Featured Application
Porphyromonas gingivalis is an oral bacterium that uses gingipains as its primary invasion factor to cause periodontitis and certain systemic diseases in humans. To study the underlying mechanisms employed by gingipains, a simpler and quick approach is required to examine the protease activity. In the present study, passive diffusion-mediated protein elution and gelatin zymography methods were demonstrated to isolate and characterize the gingipains. This study found that an eluted protein with a molecular weight of about 180 kDa exhibited both Rgp and Kgp activity. While in the gelatin zymography, the proteins with a molecular weight of approximately 50 kDa, and above 245 kDa were found to be arginine-X gingipains when specific selective inhibitors, Leupeptin and Cathepsin B inhibitor II were used. The present study protocol will allow a more efficient separation of targeted protein samples with a greater difference in isoelectric point.
Abstract
Gingipains (RgpA, RgpB, and Kgp) are major virulence factors of the periodontitis-causing bacterium Porphyromonas gingivalis. Isolation of gingipains from the crude protein sample of P. gingivalis is critical for studying the underlying invasion mechanism that contributes to periodontitis, Alzheimer’s disease, and cardiovascular disease (CVD). Chromatographic processes and molecular cloning are two standard techniques often used for gingipains isolation, which are time-consuming and costly. In this study, considerably easier methods based on passive-mediated diffusion gel elution and gelatin zymogram were used to isolate and characterize gingipains. Importantly, proteins eluted from Native-PAGE showed enzymatic activity for both Rgp and Kgp. In gelatin zymography, the proteins with a molecular size of ~50 kDa and above 245 kDa were suggested as arginine-specific gingipains. The passive diffusion-mediated gel elution method is a simpler technique to isolate gingipains from crude protein samples of P. gingivalis. By using covalent and highly specific gingipain inhibitors, gelatin zymography enabled an individual characterization of gingipain activity and inhibition. Finally, this protocol can be easily extended by adding the isoelectric focusing to further improve the protein separation and characterization.
1. Introduction
Periodontitis is a serious and persistent inflammatory illness that affects approximately 20% to 50% of the world’s population [1]. The consequences of individuals that suffered from periodontitis not only have a high risk of losing their teeth but also tend to develop certain systemic diseases like cardiovascular diseases and diabetes [1,2]. According to several studies, Porphyromonas gingivalis has also been linked to the progression of the illness, such as Alzheimer’s disease [3], rheumatoid arthritis [4,5], and cardiovascular diseases [6,7].
P. gingivalis is an anaerobic, black-pigmented oral pathogen with outer lipopolysaccharides and fimbriae that only colonize the subgingival area of humans [8,9]. These bacteria are closely linked to the development and progression of periodontitis [2,10,11]. When P. gingivalis invade and colonize subgingival areas of humans, they will tend to produce a complex bacterial biofilm with other oral bacteria and resulting in long-lasting inflammation, loss of gingival connective tissues, and resorption of alveolar bone [12,13,14].
Three forms of cysteine proteases termed gingipains are produced by P. gingivalis, which are crucial for their pathogenesis, retrieval of amino acid and heme from the host, and maturation of their fimbriae [15,16]. Gingipains are the key proteases that are involved in bacterial proteolytic activity [2,17]. There are several mechanisms are proposed to be employed by gingipains to help this oral bacterium to invade humans [18,19]. Gingipains are subdivided into two types, arginine (Rgp) and lysine gingipains (Kgp). RgpA (~95 kDa) is made up of one catalytic and several adhesion/hemagglutinin domains, which bonded non-covalently to form a complex protein [15,16]. RgpB (~48 kDa) comprises only the catalytic domain [15,16]. Kgp (~105 kDa) is similar to RgpA also consists of one catalytic and three to four adhesion/hemagglutinin domains, which differ between the strains of P. gingivalis [15,16].
A few chromatographic techniques are employed to separate the gingipains into their constituent parts in order to isolate and analyze them [20,21,22]. Besides that, cloning approach also be employed to produce gingipain of interest [17,23,24]. Nonetheless, these approaches are laborious, costly, and impracticable especially when a quick check of gingipains activity is required. In this study, relatively simpler, easier, and quick methods to isolate and characterize gingipains using passive diffusion-mediated gel elution and gelatin zymogram were demonstrated.
2. Materials and Methods
The flowchart given in Figure 1, demonstrates the simplified procedure for isolation and characterization of gingipains developed in the present study. The gingipains were first isolated using the passive diffusion-mediated gel elution method and further characterized using gelatin zymography.
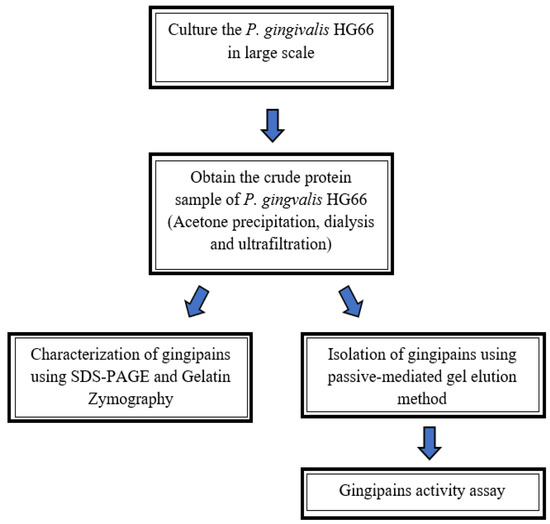
Figure 1.
An overview of the studies performed to isolate and characterize the gingipains is shown in the flowchart above.
2.1. Cultivation of P. gingivalis HG66 Strain
As per Potempa & Nguyen’s (2007) [21] protocols, P. gingivalis was grown in anaerobic conditions at 37 °C with 5% yeast extract, 1% hemin, 5% (w/v) L-cysteine, and 0.5 mg/mL menadione as supplements [21].
2.2. Preparation of Crude Protein of P. gingivalis HG66
The culture of P. gingivalis HG66 was centrifuged for 30 min at 12,000 rpm, 4 °C. The bacterial brownish supernatant was collected. The pre-chilled acetone was added slowly and made the final concentration 60%. The mixture was centrifugated for 30 min at 12,000 rpm, 4 °C to obtain protein pellet. The protein pellet was further resuspended with ice-cold 4,4′-dithiopyridine disulfide buffer [20 mM Bis-Tris, 150 mM NaCl, 0.02% NaN3, and 1.5 mM 4,4′-dithiopyridine disulfide, pH 6.8] and incubated on ice for 1 h. The protein was dialyzed against dialysis buffer [20 mM Bis-Tris, 150 mM NaCl, 5 mM CaCl2, and 0.02% NaN3, pH 6.8] overnight at 4 °C with 2 changes. The dialyzed protein was further concentrated using Amicon® PM-30 membrane with centrifugation for 30 min at 5000 rpm, 4 °C. This procedure was conducted as described by Potempa & Nguyen (2007) with slight modifications [21].
2.3. Isolation of Gingipains Using Passive Diffusion-Mediated Gel Elution Method
2.3.1. Sample Preparation
The crude protein sample was prepared as mentioned above in Section 2.2. The sample was mixed with 5× Native Gel sample loading buffer (300 mM Tris-HCl, 30% glycerol, 0.02% bromophenol, pH 6.8) [25]. The sample was mixed well and thoroughly with gentle pipetting and incubated at 20 °C for 10 min.
2.3.2. Native-PAGE Electrophoresis
All sample was loaded onto the Native-polyacrylamide gel (with the ratio of acrylamide: bisacrylamide in 29:1) using the Laemmli discontinuous buffer system. The entire gel was made up of 10% resolving gel and 6% stacking gel that was surrounded with running buffer [25 mM Tris, 190 mM glycine, and 0.1% SDS, pH 8.3]. The gel was run at 110 V constantly for 90 min [25].
2.3.3. Elution of Protein
The electrophoresed gel was cut into strips using a sterile disposable blade. One protein ladder and one separated gingipains sample gel strips were stained with Coomassie Brilliant Blue staining buffer [0.05% Coomassie Brilliant Blue R-250, 30% ethanol, and 10% acetic acid], followed by de-staining with de-staining buffer [30% ethanol and 10% acetic acid]. The stained protein gel strips were used as the reference to identify the target proteins. Using a sterile blade, the protein bands of interest were sliced out carefully from the unstained gel strip. The sliced gels were crushed using a disposable homogenizer (Takara BioMasher, Kusatsu, Japan) after adding 300 µL elution buffer [50 mM Tris-HCl, 150 mM NaCl, and 0.1 mM EDTA pH 7.5]. The obtained samples were incubated at 4 °C overnight and centrifugated for (10,000 rpm, 4 °C) for 15 min. The supernatants of eluted proteins were obtained and their gingipains activity was then examined. The aforementioned procedures were conducted as described by Burgess et al. (2009) with minor modifications [26].
2.3.4. Gingipains Activity Assay
To prepare the gingipains samples for the Rgp and Kgp activity assays, 20 µL and 50 µL of the sample were diluted in deionized water to a final volume of 90 µL, respectively. A 100 µL of 2× gingipains assay buffer [200 mM Tris-HCl, 150 mM NaCl, 5 mM CaCl2, 0.02% NaN3, pH 7.6] in a 96-wells plate, followed by adding 90 µL of prepared gingipains samples. Then, the samples were incubated for 10 min at 37 °C. After incubation, the colorimetric substrate, Bz-L-arginine-pNA (Sigma-Aldrich, St. Louis, MO, USA), and Ac-Lys-pNA (Bachem, Bubendorf, Switzerland) for testing Rgp and Kgp activity assay were added, respectively. Using a microplate reader (Infinite® 200 PRO, Tecan Group, Männedorf, Switzerland), the absorbance was measured at 405 nm [21].
2.4. Characterization of Gingipains
2.4.1. SDS-PAGE
The gingipains sample were treated with 5× of the non-reducing sample solubilizing buffer (300 mM Tris-HCl, 10% SDS, 30% glycerol, 0.02% bromophenol, pH 6.8) and boiled for 5 min at 100 °C. To the sample mixture, a 1:10 of the reducing agent, β-mercaptoethanol, was added and heated for 5 min at 100 °C. The sample was placed onto SDS-polyacrylamide gel (10% resolving gel and 6% stacking gel with acrylamide to bisacrylamide ratio: 29:1). The gel was electrophoresed at 110 V for 90 min continuously [21].
2.4.2. Gelatin Zymography
Sample Preparation
The gingipains sample were prepared as per described by Toth & Fridman (2001) [27]. The gingipains sample was mixed with 5× non-reducing sample solubilizing buffer (β-mercaptoethanol was omitted). Furthermore, the gingipains sample was incubated for 15 min at room temperature. For characterization purposes, proteases-specific inhibitors were incorporated into the sample mixture. A 200 µM of Leupeptin inhibitor, (Nacalai Tesque, Kyoto, Japan), 20 µM of Cathepsin B inhibitor II (Sigma-Aldrich), and 2 mM of TLCK (Sigma-Aldrich) was added to the gingipains sample, respectively, to test their gingipain proteases activity. This was performed in accordance with Hashimoto’s (2017) with slight modification [28].
Gelatin Zymography
The gingipains sample was loaded on the gelatin-SDS-polyacrylamide gel (with 0.1% gelatin incorporated into the 10% resolving gel) [27]. The gel was electrophoresed at 110 V constantly for 90 min.
Gel Renaturation and Incubation
The gel was removed from the electrophoretic glass plate after electrophoresis properly. The gel was first gently agitated twice for 15 min in distilled water before being washed with 2.5% (v/v) Triton X-100. After renaturation, the electrophoresed gel was treated with an incubation buffer containing 50 mM Tris-HCl, 5 mM CaCl2, and 0.02% (v/v) Triton X-100 for 2 h at 37 °C [27]. This activated the gingipains proteases.
Gel Staining
The electrophoresed gelatin-SDS-polyacrylamide was stained with Coomassie Brilliant Blue staining buffer for 30 min at room temperature with gentle agitation following the renaturation and activation processes. A de-staining buffer was used to remove the stain from the gel until clear and translucent bands could be seen.
3. Results
3.1. Purification: Passive-Diffusion Mediated Elution Method
We have demonstrated that the passive diffusion-mediated gel elution method can be an alternative method to have a quick check on the gingipain activity in the crude sample without any pre-treatment. The first trial used the passive diffusion-mediated elution method to separate the gingipain mixture into their individual components (RgpA, RgpB, and Kgp). Under the native condition of gel electrophoresis, a total of six bands were observed on the gel (Figure 2). The clear and sharp bands that were found on the gel, were selected, excised out from the gel, and further subjected to gingipains activity assay. The protein bands with a molecular weight of approximately 180 kDa gingipains were shown to have both Rgp and Kgp activities, but other eluted protein bands had very low or even no activity for either Rgp or Kgp activity, as indicated in Table 1. The eluted protein that showed little or even no gingipains activity indicated the presence of other secreted proteins from P. gingivalis.
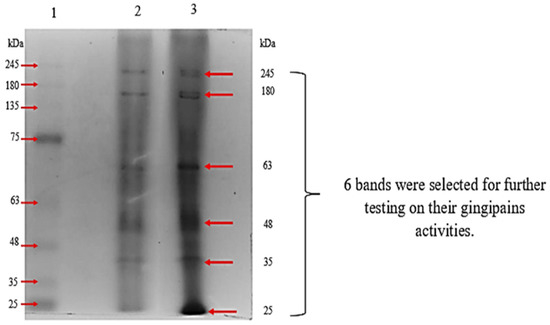
Figure 2.
Native-PAGE of crude protein samples. The gingipains samples were solubilized in non-reducing solubilizing sample buffer and electrophoresed at 110 V for 90 min. Lane 1 was BLUeye pre-stained protein standards while Lane 2 and 3 were the crude protein samples.

Table 1.
Gingipain activity assay for eluted proteins.
3.2. Characterization of Gingipains
3.2.1. SDS-PAGE
After acetone precipitation, the crude protein sample was examined with 10% polyacrylamide gel. Multiple bands were observed in the gel with a molecular size range between 17 kDa to 75 kDa. Since gingipains can be separated into RgpA, RgpB, and Kgp, three or more bands were anticipated to be observed (Figure 3).
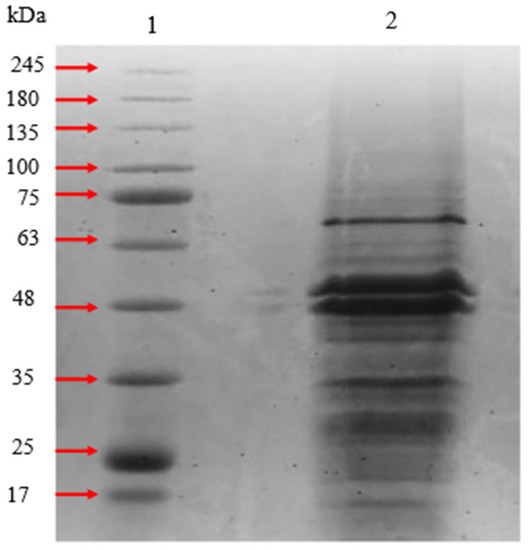
Figure 3.
SDS-PAGE of crude protein samples after acetone precipitation. The fractions were boiled in reducing solubilizing sample buffer and electrophoresed on 10% SDS polyacrylamide gel at 110 V for 90 min. Lane 1 was BLUeye pre-stained protein standards and Lane 2 was the crude gingipains sample.
3.2.2. Gelatin Zymography
Further characterization of catalytic transparent bands that observed on gelatin zymogram gel was done by incorporating selective inhibitors, Leupeptin inhibitor and Cathepsin B inhibitor II for identifying the arginine-specific gingipain (Rgp) and lysine-specific gingipains (Kgp), respectively [21,29,30]. Polyacrylamide gel electrophoresis of the partially purified gingipains under non-denaturing conditions showed two apparent bands with the molecular weight ranging from 48 kDa to 75 kDa and a band above 245 kDa (Figure 4a).
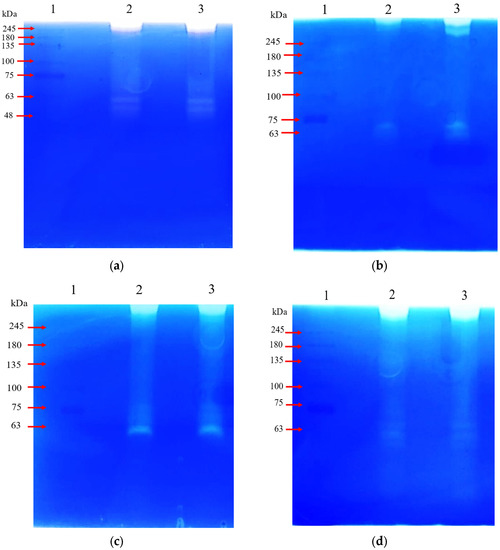
Figure 4.
Zymography gels showed the transparent bands produced by the proteases found in the crude gingipains sample of P. gingivalis HG66. All Lane 1 was BLUeye pre-stained protein standards. (a) Crude gingipains samples without any treatment (2 and 3). (b) Crude gingipains sample treated with (2) and without (3) Leupeptin inhibitor. (c) Crude gingipains sample treated with (2) and without (3) Cathepsin B inhibitor II. (d) Crude gingipains sample treated with (2) and without (3) TLCK inhibitor.
In the gelatin zymogram with leupeptin inhibitor, the bands with a molecular weight of approximately 50 kDa and 245 kDa were not observed (Figure 4b). All bands were observed in the gelatin zymogram without the inhibitor and were similarly observed in the gel for the gelatin zymogram with Cathepsin B inhibitor II (Figure 4c). In gelatin zymogram with TLCK inhibitor, there was a significant reduction of bands intensity with the molecular weight in the range of 48 kDa to 75 kDa (Figure 4d). The catalytic bands observed in gelatin zymogram for current study were consistent with those described in Potempa et al. (1995); the major catalytic zone of HG66 strains was found on the very top of the gel and in the molecular weight range between 48 kDa to 52 kDa area the gel. From this, the catalytic zones that were observed on the very top and between the molecular weight range of 48 kDa to 52 kDa in the gelatin zymogram were strongly indicated as arginine-specific gingipains (RgpA and RgpB), which are also suggested as gelatinase.
4. Discussion
To separate the gingipains into their constituents, a variety of procedures are employed. This is due to the fact that gingipains are the key invasion factor used by these bacteria to cause periodontitis [31]. Therefore, it is critical to isolate and characterize gingipains that enable the researchers to explore and understand their underlying invasion mechanisms [32,33]. In a previous study, a variety of chromatographic methods were combined to separate the gingipains into their component parts [22]. Besides chromatography techniques, other methods, including cloning the gingipain encoding gene into a bacterial vector that allows them to express the recombinant protein, are also employed for gingipain purification [24,34]. However, the methods used to isolate the gingipains in the past are time-consuming and difficult. Consequently, this study demonstrated a quick and an easy solution to solve this issue.
In the present investigation, reducing SDS-PAGE, non-reducing gelatin zymogram, and Native-PAGE all showed a significantly different amount of protein bands. The protein samples in SDS-PAGE are first reduced by β-mercaptoethanol and then further boiled at 100 °C before electrophoresis, which results in a partial or completely dissociation of non-covalently linked gingipains complexes. Multiple bands were seen to develop on the non-denatured SDS-PAGE in earlier research, which led to the conclusion that RgpA and Kgp are made up of numerous domains that are not covalently connected [21,35,36]. Whereas the protein samples that are subjected to gelatin zymogram are not reduced and heated showed a slightly higher molecular weight on the gel, which indicates that gingipains are comprised of catalytic domains bound to several hemagglutinin/adhesin domains, which remain intact as a protein complex without the treatment of a reducing agent and boiling [35]. In the present investigation, the number of protein bands and their molecular weight was identical to those found in Potempa et al. (1995) [37]. The dissociation of the gingipain’s catalytic domain and hemagglutinin/adhesion domains are thus indicated by the reduced molecular weight and appearance of numerous bands on the SDS-PAGE gel [35].
According to earlier research, gingipains are known to be produced and secreted in various forms depending on the strain of P. gingivalis [22,38]. The arginine-specific gingipain, RgpA is a complex protein with approximately 95 kDa to 110 kDa, which is composed of one catalytic domain (~50 kDa) and several non-covalently bonded adhesin chains (44 kDa, 27 kDa, 17 kDa, and 15 kDa) [39]. RgpA is mostly found in the form of single polypeptide molecule with a molecular weight in the range between 70 kDa to 90 kDa. While RgpB, the other arginine-specific gingipain, can exist either in monomeric form as catalytic domain (~48 kDa) or membrane-associated form as mt-RgpB (~70–90 kDa) [25,26,40,41]. The second major proteinase, Kgp, a lysine-specific gingipain, also consists of one catalytic domain (~60 kDa) and several non-covalently linked hemagglutinin/adhesin domains (44 kDa, 30 kDa, 27 kDa, 17 kDa) [39].
Protein elution from an electrophoresed gel using diffusion methods is a relatively simpler and cost-saving method to extract and purify the target proteins [25,26,40,41]. Native-PAGE is an excellent method for protein isolation because gel electrophoresis is performed under non-denaturing conditions, allowing the native proteins to be recovered after separation [25]. In Native-PAGE for the current study, an eluted protein with the molecular weight of approximately 180 kDa showed the gingipain activity for both Rgp and Kgp. This might be due to both Rgp and Kgp having a nearly identical hydrodynamic sizes and isoelectric point. Additionally, the important factors such as molecular size, ionic charge, and shape of native proteins fully determine their migration rate through the non-denaturing gel [25]. Thus, both arginine-X gingipains and lysine-X gingipains with similar physical and chemical properties will migrate at a similar rate through the gel [29].
From the gelatin zymogram, bands corresponding to either Rgp or Kgp were identified by incorporating the selective inhibitors during the protein sample preparation [34]. Leupeptin inhibitor is a selective inhibitor that specifically inhibits the activity of Rgp as previously described [42] while Cathepsin B inhibitor II is an artificially synthesized tripeptide that specifically inhibits Kgp activity [43]. According to the study of Grudkowska et al. (2003), the universal inhibitor of serine proteinases (e.g., PMSF) will not inhibit the serine proteases’ activities fully because of the short half-life. The incorporation of the universal inhibitor into the sample of gelatin zymogram will cause the reduction of the intensity of the transparent catalytic zone formed on the gel. On the other hand, the addition of an irreversible inhibitor (e.g., E-67) into the sample of gelatin zymogram, which binds to the active site of the enzyme covalently leads to the disappearance of the relevant transparent catalytic zone formed on the gel [44]. In Figure 4, the number of transparent bands that observed in (a) and (b) are different from those in (c) and (d), this might be due to the concentration of proteinase-containing sample loaded in the gel and the overstaining of the gelatin-SDS-polyacrylamide gel led to the loss of the sensitivity [34,36]. The molecular weight of the catalytic bands observed in the gelatin zymogram differed from those observed in SDS-PAGE due to omitting the boiling procedure during the sample preparation to ensure their structure remains intact to preserve their catalytic properties [27]. Triton X-100 was used to restore their catalytic activity because it is non-ionic and can replace the ionic SDS that surround the proteases [27]. This method was employed to characterize the different forms of gingipains because zymogram is a highly sensitive approach to detect the proteinase even in their pro-enzyme state, and due to ionic detergent, SDS treatment will cause conformational changes of proteins, which activates their catalytic activities [28]. Figure 5 outlines the detail the protocol for isolation and characterization of gingipains.
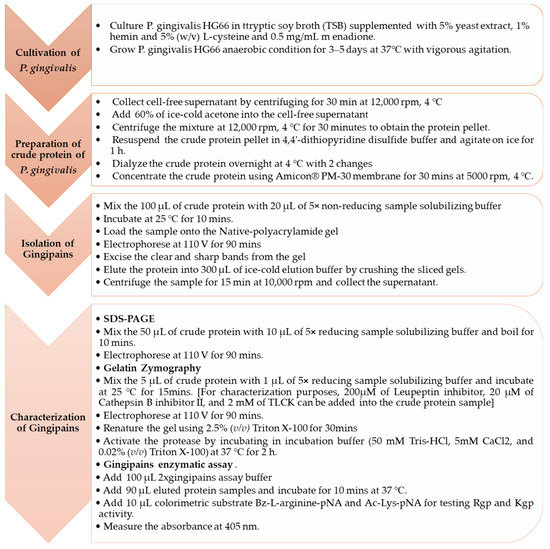
Figure 5.
Detail protocol to isolate and characterize the gingipains.
4.1. Developing and Detailing a Protocol for Isolation and Characterization of Gingipains
4.1.1. Gingipains Preparation
- 1.
- Prepare 500 mL of P. gingivalis HG66 cell-free culture supernatant by centrifuging it for 30 min at 12,000 rpm, 4 °C. Discard the pellet and preserve the supernatant on ice.
- 2.
- Add 750 mL of ice-cold acetone into the brownish supernatant slowly with vigorous agitation.
- 3.
- Obtain the protein pellet by centrifugation for 30 min at 12,000 rpm, 4 °C.
- 4.
- Resuspend the protein pellet with 10 mL of ice-cold 4,4′-dithiopyridine disulfide buffer (DTDS buffer) and incubate on ice for 1 h with agitation.
- 5.
- Transfer the protein into a dialysis bag (MWCO 12,000 to 14,000) and dialyze overnight at 4 °C with 2 changes.
- 6.
- Concentrate the protein with ultrafiltration using Amicon PM-30 membrane for 30 min at 5000 rpm, 4 °C.
- 7.
- Keep the protein sample at −20 °C for future use.
4.1.2. Isolation of Gingipains Using Passive-Diffusion Mediated Gel Elution Method
- Sample preparation
- 1.
- Transfer 100 µL of gingipains sample into the sterile 1.5 mL microcentrifuge tube.
[Note: Sample volume can be increased to obtain more purified protein]
- 2.
- Mix the gingipain sample with 20 µL of 5× non-reducing solubilizing sample buffer (without SDS and β-mercaptoethanol)
- 3.
- Load samples onto Native-polyacrylamide gel.
- Perform Gel electrophoresis
Run the gels at 110 V constantly for 90 min.
[Note: The timing of electrophoresis should be adjusted depending on the samples and gel size]
- Elution of protein
- 1.
- Cut the electrophoresed gel into 3 strips with a sterile blade.
- 2.
- Stain the protein ladder and one sample gel strip with staining buffer for 30 min with gentle agitation
[Note: Stained gel strip used as the reference to locate the target protein]
- 3.
- Cut the target protein bands using the sterile blade and transfer them into the sterile 1.5 mL microcentrifuge tube.
- 4.
- Add 300 µL of ice-cold elution buffer.
- 5.
- Crush the sliced gel with a disposable homogenizer and incubate for overnight, at 4 °C.
- 6.
- Centrifuge the sample for 15 min at 10,000 rpm and collect the supernatant.
- 7.
- Test the eluted proteins with a gingipains activity assay.
- Gingipain activity assay
- 1.
- Prepare the gingipains sample of 20 µL and 50 µL for Rgp and Kgp activity assay, respectively, and dilute the samples with deionized water, making the total volume of 90 µL.
- 2.
- Add 100 µL of 2× gingipain assay buffer to the 96-wells flat bottom microtiter plates.
- 3.
- Add 90 µL of prepared gingipains sample as test group; add 90 µL of deionized water as positive control; add 100 µL of deionized water as blank.
- 4.
- Mix the samples well by pipetting them up and down gently.
- 5.
- Incubate the samples for 10 min at 37 °C.
- 6.
- Add 10 µL colorimetric substrate, Bz-L-arginine-pNA (L-BAPNA), and Ac-Lys-pNA for Rgp and Kgp activity assay respectively.
[Note: For positive control and sample group only]
- 7.
- Measure the absorbance at 405 nm.
4.1.3. Characterization of Gingipains
SDS-PAGE
- Sample preparation
- 1.
- Transfer 50 µL of gingipains sample into the sterile 1.5 mL microcentrifuge tube.
- 2.
- Mix the gingipain sample with 10 µL of 5× non-reducing sample solubilizing buffer and 6 µL of 10% β-mercaptoethanol.
- 3.
- Boil the sample for 10 min at 100 °C.
- 4.
- Load the samples onto SDS-polyacrylamide gel.
- Perform Gel electrophoresis
Run the gels at 110 V constantly for 90 min.
[Note: The timing of electrophoresis should be adjusted depending to the samples and gel size]
- Gel staining
- 1.
- Stain the gel with Coomassie gel staining solution for 30 min at room temperature while agitating it gently.
- 2.
- De-stain the gel with the de-staining solution for 30 min until the clear and sharp, blue-colored bands are visible.
- Gel viewing
View the clear and sharp, blue-colored bands form on the SDS-polyacrylamide gel with UV transilluminator.
Gelatin Zymography
- Sample preparation
- 1.
- Transfer 5 µL of gingipains sample into the sterile 1.5 mL microcentrifuge tube.
[Note: Sample volume varies depending on target protein]
- 2.
- Mix the gingipain sample with 1 µL of 5× non-reducing sample solubilizing buffer (without β-mercaptoethanol)
- 3.
- Add 200 µM of Leupeptin inhibitor (Rgp selective inhibitor), 20 µM of Cathepsin B inhibitor II (Kgp selective inhibitor), and 2 mM of TLCK (general inhibitor) into gingipains samples separately.
[Note: In a condition to determine specific proteases, an inhibitor can be incorporated]
- 4.
- Incubate the samples for 15 min at room temperature.
[Note: Do not heat the sample because it will inactivate the proteinase’s enzymatic activities.]
- 5.
- Load samples onto SDS-gelatin-polyacrylamide gel.
- Perform Gel electrophoresis
Run the gels at 110 V constantly for 90 min.
[Note: The timing of electrophoresis should be adjusted depending to the samples and gel size]
- Gel Renaturation/Incubation
- 1.
- After electrophoresis, remove the gel from the electrophoretic gel cassette carefully.
- 2.
- Place the gel in a clean plastic container and rinse it with distilled water twice for 15 min with gentle agitation.
- 3.
- Equilibrate the gel with 100 mL of renaturation buffer for 30 min at room temperature while agitating it gently.
- 4.
- Incubate and activate the proteases present in the gel with 100 mL of incubation buffer for 2 h at 37 °C.
- Gel Staining
- 1.
- Stain the gel with Coomassie gel staining solution for 30 min at room temperature while agitating it gently.
- 2.
- De-stain the gel with de-staining buffer for 30 min until the transparent and sharp bands are observed.
- Gel viewing
View the clear bands form on the SDS-gelatin-polyacrylamide gel with the UV transilluminator.
4.1.4. Chemicals
Acetone (Merck KGaA, Darmstadt, Germany), Prestained protein standards (GenedireX, Vegas, NV, USA), Coomassie brilliant blue R-250 (Sigma-Aldrich, USA), Acetic acid (Sigma-Aldrich, USA), Ethanol (Sigma-Aldrich, USA), Gelatin (Sigma-Aldrich, USA), Triton X-100 (Sigma-Alrich, USA), Sodium dodecyl sulfate (Biobasic, Toronto, ON, Canada), Tris base (1st Base, Seri Kembangan, Selangor, Malaysia) Acrylamide (Sigma-Aldrich, USA), N,N′-methylene bis(acrylamide) (Sigma-Aldrich, USA), Glycerol (Sigma-Aldrich, USA), Bromophenol blue (Merck, Rahway, NJ, USA), Ammonium persulfate (Bio-Rad, Hercules, CA, USA), Glycine (Bio-Rad, USA), TEMED (Bio-Rad, USA), Sodium chloride (Friendemann Schmidt, Leipzig, Germany), Calcium chloride (Nacalai Tesque, Japan), EDTA (HiMedia, Thane, India), Bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane (J&K Scientific, San Jose, CA, USA), Sodium azide (Sigma-Aldrich, USA), Nα-Benzoyl-L-arginine 4-nitroanilide hydrochloride (Sigma-Aldrich, USA), Ac-Lys-pNA hydrochloride (Bachem, Bubendorf, Switzerland), Leupeptin hemisulfate (Nacalai Tesque, Japan), Cathepsin B inhibitor II (Sigma-Aldrich, USA). TLCK inhibitor (Sigma-Aldrich, USA), DMSO (Sigma-Aldrich, USA).
4.1.5. Equipment
Refrigerated centrifuge (Beckman Coulter, Brea, CA, USA), PowerPacTM Basic Power Supply (Bio-Rad, Vienna, Austria), Mini-PROTEAN® Tetra Cell (Bio-Rad, Vienna, Austria), Water bath (Memmert, Schwalbach, Germany), Orbital shaker (Biosan, Warren, MI, USA) Benchtop micro refrigerated centrifuge (Kubota, Tokyo, Japan), Microplate reader (Tecan, Switzerland, Europe).
5. Conclusions
Gingipains are one of the virulence factors that are crucial for the pathogenesis of P. gingivalis to cause infection in humans. Therefore, the isolation and characterization of gingipains are important to enable researchers to explore and understand the possible invasion mechanisms to cause the development and progression of periodontitis and certain systemic diseases. In the current study, the relatively simple and cost-saving approaches to isolate and characterize the gingipains by using the passive-diffusion mediated elution method and gelatin-zymography were demonstrated. Native-PAGE is an alternative and quick method to isolate the gingipains. Gelatin zymogram is also one of the powerful approaches to characterize and identify the catalytic domains of the gingipains by using selective inhibitors This protocol is suitable and can be extended to isolate and characterize proteins and proteinase samples with a greater difference in their isoelectric point which allows them to separate more efficiently and make ease of purification and characterization of proteins.
Author Contributions
Conceptualization, N.K.F., S.F., M.R. and P.L.; methodology, E.S.W., R.K., M.A.S., N.K.F., S.F., M.R. and P.L.; investigation, E.S.W., M.A.S., R.K., N.K.F., S.F., M.R. and P.L.; resources, E.S.W., M.A.S., R.K., N.K.F., S.F., M.R. and P.L.; data curation, E.S.W., M.A.S., R.K., N.K.F., S.F., M.R. and P.L.; writing—original draft preparation, E.S.W., M.A.S., R.K., N.K.F., S.F., M.R. and P.L.; writing—review and editing, E.S.W., M.A.S., N.K.F., S.F., M.R. and P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Higher Education (MOHE) Malaysia, MyGRANTS, grant number FRGS/1/2018/SKK14/AIMST/01/1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors would like to thank the Ministry of Higher Education (Ref: FRGS/1/2018/SKK14/AIMST/01/1) and AIMST University for financial support and assistance to successfully complete this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nazir, M.A. Prevalence of Periodontal Disease, Its Association with Systemic Diseases and Prevention. Int. J. Health Sci. 2017, 11, 72–80. [Google Scholar]
- Fitzpatrick, R.E.; Wijeyewickrema, L.C.; Pike, R.N. The Gingipains: Scissors and Glue of the Periodontal Pathogen, Porphyromonas gingivalis. Future Microbiol. 2009, 4, 471–487. [Google Scholar] [CrossRef]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Griffin, C.; et al. Porphyromonas gingivalis in Alzheimer’s Disease Brains: Evidence for Disease Causation and Treatment with Small-Molecule Inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Peng, H.-Y.; Chen, S.-Y.; Siao, S.-H.; Chang, J.T.; Xue, T.-Y.; Lee, Y.-H.; Jan, M.-S.; Tsay, G.J.; Zouali, M. Targeting a Cysteine Protease from a Pathobiont Alleviates Experimental Arthritis. Arthritis Res. Ther. 2020, 22, 114. [Google Scholar] [CrossRef]
- Gómez-Bañuelos, E.; Mukherjee, A.; Darrah, E.; Andrade, F. Rheumatoid Arthritis-Associated Mechanisms of Porphyromonas gingivalis and Aggregatibacter Actinomycetemcomitans. J. Clin. Med. 2019, 8, 1309. [Google Scholar] [CrossRef]
- Haraszthy, V.; Zambon, J.; Trevisan, M.; Zeid, M.; Genco, R. Identification of Periodontal Pathogens in Atheromatous Plaques. J. Periodontol. 2000, 71, 1554–1560. [Google Scholar] [CrossRef]
- Szulc, M.; Kustrzycki, W.; Janczak, D.; Michalowska, D.; Baczynska, D.; Radwan-Oczko, M. Presence of Periodontopathic Bacteria DNA in Atheromatous Plaques from Coronary and Carotid Arteries. BioMed Res. Int. 2015, 2015, e825397. [Google Scholar] [CrossRef]
- Bostanci, N.; Belibasakis, G.N. Porphyromonas gingivalis: An Invasive and Evasive Opportunistic Oral Pathogen. FEMS Microbiol. Lett. 2012, 333, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K. Porphyromonas gingivalis and Related Bacteria: From Colonial Pigmentation to the Type IX Secretion System and Gliding Motility. J. Periodontal Res. 2014, 50, 12255. [Google Scholar] [CrossRef]
- Gruss, A.; Borezée-Durant, E.; Lechardeur, D. Chapter Three—Environmental Heme Utilization by Heme-Auxotrophic Bacteria. In Advances in Microbial Physiology; Poole, R.K., Ed.; Advances in Bacterial Respiratory Physiology; Academic Press: Cambridge, MA, USA, 2012; Volume 61, pp. 69–124. [Google Scholar]
- Zhang, Z.; Liu, D.; Liu, S.; Zhang, S.; Pan, Y. The Role of Porphyromonas gingivalis Outer Membrane Vesicles in Periodontal Disease and Related Systemic Diseases. Front. Cell. Infect. Microbiol. 2021, 10, 585917. [Google Scholar] [CrossRef]
- Singhrao, S.K.; Harding, A.; Poole, S.; Kesavalu, L.; Crean, S. Porphyromonas gingivalis Periodontal Infection and Its Putative Links with Alzheimer’s Disease. Mediat. Inflamm. 2015, 2015, 137357. [Google Scholar] [CrossRef]
- Kavitha, R.; Sa’ad, M.A.; Fuloria, S.; Fuloria, N.K.; Ravichandran, M.; Lalitha, P. Synthesis, Characterization, Cytotoxicity Analysis and Evaluation of Novel Heterocyclic Derivatives of Benzamidine against Periodontal Disease Triggering Bacteria. Antibiotics 2023, 12, 306. [Google Scholar] [CrossRef]
- Bao, K.; Belibasakis, G.N.; Thurnheer, T.; Aduse-Opoku, J.; Curtis, M.A.; Bostanci, N. Role of Porphyromonas gingivalis Gingipains in Multi-Species Biofilm Formation. BMC Microbiol. 2014, 14, 258. [Google Scholar] [CrossRef]
- Imamura, T. The Role of Gingipains in the Pathogenesis of Periodontal Disease. J. Periodontol. 2003, 74, 111–118. [Google Scholar] [CrossRef]
- Kadowaki, T. Enzymatic Characteristics and Activities of Gingipains from Porphyromonas gingivalis. Methods Mol. Biol. 2021, 2210, 97–112. [Google Scholar] [CrossRef]
- Veillard, F.; Sztukowska, M.; Nowakowska, Z.; Mizgalska, D.; Thøgersen, I.B.; Enghild, J.J.; Bogyo, M.; Potempa, B.; Nguyen, K.-A.; Potempa, J. Proteolytic Processing and Activation of Gingipain Zymogens Secreted by T9SS of Porphyromonas gingivalis. Biochimie 2019, 166, 161–172. [Google Scholar] [CrossRef]
- Sugawara, S.; Nemoto, E.; Tada, H.; Miyake, K.; Imamura, T.; Takada, H. Proteolysis of Human Monocyte CD14 by Cysteine Proteinases (Gingipains) from Porphyromonas gingivalis Leading to Lipopolysaccharide Hyporesponsiveness. J. Immunol. 2000, 165, 411–418. [Google Scholar] [CrossRef]
- Jia, L.; Han, N.; Du, J.; Guo, L.; Luo, Z.; Liu, Y. Pathogenesis of Important Virulence Factors of Porphyromonas gingivalis via Toll-Like Receptors. Front. Cell. Infect. Microbiol. 2019, 9, 262. [Google Scholar] [CrossRef]
- Chen, Z.; Potempa, J.; Polanowski, A.; Wikstrom, M.; Travis, J. Purification and Characterization of a 50-KDa Cysteine Proteinase (Gingipain) from Porphyromonas gingivalis. J. Biol. Chem. 1992, 267, 18896–18901. [Google Scholar] [CrossRef] [PubMed]
- Potempa, J.; Nguyen, K.-A. Purification and Characterization of Gingipains. Curr. Protoc. Protein. Sci. 2007, 21, Unit 21.20. [Google Scholar] [CrossRef]
- McGraw, W.T.; Potempa, J.; Farley, D.; Travis, J. Purification, Characterization, and Sequence Analysis of a Potential Virulence Factor from Porphyromonas gingivalis, Peptidylarginine Deiminase. Infect. Immun. 1999, 67, 3248–3256. [Google Scholar] [CrossRef]
- Pavloff, N.; Pemberton, P.A.; Potempa, J.; Chen, W.C.; Pike, R.N.; Prochazka, V.; Kiefer, M.C.; Travis, J.; Barr, P.J. Molecular Cloning and Characterization of Porphyromonas gingivalis Lysine-Specific Gingipain. A New Member of an Emerging Family of Pathogenic Bacterial Cysteine Proteinases. J. Biol. Chem. 1997, 272, 1595–1600. [Google Scholar] [CrossRef]
- Margetts, M.B.; Barr, I.G.; Webb, E.A. Overexpression, Purification, and Refolding of a Porphyromonas gingivalis Cysteine Protease from Escherichia Coli. Protein Expr. Purif. 2000, 18, 262–268. [Google Scholar] [CrossRef]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bachmann, M. Native Polyacrylamide Gels. In Electrophoretic Separation of Proteins: Methods and Protocols; Kurien, B.T., Scofield, R.H., Eds.; Methods in Molecular Biology; Springer: New York, NY, 2019; pp. 87–91. ISBN 978-1-4939-8793-1. [Google Scholar]
- Burgess, R.R. Elution of Proteins from Gels. Methods Enzym. 2009, 463, 565–572. [Google Scholar] [CrossRef]
- Toth, M.; Fridman, R. Assessment of Gelatinases (MMP-2 and MMP-9) by Gelatin Zymography. Methods Mol. Med. 2001, 57, 163. [Google Scholar] [CrossRef]
- Hashimoto, Y. Gelatin Zymography Using Leupeptin for the Detection of Various Cathepsin L Forms. Methods Mol. Biol. 2017, 1594, 243–254. [Google Scholar] [CrossRef]
- Potempa, J.; Mikolajczyk-Pawlinska, J.; Brassell, D.; Nelson, D.; Thøgersen, I.B.; Enghild, J.J.; Travis, J. Comparative Properties of Two Cysteine Proteinases (Gingipains R), the Products of Two Related but Individual Genes of Porphyromonas Gingivalis. J. Biol. Chem. 1998, 273, 21648–21657. [Google Scholar] [CrossRef]
- Platt, M.O.; Ankeny, R.F.; Jo, H. Laminar Shear Stress Inhibits Cathepsin L Activity in Endothelial Cells. Arter. Thromb. Vasc. Biol. 2006, 26, 1784–1790. [Google Scholar] [CrossRef]
- Jung, Y.-J.; Jun, H.-K.; Choi, B.-K. Porphyromonas Gingivalis Suppresses Invasion of Fusobacterium Nucleatum into Gingival Epithelial Cells. J. Oral Microbiol. 2017, 9, 1320193. [Google Scholar] [CrossRef]
- Mei, F.; Xie, M.; Huang, X.; Long, Y.; Lu, X.; Wang, X.; Chen, L. Porphyromonas gingivalis and Its Systemic Impact: Current Status. Pathogens 2020, 9, 944. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L. Porphyromonas gingivalis. Trends Microbiol. 2021, 29, 376–377. [Google Scholar] [CrossRef]
- Leonard, A.K.; Loughran, E.A.; Klymenko, Y.; Liu, Y.; Kim, O.; Asem, M.; McAbee, K.; Ravosa, M.J.; Stack, M.S. Chapter 4—Methods for the Visualization and Analysis of Extracellular Matrix Protein Structure and Degradation. In Methods in Cell Biology; Mecham, R.P., Ed.; Methods in Extracellular Matrix Biology; Academic Press: Cambridge, MA, USA, 2018; Volume 143, pp. 79–95. [Google Scholar]
- Ciborowski, P.; Nishikata, M.; Allen, R.D.; Lantz, M.S. Purification and Characterization of Two Forms of a High-Molecular-Weight Cysteine Proteinase (Porphypain) from Porphyromonas gingivalis. J. Bacteriol. 1994, 176, 4549–4557. [Google Scholar] [CrossRef]
- Hočevar, K.; Vizovišek, M.; Wong, A.; Kozieł, J.; Fonović, M.; Potempa, B.; Lamont, R.J.; Potempa, J.; Turk, B. Proteolysis of Gingival Keratinocyte Cell Surface Proteins by Gingipains Secreted From Porphyromonas gingivalis—Proteomic Insights Into Mechanisms Behind Tissue Damage in the Diseased Gingiva. Front. Microbiol. 2020, 11, 722. [Google Scholar] [CrossRef]
- Potempa, J.; Pike, R.; Travis, J. The Multiple Forms of Trypsin-like Activity Present in Various Strains of Porphyromonas gingivalis Are Due to the Presence of Either Arg-Gingipain or Lys-Gingipain. Infect. Immun. 1995, 63, 1176–1182. [Google Scholar] [CrossRef]
- Sa’ad, M.A.; Kavitha, R.; Fuloria, N.K.; Fuloria, S.; Ravichandran, M.; Lalitha, P. The virulence system of Porphyromonas gingivalis: Genes, mechanism and potential role of gingipains inhibitors. Malays. J. Microbiol. 2021, 17, 212–226. [Google Scholar] [CrossRef]
- Curtis, M.A.; Aduse-Opoku, J.; Rangarajan, M. Cysteine Proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 2001, 12, 192–216. [Google Scholar] [CrossRef]
- Kim, R. Native Agarose Gel Electrophoresis of Multiprotein Complexes. Cold Spring Harb. Protoc. 2011, 2011, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Kurien, B.T.; Scofield, R.H. Extraction of Proteins from Gels—A Brief Review. Methods Mol. Biol. 2012, 869, 403–405. [Google Scholar] [CrossRef]
- Hosn, K.N.; Jefferson, M.M.; Leding, C.; Shokouh-Amiri, S.; Thomas, E.L. Inhibitors of Bacterial Protease Enzymes for Periodontal Therapy. Clin. Exp. Dent. Res. 2015, 1, 18–25. [Google Scholar] [CrossRef]
- Grenier, D.; Imbeault, S.; Plamondon, P.; Grenier, G.; Nakayama, K.; Mayrand, D. Role of Gingipains in Growth of Porphyromonas gingivalis in the Presence of Human Serum Albumin. Infect. Immun. 2001, 69, 5166–5172. [Google Scholar] [CrossRef] [PubMed]
- Grudkowska, M.; Lisik, P.; Rybka, K. Two-Dimensional Zymography in Detection of Proteolytic Enzymes in Wheat Leaves. Acta Physiol. Plant 2013, 35, 3477–3482. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).