Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Tandem Mass Spectrometry
2.2. Spectrum Graph
2.3. Convolutional Neural Networks
2.4. Graph Convolutional Neural Networks
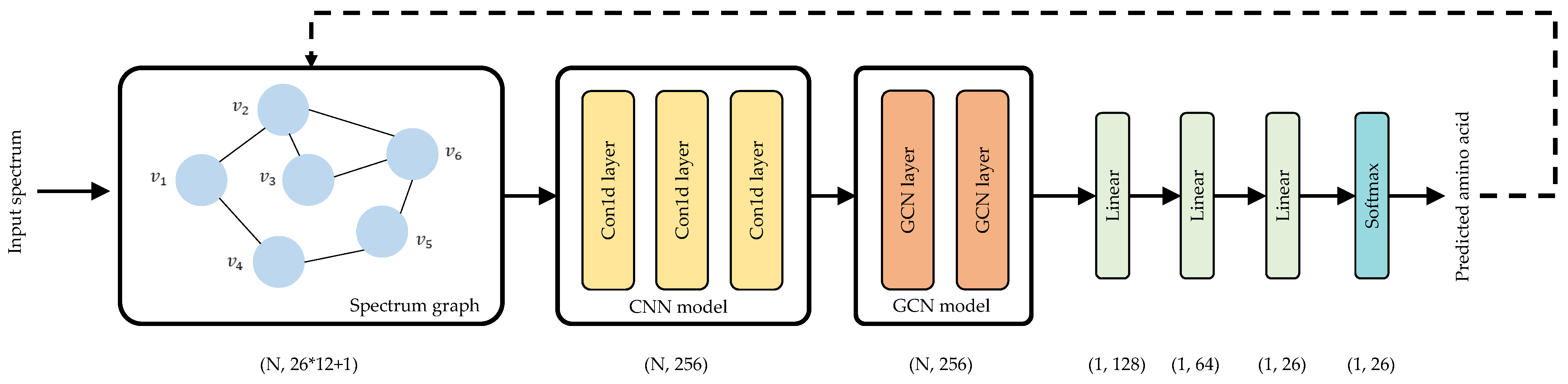
2.5. Denovo-GCN Network
2.6. Data Sets and Evaluation Metrics
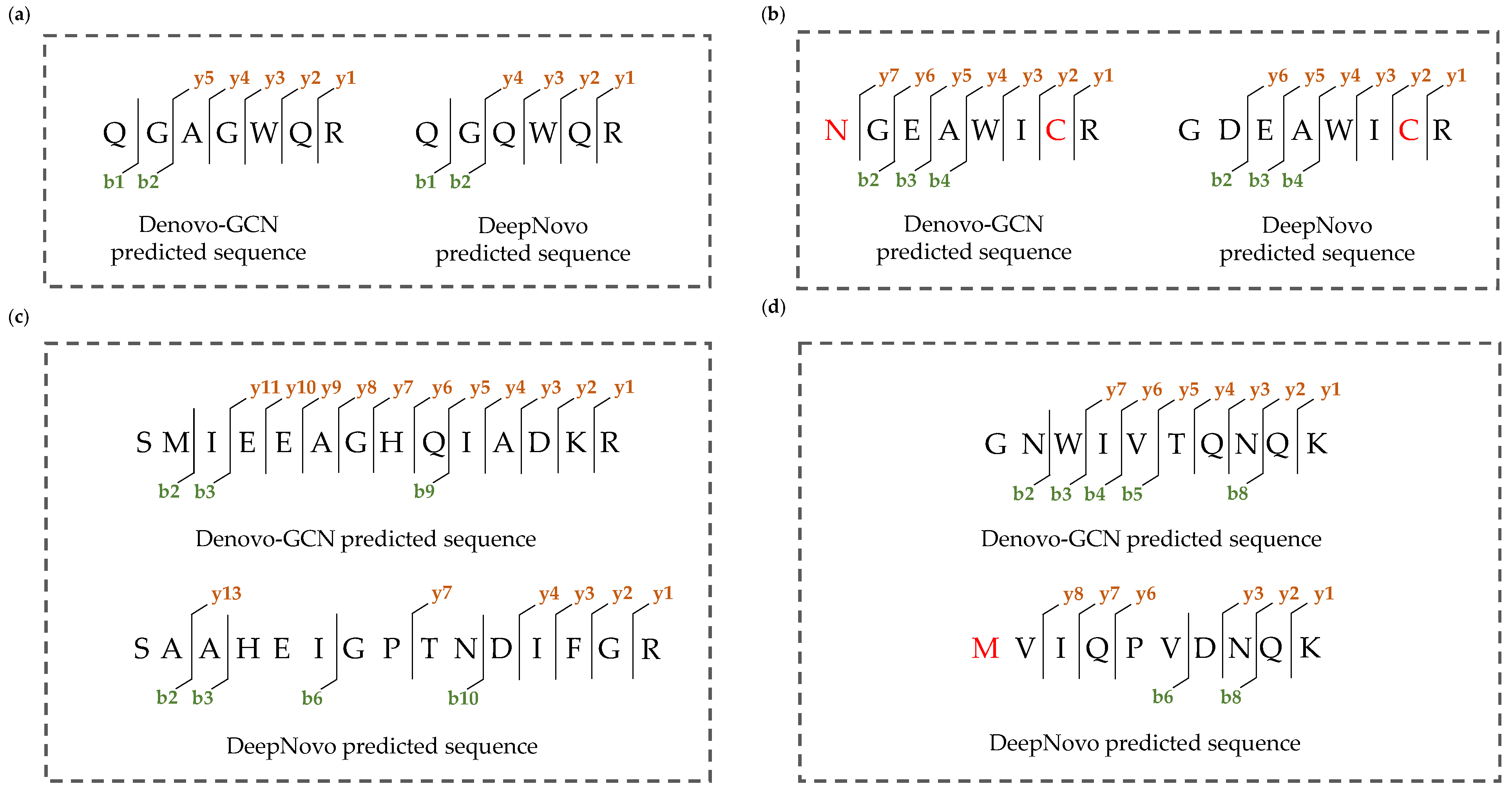
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakurai, T.; Matsuo, T.; Matsuda, H.; Katakuse, I. PAAS 3: A computer program to determine probable sequence of peptides from mass spectrometric data. J. Mass Spectrom. 1984, 11, 396–399. [Google Scholar] [CrossRef]
- Ishikawa, K.; Niwa, Y. Computer-aided peptide sequencing by fast atom bombardment mass spectrometry. J. Mass Spectrom. 1986, 13, 373–380. [Google Scholar] [CrossRef]
- Yates, J.R.; Griffin, P.; Hood, L.; Zhou, J. Computer-Aided Interpretation of Low-Energy MS/MS Mass-Spectra of Peptides. Tech. Protein Chem. Second edition. 1991, 46, 477–485. [Google Scholar]
- Tran, N.H.; Rahman, M.Z.; He, L.; Xin, L.; Shan, B.; Li, M. Complete de novo assembly of monoclonal antibody sequences. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Faridi, P.; Li, C.; Ramarathinam, S.H.; Vivian, J.P.; Illing, P.T.; Mifsud, N.A.; Ayala, R.; Song, J.; Gearing, L.J.; Hertzog, P.J.; et al. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 2018, 3, eaar3947. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Laumont, C.M.; Vincent, K.; Hesnard, L.; Audemard, É.; Bonneil, É.; Laverdure, J.-P.; Gendron, P.; Courcelles, M.; Hardy, M.-P.; Côté, C.; et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 2018, 10, eaau5516. [Google Scholar] [CrossRef]
- Taylor, J.A.; Johnson, R.S. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 2001, 73, 2594–2604. [Google Scholar] [CrossRef]
- Taylor, J.A.; Johnson, R.S. Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 1997, 11, 1067–1075. [Google Scholar] [CrossRef]
- Johnson, R.S.; Taylor, J.A. Searching sequence databases via de novo peptide sequencing by tandem mass spectrometry. Methods Mol. Biol. 2002, 22, 301–315. [Google Scholar] [CrossRef]
- Fernandez-de-Cossio, J.; Gonzalez, J.; Betancourt, L.; Besada, V.; Padron, G.; Shimonishi, Y.; Takao, T. Automated interpretation of high-energy collision-induced dissociation spectra of singly protonated peptides by ‘SeqMS’, a software aid for de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 1998, 12, 1867–1878. [Google Scholar] [CrossRef]
- Dančík, V.; Addona, T.A.; Clauser, K.R.; Vath, J.E.; Pevzner, P.A. De Novo Peptide Sequencing via Tandem Mass Spectrometry. J. Comput. Biol. 1999, 6, 327–342. [Google Scholar] [CrossRef]
- Frank, A.; Pevzner, P. PepNovo: De novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005, 77, 964–973. [Google Scholar] [CrossRef]
- Grossmann, J.; Roos, F.F.; Cieliebak, M.; Lipták, Z.; Mathis, L.K.; Müller, M.; Gruissem, W.; Baginsky, S. AUDENS: A tool for automated peptide de novo sequencing. J. Proteome Res. 2005, 4, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Sun, R.-X.; Yang, B.; Song, C.Q.; Wang, L.H.; Liu, C.; Fu, Y.; Yuan, Z.-F.; Wang, H.-P.; He, S.-M.; et al. pNovo: De novo peptide sequencing and identification using HCD spectra. J. Proteome Res. 2010, 9, 2713–2724. [Google Scholar] [CrossRef]
- Chi, H.; Chen, H.; He, K.; Wu, L.; Yang, B.; Sun, R.-X.; Liu, J.; Zeng, W.-F.; Song, C.-Q.; He, S.-M.; et al. pNovo+: De novo peptide sequencing using complementary HCD and ETD tandem mass spectra. J. Proteome Res. 2013, 12, 615–625. [Google Scholar] [CrossRef]
- Ciresan, D.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. Neural Inf. Process. Syst. 2012, 25, 2843–2851. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q. Sequence to sequence learning with neural networks. Neural Inf. Process. Syst. 2014, 27, 3104–3112. [Google Scholar]
- Karpathy, A.; Li, F.F. Deep Visual-Semantic Alignments for Generating Image Description. In Proceedings of the Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3128–3137. [Google Scholar]
- Vinyals, O.; Toshev, A.; Bengio, S.; Erhan, D. Show and tell: A Neural Image Caption Generator. In Proceedings of the Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3156–3164. [Google Scholar]
- Ma, B. Novor: Real-Time Peptide de Novo Sequencing Software. J. Am. Soc. Mass Spectrom. 2015, 11, 1885–1894. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Tran, N.H.; Zhang, X.; Lei, X.; Shan, B.; Ming, L. De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. USA 2017, 114, 8247–8252. [Google Scholar] [CrossRef]
- Muth, T.; Renard, B.Y. Evaluating de novo sequencing in proteomics: Already an accurate alternative to database-driven peptide identification? Brief Bioinform. 2018, 19, 954–970. [Google Scholar] [CrossRef]
- Qiao, R.; Tran, N.H.; Xin, L.; Chen, X.; Li, M.; Shan, B.; Ghodsi, A. Computationally instrument-resolution-independent de novo peptide sequencing for high-resolution devices. Nat. Mach. Intell. 2021, 3, 420–425. [Google Scholar] [CrossRef]
- Thomas, N.K.; Welling, M. Semi-Supervised Classification with Graph Convolution Networks. In Proceedings of the Machine Learning. Palais des Congrès Neptune, Toulon, France, 9 September 2016. [Google Scholar] [CrossRef]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive representation learning on large graphs. Neural Inf. Process. Syst. (NIPS) 2017. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. In Proceedings of the Machine Learning, Vancouver, BC, Canada, 4 February 2018. [Google Scholar]
- Guo, K.; Wang, P.; Shi, P.; He, C.; Wei, C. A New Partitioned Spatial–Temporal Graph Attention Convolution Network for Human Motion Recognition. Appl. Sci. 2023, 13, 1647. [Google Scholar] [CrossRef]
- Wan, H.; Tang, P.; Tian, B.; Yu, H.; Jin, C.; Zhao, B.; Wang, H. Water Extraction in PolSAR Image Based on Superpixel and Graph Convolutional Network. Appl. Sci. 2023, 13, 2610. [Google Scholar] [CrossRef]
- Bioinformatics Solutions Inc. PEAKS Studio, Version 8.0; Bioinformatics Solutions Inc.: Waterloo, ON, Canada, 2017. [Google Scholar]


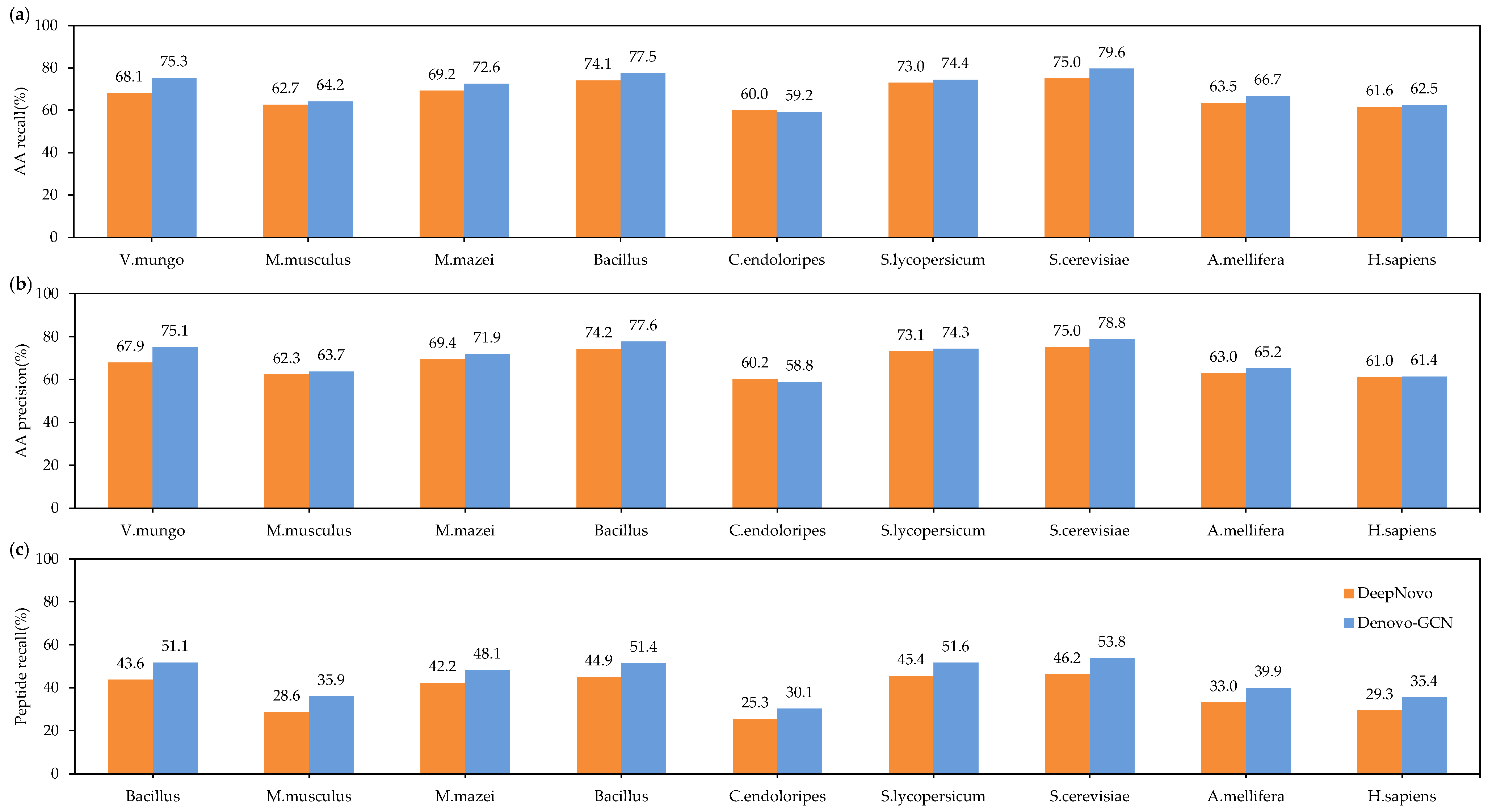



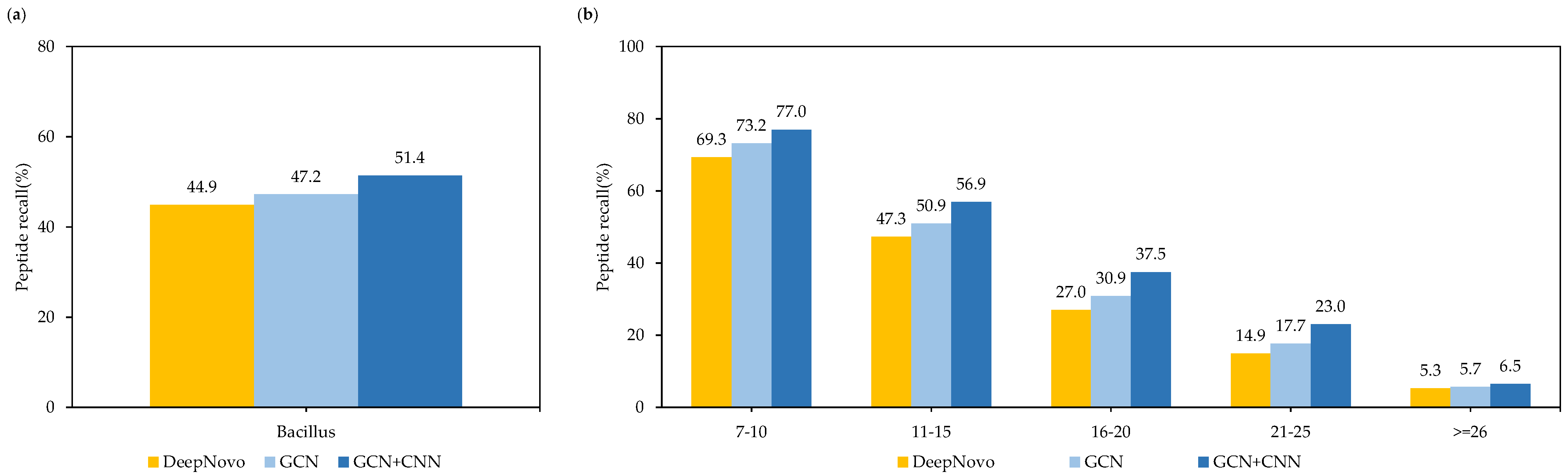
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Zhang, X.; Wang, R.; Wang, H. Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks. Appl. Sci. 2023, 13, 4604. https://doi.org/10.3390/app13074604
Wu R, Zhang X, Wang R, Wang H. Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks. Applied Sciences. 2023; 13(7):4604. https://doi.org/10.3390/app13074604
Chicago/Turabian StyleWu, Ruitao, Xiang Zhang, Runtao Wang, and Haipeng Wang. 2023. "Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks" Applied Sciences 13, no. 7: 4604. https://doi.org/10.3390/app13074604
APA StyleWu, R., Zhang, X., Wang, R., & Wang, H. (2023). Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks. Applied Sciences, 13(7), 4604. https://doi.org/10.3390/app13074604



