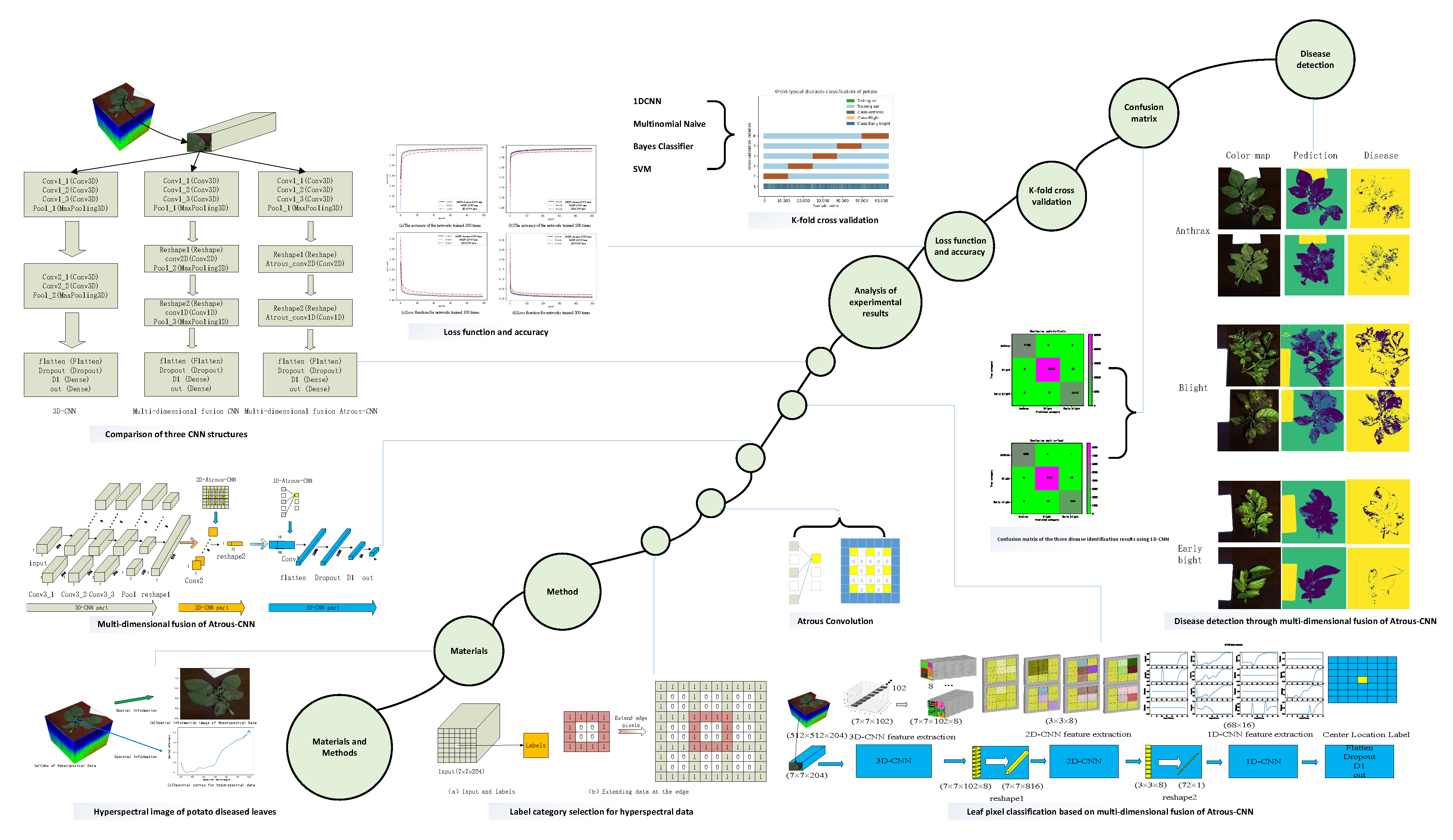
2.2.3. Multidimensional Fusion Atrous-CNN
2.2.4. Leaf Pixel Classification Based on Multidimensional Fusion Atrous-CNN
2.2.5. Disease Classification Method: 1D-CNN
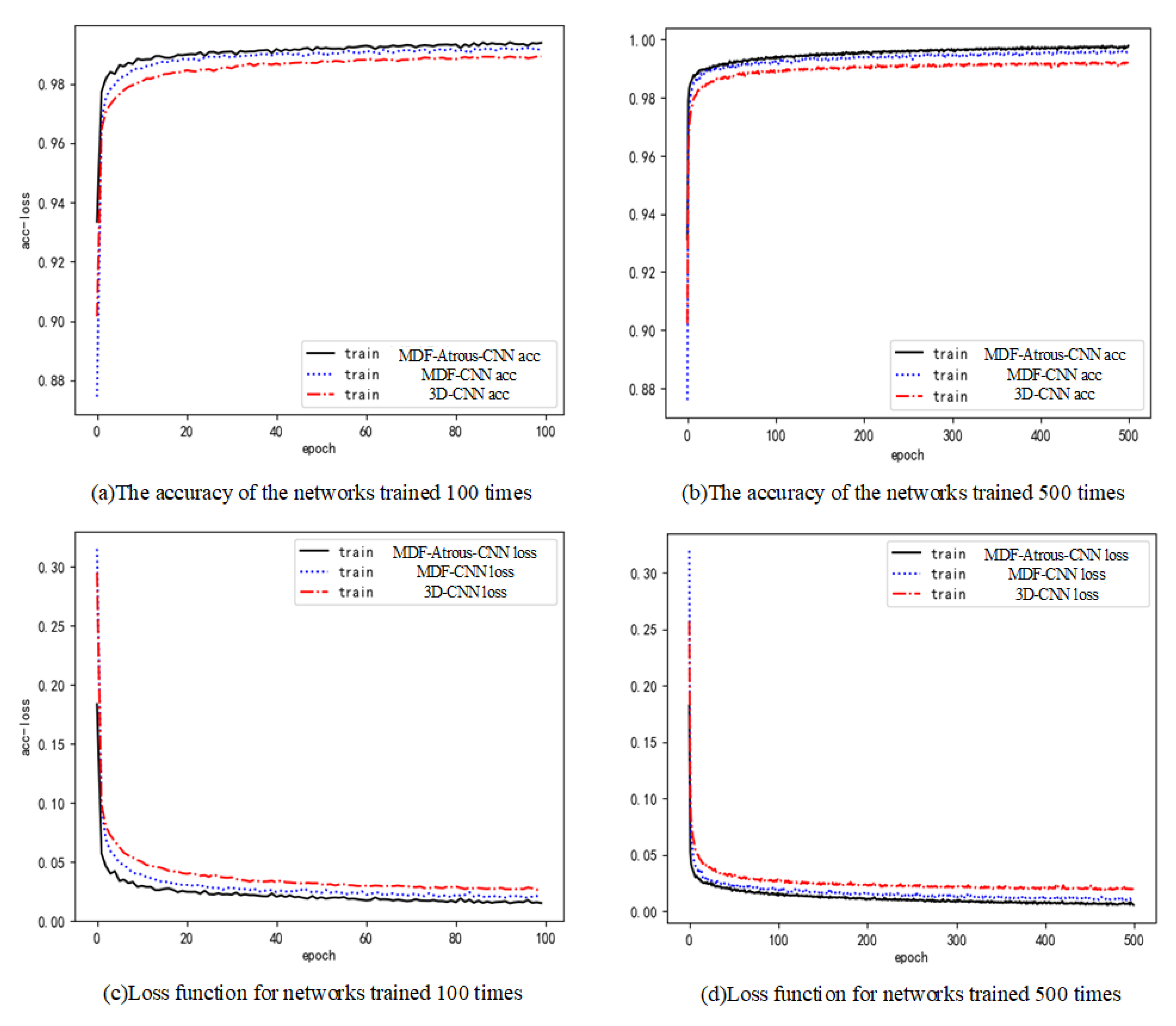
3. Analysis of Experimental Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Fen, X.; Yu, W.; Hu, H.H.; Dai, X.F. Progress of potato staple food research and industry development in China. J. Integr. Agric. 2017, 16, 2924–2932. [Google Scholar] [CrossRef]
- Bruckner, M.; Wood, R.; Moran, D.; Kuschnig, N.; Wieland, H.; Maus, V.; Börner, J. FABIO—The construction of the food and agriculture biomass input–output model. Environ. Sci. Technol. 2019, 53, 11302–11312. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.; Sharma, K.; Parker, M.L.; Secor, G.A.; Elphinstone, J. Bacterial diseases of potato. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 351–388. [Google Scholar]
- Waleron, M.; Misztak, A.; Jońca, J.; Waleron, K. First report of Pectobacterium polaris causing soft rot of potato in Poland. Plant Dis. 2019, 103, 144. [Google Scholar] [CrossRef]
- Bergsma-Vlami, M.; Saddler, G.; Hélias, V.; Tsror, L.; Yedida, I.; Pirhonen, M.; Degefu, Y.; Tuomisto, J.; Lojkowska, E.; Li, S.; et al. Assessment of Dickeya and Pectobacterium spp. on Vegetables and Ornamentals (Soft Rot); Zenodo: Honolulu, HI, USA, 2020. [Google Scholar]
- Hadizadeh, I.; Peivastegan, B.; Hannukkala, A.; Van der Wolf, J.; Nissinen, R.; Pirhonen, M. Biological control of potato soft rot caused by Dickeya solani and the survival of bacterial antagonists under cold storage conditions. Plant Pathol. 2019, 68, 297–311. [Google Scholar] [CrossRef]
- Stark, J.C.; Thornton, M.; Nolte, P. Potato Production Systems; Springer Nature: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Shukla, A.; Ratan, V. Management of Early Blight of Potato by Using Different Bioagents as Tuber Dressing and its Effect on Germination and Growth. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1965–1970. [Google Scholar] [CrossRef]
- Landschoot, S.; Vandecasteele, M.; De Baets, B.; Höfte, M.; Audenaert, K.; Haesaert, G. Identification of A. arborescens, A. grandis, and A. protenta as new members of the European Alternaria population on potato. Fungal Biol. 2017, 121, 172–188. [Google Scholar] [CrossRef]
- Abuley, I.K.; Hansen, J.G. An epidemiological analysis of the dilemma of plant age and late blight (Phytophthora infestans) susceptibility in potatoes. Eur. J. Plant Pathol. 2021, 161, 645–663. [Google Scholar] [CrossRef]
- Degefu, Y. Co-occurrence of latent Dickeya and Pectobacterium species in potato seed tuber samples from northern Finland: Co-colonization of latent Dickeya and Pectobacterium species in potato seed lots. Agric. Food Sci. 2021, 30, 1–7. [Google Scholar] [CrossRef]
- Meno, L.; Escuredo, O.; Rodríguez-Flores, M.S.; Seijo, M.C. Looking for a sustainable potato crop. Field assessment of early blight management. Agric. For. Meteorol. 2021, 308, 108617. [Google Scholar] [CrossRef]
- Peters, R.; Sturz, A.; Carter, M.; Sanderson, J. Influence of crop rotation and conservation tillage practices on the severity of soil-borne potato diseases in temperate humid agriculture. Can. J. Soil Sci. 2004, 84, 397–402. [Google Scholar] [CrossRef]
- Adolf, B.; Andrade-Piedra, J.; Bittara Molina, F.; Przetakiewicz, J.; Hausladen, H.; Kromann, P.; Lees, A.; Lindqvist-Kreuze, H.; Perez, W.; Secor, G.A. Fungal, oomycete, and plasmodiophorid diseases of potato. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 307–350. [Google Scholar]
- Kolychikhina, M.; Beloshapkina, O.; Phiri, C. Change in potato productivity under the impact of viral diseases. IOP Conf. Ser. Earth Environ. Sci. 2021, 663, 012035. [Google Scholar] [CrossRef]
- Garhwal, A.S.; Pullanagari, R.R.; Li, M.; Reis, M.M.; Archer, R. Hyperspectral imaging for identification of Zebra Chip disease in potatoes. Biosyst. Eng. 2020, 197, 306–317. [Google Scholar] [CrossRef]
- Iftikhar, S.; Shahid, A.A.; Halim, S.A.; Wolters, P.J.; Vleeshouwers, V.G.; Khan, A.; Al-Harrasi, A.; Ahmad, S. Discovering novel Alternaria solani succinate dehydrogenase inhibitors by in silico modeling and virtual screening strategies to combat early blight. Front. Chem. 2017, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yin, X. Image recognition of typical potato diseases and insect pests using deep learning. Fresenius Environ. Bull. 2021, 30, 9956–9965. [Google Scholar]
- Gold, K.M.; Townsend, P.A.; Herrmann, I.; Gevens, A.J. Investigating potato late blight physiological differences across potato cultivars with spectroscopy and machine learning. Plant Sci. 2020, 295, 110316. [Google Scholar] [CrossRef]
- Zheng, C.; Abd-Elrahman, A.; Whitaker, V. Remote sensing and machine learning in crop phenotyping and management, with an emphasis on applications in strawberry farming. Remote Sens. 2021, 13, 531. [Google Scholar] [CrossRef]
- Singh, A.; Kaur, H. Potato plant leaves disease detection and classification using machine learning methodologies. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1022, 012121. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Talukder, K.H. Detection of potato disease using image segmentation and machine learning. In Proceedings of the 2020 International Conference on Wireless Communications Signal Processing and Networking (WiSPNET), Chennai, India, 4–6 August 2020; pp. 43–47. [Google Scholar]
- Teke, M.; Deveci, H.S.; Haliloğlu, O.; Gürbüz, S.Z.; Sakarya, U. A short survey of hyperspectral remote sensing applications in agriculture. In Proceedings of the 2013 6th International Conference on Recent Advances in Space Technologies (RAST), Istanbul, Turkey, 12–14 June 2013; pp. 171–176. [Google Scholar]
- Agilandeeswari, L.; Prabukumar, M.; Radhesyam, V.; Phaneendra, K.L.B.; Farhan, A. Crop classification for agricultural applications in hyperspectral remote sensing images. Appl. Sci. 2022, 12, 1670. [Google Scholar] [CrossRef]
- Sulaiman, N.; Che’Ya, N.N.; Mohd Roslim, M.H.; Juraimi, A.S.; Mohd Noor, N.; Fazlil Ilahi, W.F. The application of Hyperspectral Remote Sensing Imagery (HRSI) for weed detection analysis in rice fields: A review. Appl. Sci. 2022, 12, 2570. [Google Scholar] [CrossRef]
- Zhang, F.; Li, X.; Qiu, S.; Feng, J.; Wang, D.; Wu, X.; Cheng, Q. Hyperspectral imaging combined with convolutional neural network for outdoor detection of potato diseases. In Proceedings of the 2021 6th International Symposium on Computer and Information Processing Technology (ISCIPT), Changsha, China, 11–13 June 2021; pp. 846–850. [Google Scholar]
- Martinez-Nolasco, C.; Padilla-Medina, J.A.; Nolasco, J.J.M.; Guevara-Gonzalez, R.G.; Barranco-Gutiérrez, A.I.; Diaz-Carmona, J.J. Non-Invasive Monitoring of the Thermal and Morphometric Characteristics of Lettuce Grown in an Aeroponic System through Multispectral Image System. Appl. Sci. 2022, 12, 6540. [Google Scholar] [CrossRef]
- Leng, J.; Li, T.; Bai, G.; Dong, Q.; Dong, H. Cube-CNN-SVM: A novel hyperspectral image classification method. In Proceedings of the 2016 IEEE 28th International Conference on Tools with Artificial Intelligence (ICTAI), San Jose, CA, USA, 6–8 November 2016; pp. 1027–1034. [Google Scholar]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–spatial classification of hyperspectral imagery with 3D convolutional neural network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.Q.; Chan, J.C.W.; Xiao, L. A multi-scale wavelet 3D-CNN for hyperspectral image super-resolution. Remote Sens. 2019, 11, 1557. [Google Scholar] [CrossRef]
- Firat, H.; Hanbay, D. Classification of hyperspectral images using 3d cnn based resnet50. In Proceedings of the 2021 29th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 9–11 June 2021; pp. 1–4. [Google Scholar]
- Sabokrou, M.; Fayyaz, M.; Fathy, M.; Klette, R. Deep-cascade: Cascading 3d deep neural networks for fast anomaly detection and localization in crowded scenes. IEEE Trans. Image Process. 2017, 26, 1992–2004. [Google Scholar] [CrossRef]
- Yu, C.; Han, R.; Song, M.; Liu, C.; Chang, C.I. A simplified 2D-3D CNN architecture for hyperspectral image classification based on spatial–spectral fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2485–2501. [Google Scholar] [CrossRef]
- Bechar, A.; Vigneault, C. Agricultural robots for field operations: Concepts and components. Biosyst. Eng. 2016, 149, 94–111. [Google Scholar] [CrossRef]
- Polder, G.; Blok, P.M.; Villiers, H.; Wolf, J.; Kamp, J. Potato Virus Y Detection in Seed Potatoes Using Deep Learning on Hyperspectral Images. Front. Plant Sci. 2019, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. New Ser. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Atherton, D.; Watson, D.G.; Zhang, M.; Qin, Z.; Liu, X. Hyperspectral Spectroscopy for Detection of Early Blight (Alternaria solani) Disease in Potato (Solanum tuberosum) Plants at Two Different Growth Stages. In Proceedings of the 2015 ASABE Annual International Meeting, New Orleans, LA, USA, 26–29 July 2015. [Google Scholar]
- Atherton, D.; Choudhary, R.; Watson, D. Hyperspectral Remote Sensing for Advanced Detection of Early Blight (Alternaria solani) Disease in Potato (Solanum tuberosum) Plants. In Proceedings of the 2017 ASABE Annual International Meeting Spokane, Washington, DC, USA, 16–19 July 2017. [Google Scholar]
- Ray, S.S.; Jain, N.; Arora, R.K.; Chavan, S.; Panigrahy, S. Utility of Hyperspectral Data for Potato Late Blight Disease Detection. J. Indian Soc. Remote Sens. 2011, 39, 161–169. [Google Scholar] [CrossRef]
- Hu, Y.H.; Ping, X.W.; Xu, M.Z.; Shan, W.X.; He, Y. Detection of Late Blight Disease on Potato Leaves Using Hyperspectral Imaging Technique. Spectrosc. Spec. Anal. 2016, 36, 515–519. [Google Scholar]
- Griffel, L.M.; Delparte, D.; Edwards, J. Using Support Vector Machines classification to differentiate spectral signatures of potato plants infected with Potato Virus Y. Comput. Electron. Agric. 2018, 153, 318–324. [Google Scholar] [CrossRef]
- Kang, F.; Li, J.; Wang, C.; Wang, F. A Lightweight Neural Network-Based Method for Identifying Early-Blight and Late-Blight Leaves of Potato. Appl. Sci. 2023, 13, 1487. [Google Scholar] [CrossRef]
- Shi, Y.; Han, L.; Kleerekoper, A.; Chang, S.; Hu, T. A Novel CropdocNet for Automated Potato Late Blight Disease Detection from the Unmanned Aerial Vehicle-based Hyperspectral Imagery. arXiv 2021, arXiv:2107.13277. [Google Scholar] [CrossRef]
- Gao, J.; Westergaard, J.C.; Sundmark, E.; Bagge, M.; Alexandersson, E. Automatic late blight lesion recognition and severity quantification based on field imagery of diverse potato genotypes by deep learning. Knowl.-Based Syst. 2021, 214, 106723. [Google Scholar] [CrossRef]
- Qi, C.; Sandroni, M.; Westergaard, J.C.; Sundmark, E.; Bagge, M.; Alexandersson, E.; Gao, J. In-field early disease recognition of potato late blight based on deep learning and proximal hyperspectral imaging. arXiv 2021, arXiv:2111.12155. [Google Scholar] [CrossRef]
- Chen, J.; Deng, X.; Wen, Y.; Chen, W.; Zeb, A.; Zhang, D. Weakly-supervised learning method for the recognition of potato leaf diseases. In Artificial Intelligence Review; Springer Nature: Berlin/Heidelberg, Germany, 2022; pp. 1–18. [Google Scholar]
- Chen, Y. Convolutional Neural Network for Sentence Classification. Master’s Thesis, University of Waterloo, Waterloo, ON, Canada, 2015. [Google Scholar]
- Huang, Y.; Wang, Q.; Jia, W.; Lu, Y.; Li, Y.; He, X. See more than once: Kernel-sharing atrous convolution for semantic segmentation. Neurocomputing 2021, 443, 26–34. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Qiao, S.; Chen, L.C.; Yuille, A. Detectors: Detecting objects with recursive feature pyramid and switchable atrous convolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 10213–10224. [Google Scholar]

| Layer | Type | Output Shape | Param | Connected to |
|---|---|---|---|---|
| input | InputLayer | (None, 7, 7, 204, 1) | 0 | |
| Conv3_1 | Conv3D | (None, 7, 7, 204, 8) | 224 | input |
| Conv3_2 | Conv3D | (None, 7, 7, 204, 8) | 1736 | Conv3_1 |
| Conv3_3 | Conv3D | (None, 7, 7,204, 8) | 1736 | Conv3_2 |
| Pool | MaxPooling3D | (None, 7, 7, 102, 8) | 0 | Conv3_3 |
| reshape1 | Reshape | (None, 7, 7, 816) | 0 | Pool3 |
| Conv2 | Conv2D | (None, 3, 3, 8) | 58, 760 | reshape1 |
| reshape2 | Reshape | (None, 72, 1) | 0 | Conv2 |
| Conv1 | Conv1D | (None, 68, 16) | 64 | reshape2 |
| flatten | Flatten | (None, 1088) | 0 | Conv1 |
| Dropout | Dropout | (None, 1088) | 0 | flatten |
| D1 | Dense | (None, 50) | 54,450 | Dropout |
| out | Dense | (None, 4) | 204 | D1 |
| Layer | Type | Output Shape | Param | Connected to |
|---|---|---|---|---|
| input | InputLayer | (None, 1, 204, 1) | 0 | |
| Conv1_1 | Conv1D | (None, 204, 32) | 224 | input |
| Pool1 | MaxPooling1D | (None, 51, 32) | 0 | Conv1_1 |
| Conv2_1 | Conv1D | (None, 51, 64) | 12,352 | Pool1 |
| Pool2 | MaxPooling1D | (None, 26, 64) | 0 | Conv2_1 |
| Conv3_1 | Conv1D | (None, 26, 128) | 49,280 | Pool2 |
| Pool3 | MaxPooling1D | (None, 13, 128) | 0 | Conv3_1 |
| flatten | Flatten | (None, 1664) | 0 | Pool3 |
| D1 | Dense | (None, 128) | 213,120 | flatten |
| out | Dense | (None, 3) | 387 | D1 |
| Datasets | Assessment Metrics | Models | ||
|---|---|---|---|---|
| 3D-CNN | Multidimensional Fusion CNN | Multidimensional Fusion Atrous-CNN | ||
| Train-100 | ||||
| Time-100 | 2:15:50 | 2:02:11 | 2:07:57 | |
| Train | Loss-100 | 0.0259 | 0.0201 | 0.0141 |
| Precision-100 | 98.92% | 99.16% | 99.41% | |
| Val | Loss-100 | 0.0231 | 0.0336 | 0.0233 |
| Precision-100 | 99.07% | 98.44% | 99.13% | |
| Train-500 | ||||
| Time-500 | 11:42:40 | 10:52:13 | 10:59:43 | |
| Train | Loss-500 | 0.0195 | 0.0106 | 0.0054 |
| Precision-500 | 99.18% | 99.57% | 99.78% | |
| Val | Loss-500 | 0.0214 | 0.0254 | 0.0226 |
| Precision-500 | 99.16% | 98.86% | 99.31% | |
| Category Labels | Models | |||||
|---|---|---|---|---|---|---|
| 3D-CNN | Multidimensional Fusion CNN | Multidimensional Fusion Atrous-CNN | ||||
| Correct Pixels | Precision | Correct Pixels | Precision | Correct Pixels | Precision | |
| Healthy leaf pixels (16,970) | 16,894 | 99.55% | 16,499 | 97.22% | 16,934 | 99.79% |
| Diseased leaves pixels (3173) | 2940 | 92.66% | 3111 | 98.05% | 3165 | 99.75% |
| Background pixels (29,773) | 29,756 | 99.94% | 29,752 | 99.93% | 29,758 | 99.95% |
| Whiteboard pixels (2513) | 2510 | 99.88% | 2509 | 99.84% | 2511 | 99.92% |
| Total (52,429) | 52,100 | 99.37% | 51871 | 98.94% | 52,368 | 99.88% |
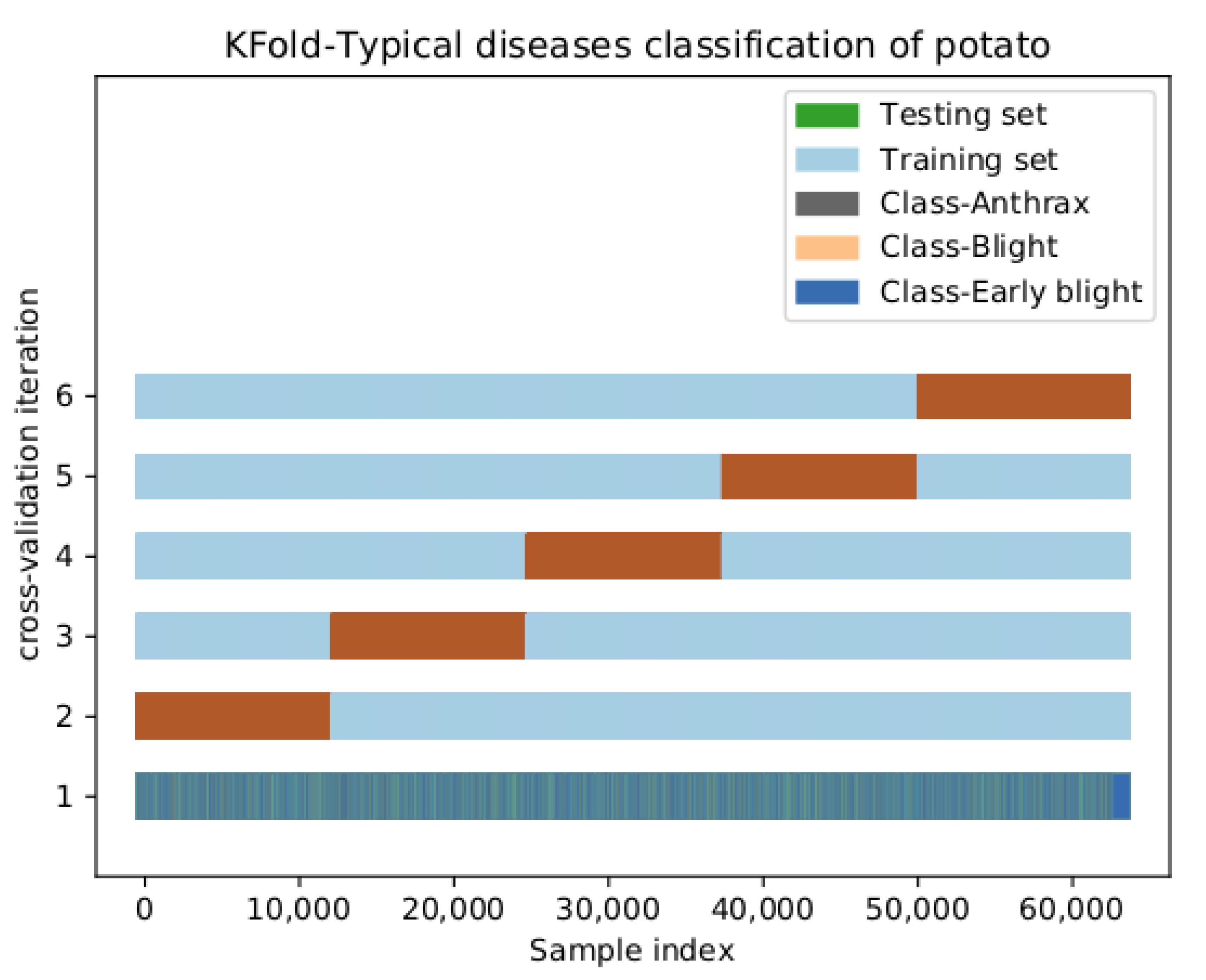
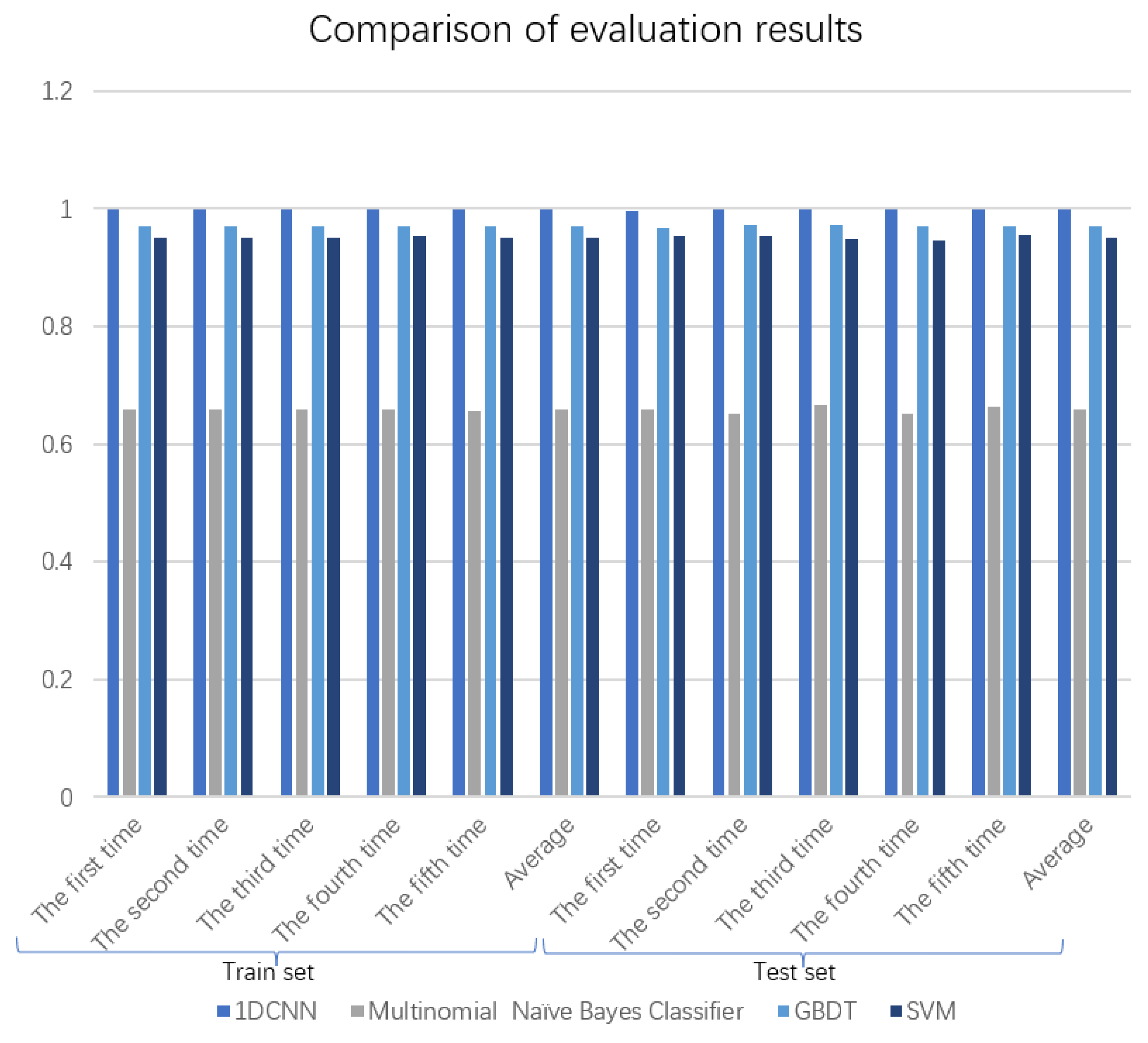
| Dataset | K-Fold cross Validation | 1DCNN | Multinomial Naive Bayes Classifier | GBDT | SVM |
|---|---|---|---|---|---|
| Train set | The first time | 0.9979 | 0.6582 | 0.9707 | 0.9508 |
| The second time | 0.9987 | 0.6583 | 0.9706 | 0.9509 | |
| The third time | 0.9989 | 0.6592 | 0.9708 | 0.9517 | |
| The fourth time | 0.9978 | 0.6581 | 0.9706 | 0.9522 | |
| The fifth time | 0.9982 | 0.6572 | 0.9706 | 0.9510 | |
| Average | 0.9983 | 0.6582 | 0.9707 | 0.9513 | |
| Test set | The first time | 0.9967 | 0.6577 | 0.9682 | 0.9527 |
| The second time | 0.9980 | 0.6516 | 0.9735 | 0.9528 | |
| The third time | 0.9990 | 0.6664 | 0.9718 | 0.9482 | |
| The fourth time | 0.9976 | 0.6526 | 0.9711 | 0.9470 | |
| The fifth time | 0.9997 | 0.6627 | 0.9688 | 0.9554 | |
| Average | 0.9982 | 0.6582 | 0.9707 | 0.9512 |
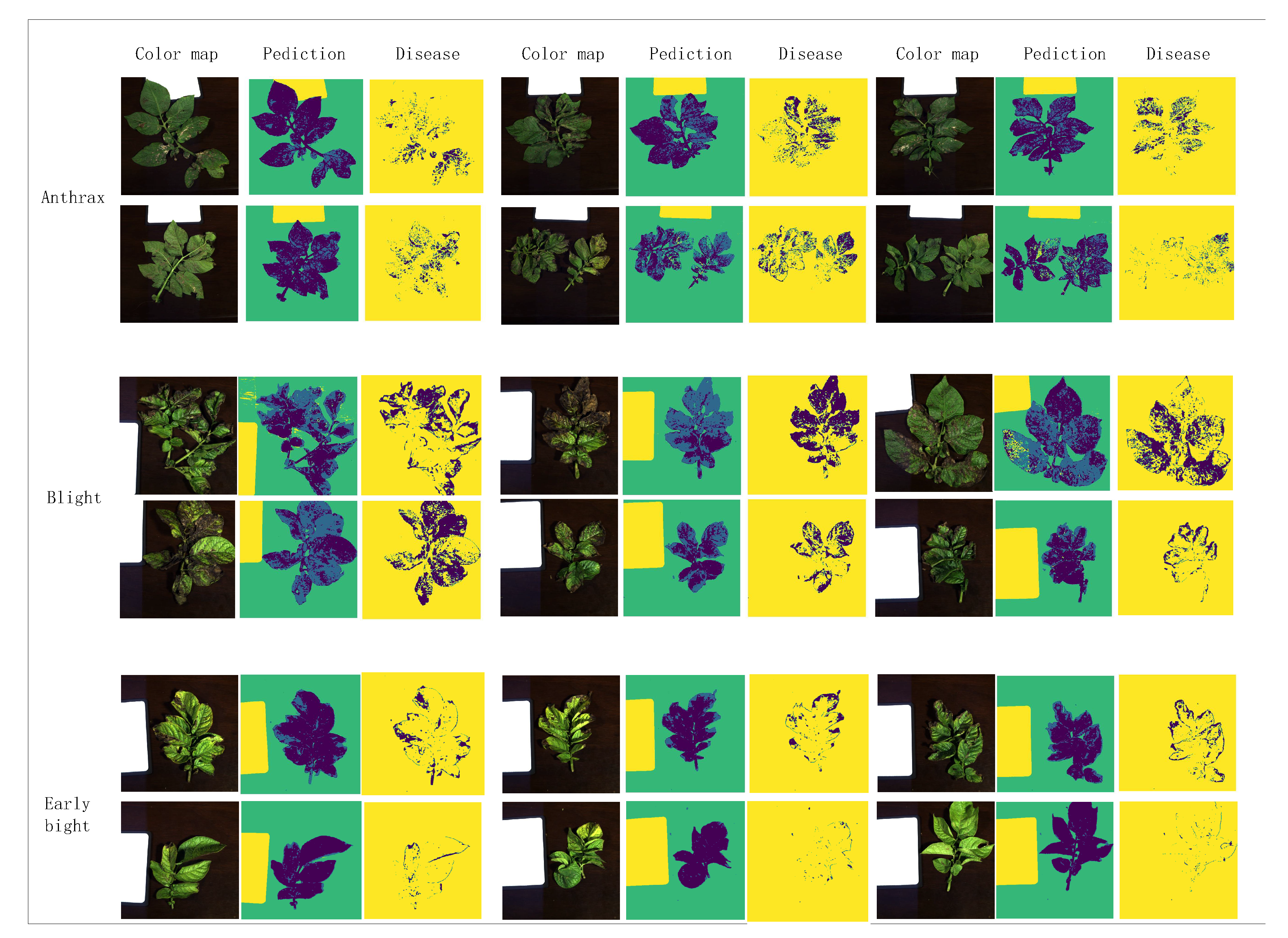
| Datasets | Accuracy and Recall | Disease Category | ||
|---|---|---|---|---|
| Anthrax | Blight | Early Blight | ||
| Train | Accuracy | 1 | 0.9969 | 0.9979 |
| Recall | 1 | 0.9992 | 0.9923 | |
| Test | Accuracy | 0.9987 | 0.9895 | 0.9842 |
| Recall | 0.9997 | 0.9942 | 0.971 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, W.; Xiao, Z.; Bao, T. Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data. Appl. Sci. 2023, 13, 5023. https://doi.org/10.3390/app13085023
Gao W, Xiao Z, Bao T. Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data. Applied Sciences. 2023; 13(8):5023. https://doi.org/10.3390/app13085023
Chicago/Turabian StyleGao, Wenqiang, Zhiyun Xiao, and Tengfei Bao. 2023. "Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data" Applied Sciences 13, no. 8: 5023. https://doi.org/10.3390/app13085023
APA StyleGao, W., Xiao, Z., & Bao, T. (2023). Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data. Applied Sciences, 13(8), 5023. https://doi.org/10.3390/app13085023






