Comparison of Algorithms to Compute Relaxation Time Maps in Magnetic Resonance Imaging
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
- Inversion recovery (IR) images from a healthy mouse.
- A series of MRI human brain images, simulated using a web-based software available at http://brainweb.bic.mni.mcgill.ca/brainweb (accessed on 10 March 2018) [27,28].
- Multi-slice multi-echo spin-echo images of a phantom consisting of scaffolds in which cells were grown, filled with phosphate-buffered saline.
- Multiple minima: number of pixels (as a percentage of total pixels) for which the fitting error as a function of Trel after linear parameters have been fitted shows more than one local minimum.
- Pixels: The number of pixels (as a percentage of total pixels), for which the Trel difference between the two fits is above a threshold and εa < εb. No pixels for which the Trel difference between the two fits is above the threshold and εa > εb or εa = εb were found. Therefore, the remaining pixels (up to 100%) are pixels for which the Trel difference between the two fits is below the threshold. The threshold exists because if it did not exist, it could be possible to conclude that the same solution is two different solutions due only to rounding errors. The threshold must be small enough not to allow two different minima to be confused. For these reasons, it was set arbitrarily as 1 ms.
- Relaxation time: Average Trel value given by fit a (the RD-NLS algorithm) and fit b (the algorithm it is compared to).
3. Results
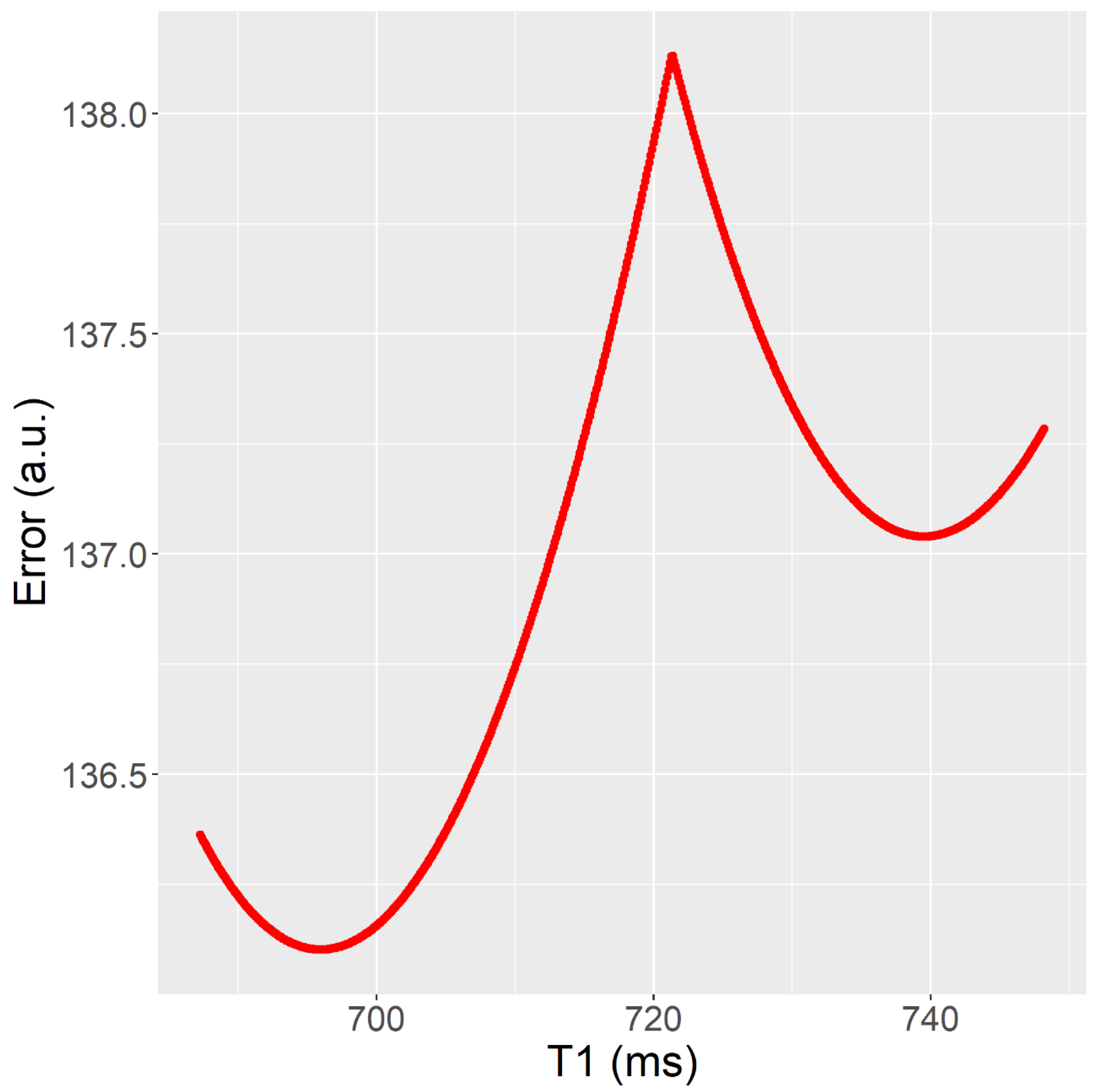
- The T1 fits (Tests 1–10) show many pixels in which error vs. T1 has more than one local minimum, in the 9–70% range. Having more than one local minimum means that a local search algorithm could get trapped in a local minimum which is not a global minimum (see Figure 3). This is not the case for the T2 fits (Tests 11–14), where error vs. T2 has only one local minimum.
- The Trel value obtained by the RD-NLS algorithm always corresponds to the global optimum.
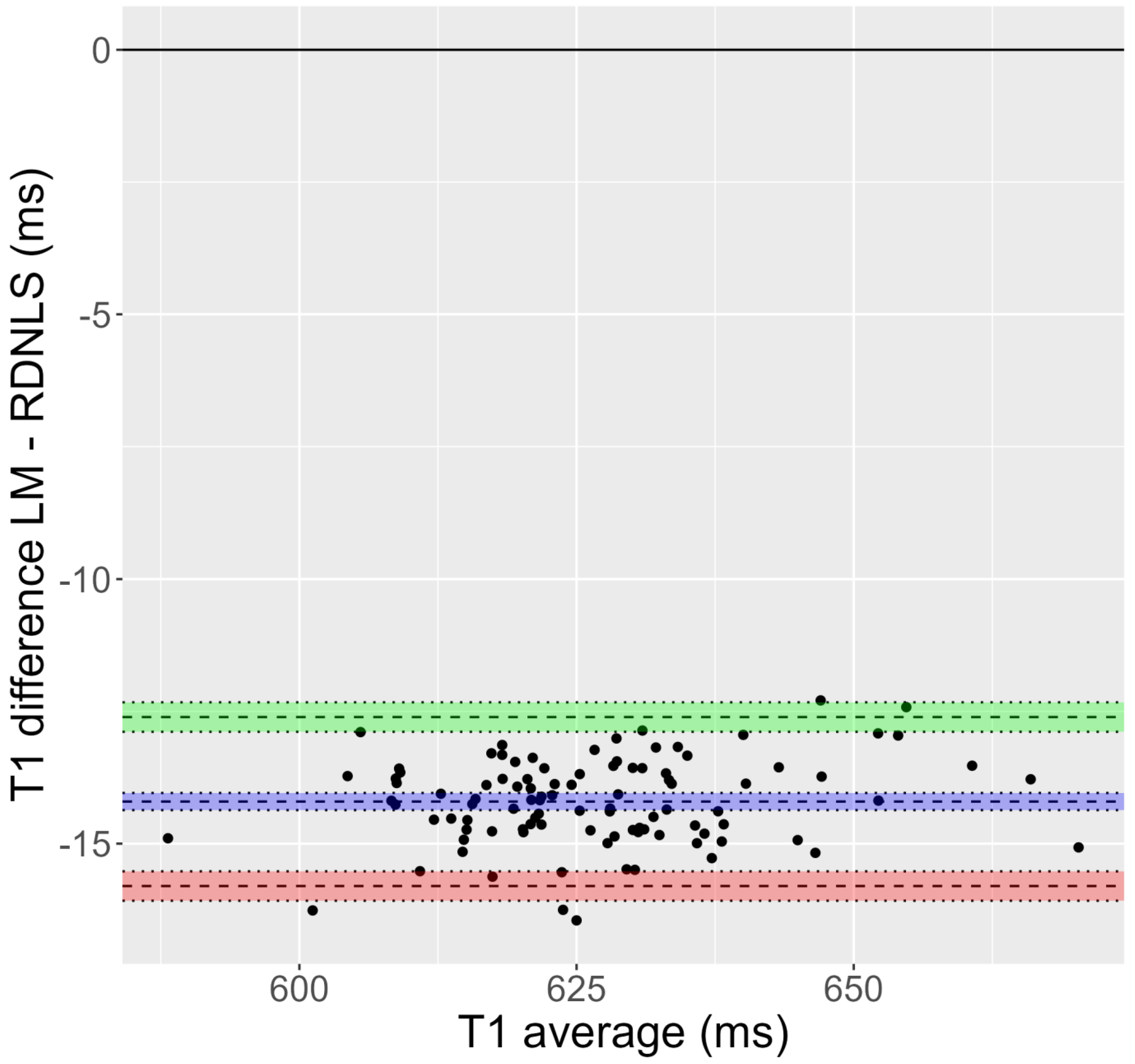
- Comparing Bruker and Matlab fit versus RD-NLS for the T1 maps (Tests 1–10), the fit is different in a significant number of pixels, in the range of 5–97%. However, this is not true for the T2 maps (Tests 11–14).
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kellman, P.; Hansen, M.S. T1-Mapping in the Heart: Accuracy and Precision. J. Cardiovasc. Magn. Reson. Off. J. Soc. Cardiovasc. Magn. Reson. 2014, 16, 2. [Google Scholar] [CrossRef]
- Ugander, M.; Bagi, P.S.; Oki, A.J.; Chen, B.; Hsu, L.-Y.; Aletras, A.H.; Shah, S.; Greiser, A.; Kellman, P.; Arai, A.E. Myocardial Edema as Detected by Pre-Contrast T1 and T2 CMR Delineates Area at Risk Associated with Acute Myocardial Infarction. JACC Cardiovasc. Imaging 2012, 5, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Lescher, S.; Jurcoane, A.; Veit, A.; Bähr, O.; Deichmann, R.; Hattingen, E. Quantitative T1 and T2 Mapping in Recurrent Glioblastomas under Bevacizumab: Earlier Detection of Tumor Progression Compared to Conventional MRI. Neuroradiology 2015, 57, 11–20. [Google Scholar] [CrossRef]
- Truhn, D.; Sondern, B.; Oehrl, S.; Tingart, M.; Knobe, M.; Merhof, D.; Kuhl, C.; Thüring, J.; Nebelung, S. Differentiation of Human Cartilage Degeneration by Functional MRI Mapping-an Ex Vivo Study. Eur. Radiol. 2019, 29, 6671–6681. [Google Scholar] [CrossRef]
- Gracien, R.-M.; Nürnberger, L.; Hok, P.; Hof, S.-M.; Reitz, S.C.; Rüb, U.; Steinmetz, H.; Hilker-Roggendorf, R.; Klein, J.C.; Deichmann, R.; et al. Evaluation of Brain Ageing: A Quantitative Longitudinal MRI Study over 7 Years. Eur. Radiol. 2017, 27, 1568–1576. [Google Scholar] [CrossRef]
- Mariette, F.; Collewet, G.; Davenel, A.; Lucas, T.; Musse, M. Quantitative MRI in Food Science & Food Engineering. In Encyclopedia of Magnetic Resonance; Harris, R.K., Ed.; John Wiley & Sons, Ltd: Chichester, UK, 2012; p. emrstm1272. ISBN 978-0-470-03459-0. [Google Scholar]
- Bidhult, S.; Kantasis, G.; Aletras, A.H.; Arheden, H.; Heiberg, E.; Hedström, E. Validation of T1 and T2 Algorithms for Quantitative MRI: Performance by a Vendor-Independent Software. BMC Med. Imaging 2016, 16, 46. [Google Scholar] [CrossRef]
- Messroghli, D.R.; Moon, J.C.; Ferreira, V.M.; Grosse-Wortmann, L.; He, T.; Kellman, P.; Mascherbauer, J.; Nezafat, R.; Salerno, M.; Schelbert, E.B.; et al. Clinical Recommendations for Cardiovascular Magnetic Resonance Mapping of T1, T2, T2* and Extracellular Volume: A Consensus Statement by the Society for Cardiovascular Magnetic Resonance (SCMR) Endorsed by the European Association for Cardiovascular Imaging (EACVI). J. Cardiovasc. Magn. Reson. Off. J. Soc. Cardiovasc. Magn. Reson. 2017, 19, 75. [Google Scholar] [CrossRef]
- Salerno, M.; Kramer, C.M. Advances in Parametric Mapping with CMR Imaging. JACC Cardiovasc. Imaging 2013, 6, 806–822. [Google Scholar] [CrossRef]
- Taylor, A.J.; Salerno, M.; Dharmakumar, R.; Jerosch-Herold, M. T1 Mapping: Basic Techniques and Clinical Applications. JACC Cardiovasc. Imaging 2016, 9, 67–81. [Google Scholar] [CrossRef]
- Brown, R.W.; Cheng, Y.-C.N.; Haacke, E.M.; Thompson, M.R.; Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; ISBN 978-0-471-72085-0. [Google Scholar]
- Vlaardingerbroek, M.T.; den Boer, J.A.; Luiten, A. Magnetic Resonance Imaging: Theory and Practice; with a Historical Introduction by André Luiten; with 57 Image Sets; Physics and Astronomy online Library, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 978-3-642-07823-1. [Google Scholar]
- Bernstein, M.A.; King, K.F.; Zhou, X.J. Handbook of MRI Pulse Sequences; Elsevier Academic Press: Amsterdam, The Netherlands, 2004; ISBN 978-0-12-092861-3. [Google Scholar]
- Deoni, S.C.L.; Rutt, B.K.; Peters, T.M. Rapid Combined T1 and T2 Mapping Using Gradient Recalled Acquisition in the Steady State. Magn. Reson. Med. 2003, 49, 515–526. [Google Scholar] [CrossRef]
- Deoni, S.C.L.; Peters, T.M.; Rutt, B.K. Determination of Optimal Angles for Variable Nutation Proton Magnetic Spin-Lattice, T1, and Spin-Spin, T2, Relaxation Times Measurement. Magn. Reson. Med. 2004, 51, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Pirastru, A.; Chen, Y.; Pelizzari, L.; Baglio, F.; Clerici, M.; Haacke, E.M.; Laganà, M.M. Quantitative MRI Using STrategically Acquired Gradient Echo (STAGE): Optimization for 1.5 T Scanners and T1 Relaxation Map Validation. Eur. Radiol. 2021, 31, 4504–4513. [Google Scholar] [CrossRef] [PubMed]
- Haacke, E.M.; Chen, Y.; Utriainen, D.; Wu, B.; Wang, Y.; Xia, S.; He, N.; Zhang, C.; Wang, X.; Lagana, M.M.; et al. STrategically Acquired Gradient Echo (STAGE) Imaging, Part III: Technical Advances and Clinical Applications of a Rapid Multi-Contrast Multi-Parametric Brain Imaging Method. Magn. Reson. Imaging 2020, 65, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Llorden, G.; Vegas-Sanchez-Ferrero, G.; Bjork, M.; Vanhevel, F.; Parizel, P.M.; San Jose Estepar, R.; den Dekker, A.J.; Sijbers, J. NOVIFAST: A Fast Algorithm for Accurate and Precise VFA MRI Mapping. IEEE Trans. Med. Imaging 2018, 37, 2414–2427. [Google Scholar] [CrossRef] [PubMed]
- Levenberg, K. A Method for the Solution of Certain Non-Linear Problems in Least Squares. Q. Appl. Math. 1944, 2, 164–168. [Google Scholar] [CrossRef]
- Marquardt, D. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. SIAM J. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Nelder, J.A.; Mead, R. A Simplex Method for Function Minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
- Golub, G.H.; Pereyra, V. The Differentiation of Pseudo-Inverses and Nonlinear Least Squares Problems Whose Variables Separate. SIAM J. Numer. Anal. 1973, 10, 413–432. [Google Scholar] [CrossRef]
- Nonlinear Data-Fitting—MATLAB & Simulink—MathWorks. Available online: https://www.mathworks.com/help/optim/ug/nonlinear-data-fitting-example.html#datdemo-4 (accessed on 6 November 2020).
- Barral, J.K.; Gudmundson, E.; Stikov, N.; Etezadi-Amoli, M.; Stoica, P.; Nishimura, D.G. A Robust Methodology for in Vivo T1 Mapping. Magn. Reson. Med. 2010, 64, 1057–1067. [Google Scholar] [CrossRef]
- Messroghli, D.R.; Radjenovic, A.; Kozerke, S.; Higgins, D.M.; Sivananthan, M.U.; Ridgway, J.P. Modified Look-Locker Inversion Recovery (MOLLI) for High-Resolution T1 Mapping of the Heart. Magn. Reson. Med. 2004, 52, 141–146. [Google Scholar] [CrossRef]
- Nekolla, S.; Gneiting, T.; Syha, J.; Deichmann, R.; Haase, A. T1 Maps by K-Space Reduced Snapshot-FLASH MRI. J. Comput. Assist. Tomogr. 1992, 16, 327–332. [Google Scholar] [PubMed]
- Kwan, R.K.; Evans, A.C.; Pike, G.B. MRI Simulation-Based Evaluation of Image-Processing and Classification Methods. IEEE Trans. Med. Imaging 1999, 18, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.L.; Zijdenbos, A.P.; Kollokian, V.; Sled, J.G.; Kabani, N.J.; Holmes, C.J.; Evans, A.C. Design and Construction of a Realistic Digital Brain Phantom. IEEE Trans. Med. Imaging 1998, 17, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M.; Altman, D.G. Statistical Methods for Assessing Agreement between Two Methods of Clinical Measurement. Lancet Lond. Engl. 1986, 1, 307–310. [Google Scholar]
- Kim, Y.-C.; Kim, K.R.; Lee, H.; Choe, Y.H. Fast Calculation Software for Modified Look-Locker Inversion Recovery (MOLLI) T1 Mapping. BMC Med. Imaging 2021, 21, 26. [Google Scholar] [CrossRef]

| Name | Equation |
|---|---|
| I. Inversion Recovery | |
| II. Inversion Recovery or MOLLI | |
| III. Simplified Spin Echo or Simplified Gradient Echo |
| Test Number | Test Model | Algorithm to Compare | Software to Compare | Data |
|---|---|---|---|---|
| 1 | I | Levenberg-Marquardt | ISA | mouse |
| 2 | I | Levenberg-Marquardt | Matlab | mouse |
| 3 | I | Nelder-Mead | Matlab | mouse |
| 4 | I | Split | Matlab | mouse |
| 5 | II | Levenberg-Marquardt | Matlab | mouse |
| 6 | II | Nelder-Mead | Matlab | mouse |
| 7 | II | Split | Matlab | mouse |
| 8 | II | Levenberg-Marquardt | Matlab | simulated |
| 9 | II | Nelder-Mead | Matlab | simulated |
| 10 | II | Split | Matlab | simulated |
| 11 | III | Levenberg-Marquardt | ISA | phantom |
| 12 | III | Levenberg-Marquardt | Matlab | phantom |
| 13 | III | Nelder-Mead | Matlab | phantom |
| 14 | III | Split | Matlab | phantom |
| Test Number | Minima (%) | Pixels | Relaxation Time (ms) |
|---|---|---|---|
| 1 | 9.2% | 97.0% | 622/636 |
| 2 | 9.2% | 4.1% | 622/623 |
| 3 | 9.2% | 0.6% | 622/622 |
| 4 | 9.2% | 4.1% | 622/623 |
| 5 | 18.6% | 4.8% | 807/805 |
| 6 | 18.6% | 2.0% | 807/807 |
| 7 | 18.6% | 2.7% | 807/806 |
| 8 | 68.1% | 15.7% | 823/842 |
| 9 | 68.1% | 10.3% | 823/841 |
| 10 | 68.1% | 10.8% | 823/823 |
| 11 | 0.0% | 0.2% | 113/113 |
| 12 | 0.0% | 0.0% | 113/113 |
| 13 | 0.0% | 0.0% | 113/113 |
| 14 | 0.0% | 0.2% | 113/113 |
| Test Number | Bias (ms) | Limits of Agreement (ms) |
|---|---|---|
| 1 | 14.07 | −1.63 to 29.76 |
| 2 | 0.88 | −13.11 to 14.86 |
| 3 | 0.07 | −5.22 to 5.36 |
| 4 | 0.92 | −13.19 to 15.03 |
| 5 | −1.87 | −27.21 to 23.47 |
| 6 | −0.61 | −19.67 to 18.45 |
| 7 | −1.32 | −22.08 to 19.45 |
| 8 | 18.20 | −497.54 to 533.95 |
| 9 | 17.34 | −472.89 to 507.57 |
| 10 | −0.63 | −449.79 to 448.53 |
| 11 | −0.01 | −3.32 to 3.31 |
| 12 | 0.01 | −2.76 to 2.78 |
| 13 | 0.01 | −2.75 to 2.78 |
| 14 | 0.02 | −2.82 to 2.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez, I.; Izquierdo-Garcia, J.L.; Yazdanparast, E.; Castejón, D.; Ruiz-Cabello, J. Comparison of Algorithms to Compute Relaxation Time Maps in Magnetic Resonance Imaging. Appl. Sci. 2023, 13, 4083. https://doi.org/10.3390/app13074083
Rodriguez I, Izquierdo-Garcia JL, Yazdanparast E, Castejón D, Ruiz-Cabello J. Comparison of Algorithms to Compute Relaxation Time Maps in Magnetic Resonance Imaging. Applied Sciences. 2023; 13(7):4083. https://doi.org/10.3390/app13074083
Chicago/Turabian StyleRodriguez, Ignacio, Jose Luis Izquierdo-Garcia, Ehsan Yazdanparast, David Castejón, and Jesús Ruiz-Cabello. 2023. "Comparison of Algorithms to Compute Relaxation Time Maps in Magnetic Resonance Imaging" Applied Sciences 13, no. 7: 4083. https://doi.org/10.3390/app13074083
APA StyleRodriguez, I., Izquierdo-Garcia, J. L., Yazdanparast, E., Castejón, D., & Ruiz-Cabello, J. (2023). Comparison of Algorithms to Compute Relaxation Time Maps in Magnetic Resonance Imaging. Applied Sciences, 13(7), 4083. https://doi.org/10.3390/app13074083







