The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Collection of the Samples and the Isolation of the Bacteria
2.3. Screening of the Bacterial Isolates for PET Degradation
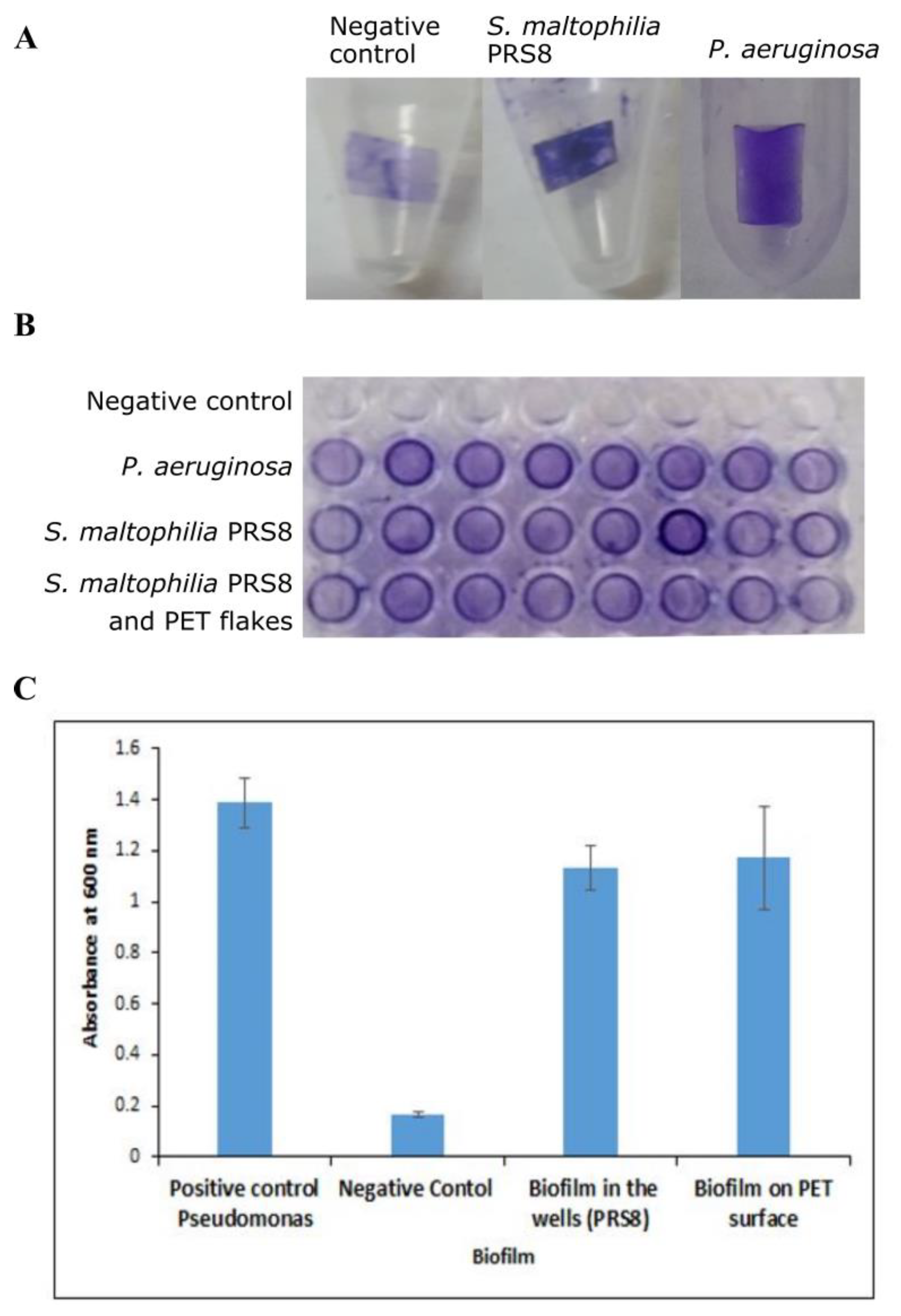
2.4. Biofilm Assay for Bacterial Strain
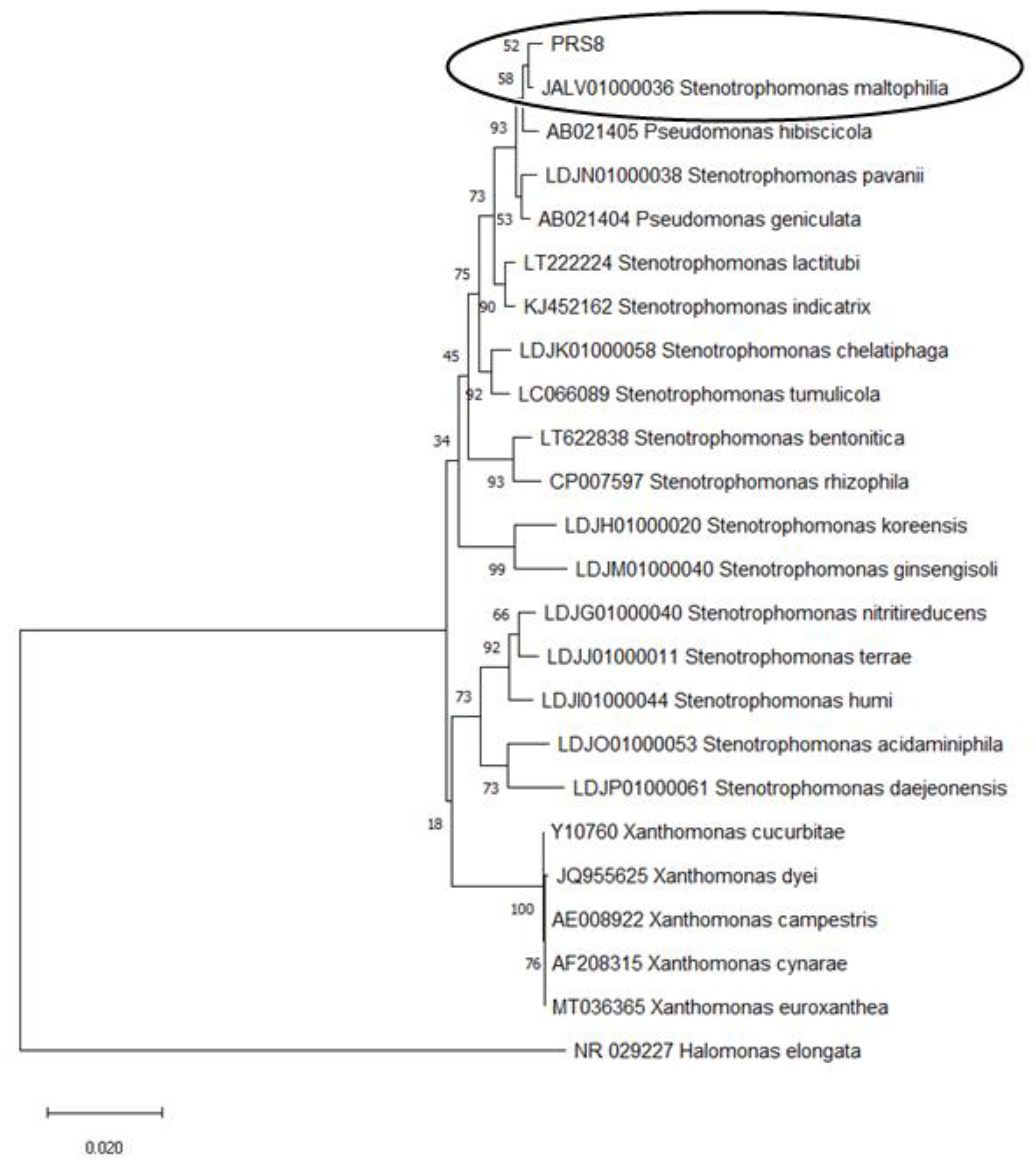
2.5. Identification of the Bacterial Strain PRS8
2.6. Polycaprolactone Agar for Cutinase Activity
2.7. Biodegradation of PET by S. maltophilia PRS8
2.8. Analysis of the Degradation of PET by S. maltophilia PRS8
2.8.1. Determination of the Dry Weight of the Residual PET
2.8.2. Fourier Transform Infrared (FTIR) Spectroscopy
2.8.3. Scanning Electron Microscopy (SEM)
2.8.4. Preparation of the PET Powder
2.9. Standard Assay of the Esterase/Cutinase Activity
2.10. Optimization of the Physicochemical Parameters for the Cutinase Production in S. maltophilia PRS8
2.11. Bulk Production of the Crude Cutinase-like Enzyme
2.12. Purification of the Cutinase-like Enzyme
2.13. Zymography
2.14. Effect of the Temperature and pH on the Cutinase Activity and Stability
2.15. Effect of the Metals Ions
2.16. Effect of the Organic Solvents and the Surfactant
2.17. Enzymatic Depolymerization of the PET Flakes
2.18. Analysis of the Enzymatic Depolymerization Products
2.19. Specifications for the LC-MS Equipment
2.20. DSC Analysis of the Residual PET
3. Results
3.1. Isolation and Identification of the PET-degrading Bacteria S. maltophilia PRS8
3.2. Biofilm Assay of S. maltophilia PRS8
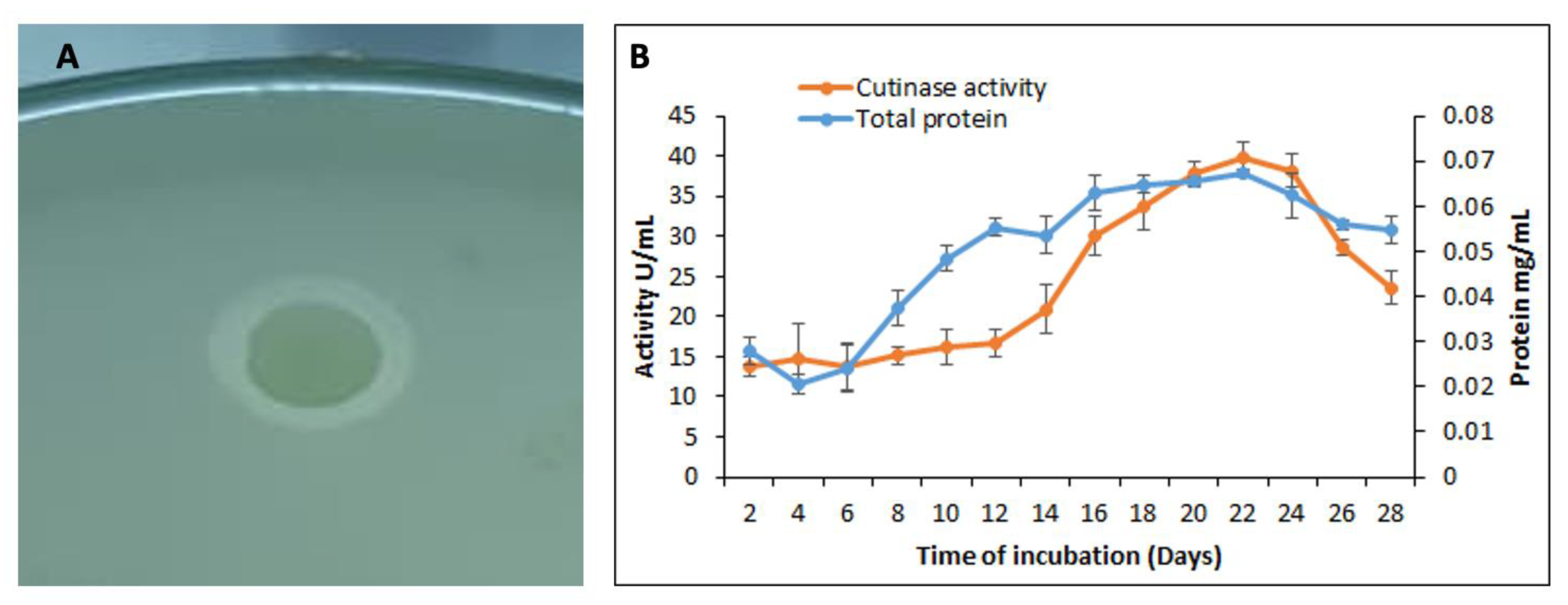
3.3. Qualitative and Quantitative Assay for the Cutinase Activity by S. maltophilia PRS8
3.4. Analysis of PET Depolymerization by S. maltophilia PRS8
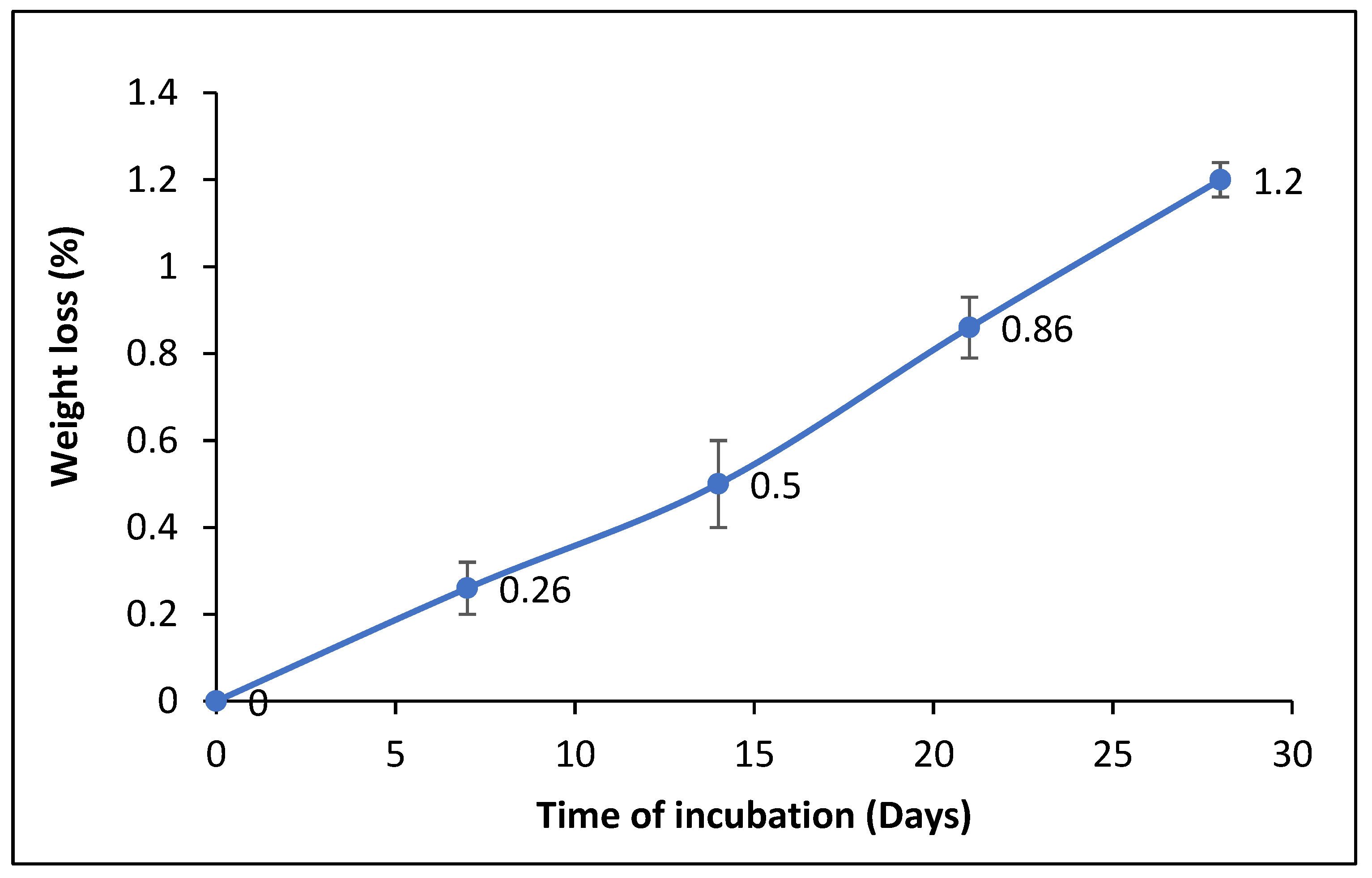
3.4.1. Determination of the Dry Weight of the Residual PET after Film Degradation
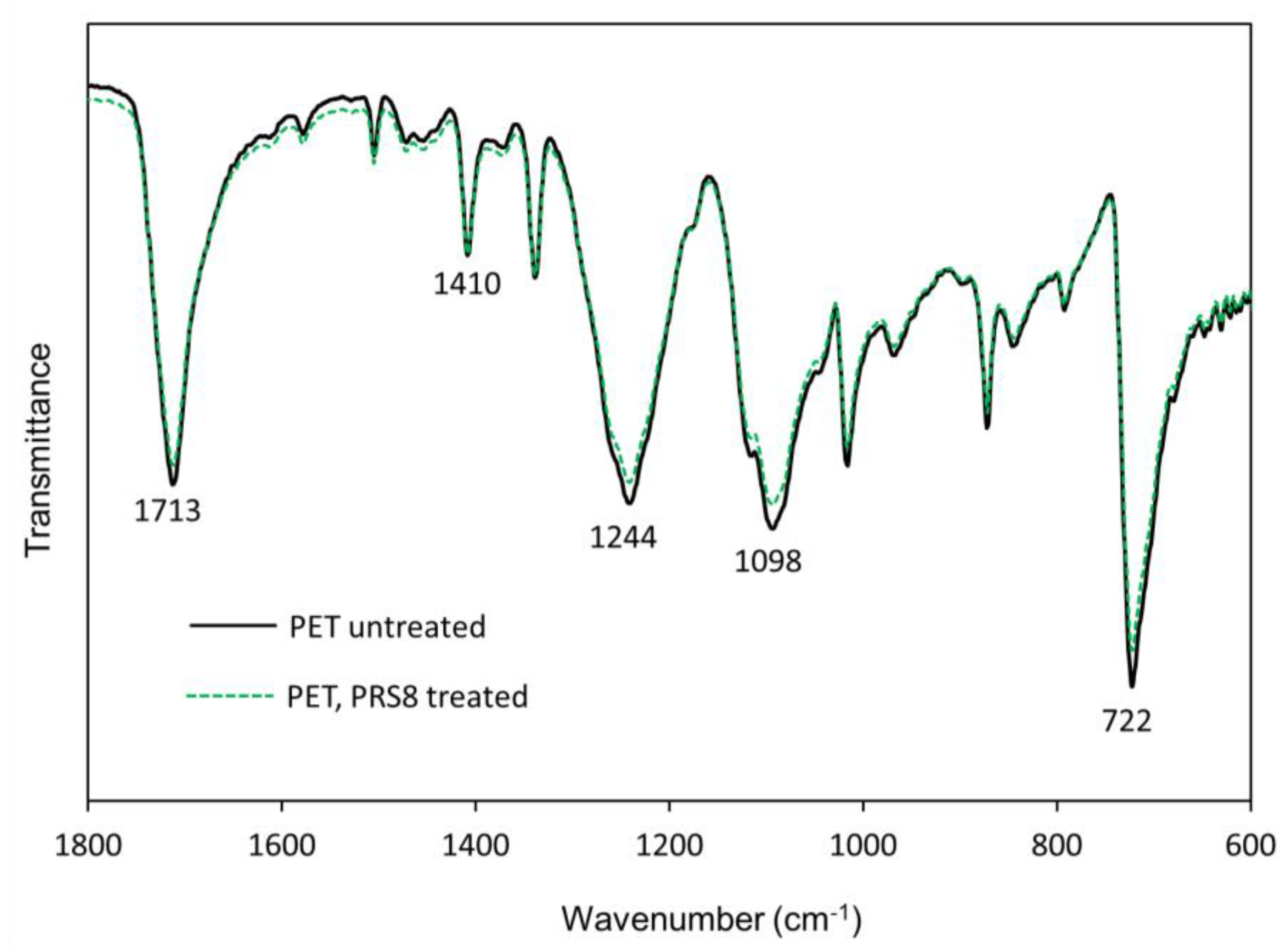
3.4.2. Fourier-Transform Infrared (FTIR) Spectroscopy
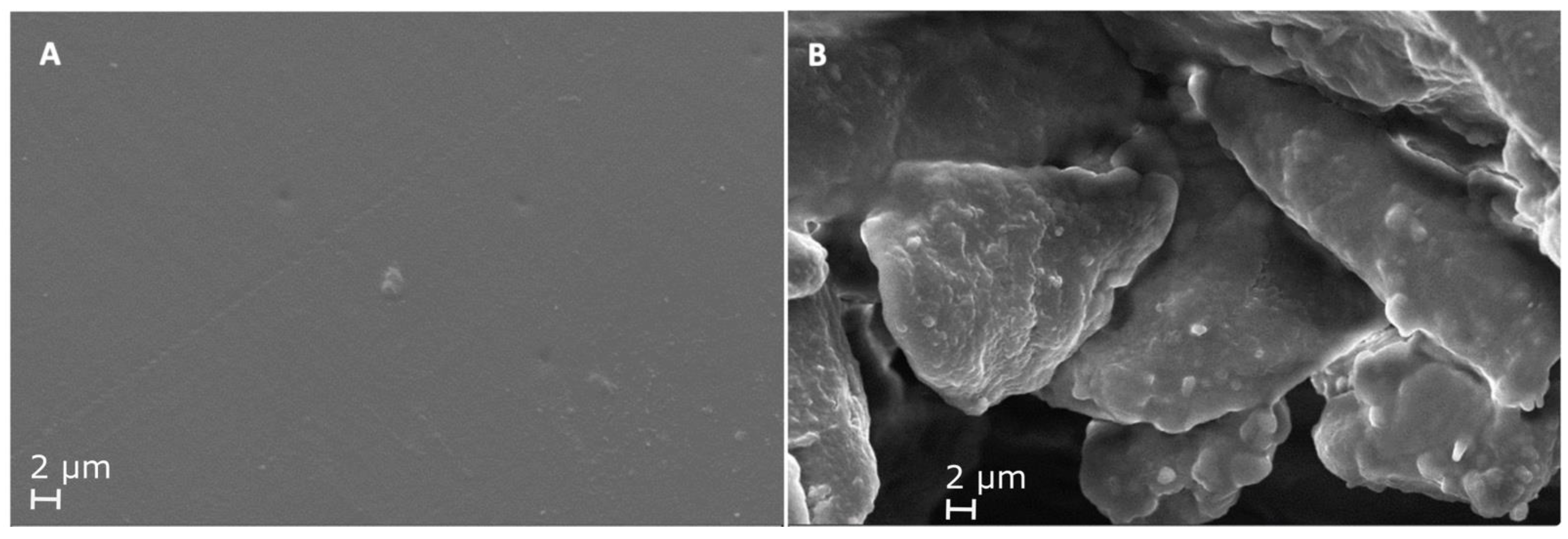
3.4.3. Scanning Electron Microscopy (SEM)
3.5. Parameter Optimization for Enzyme Production by S. maltophila PRS8
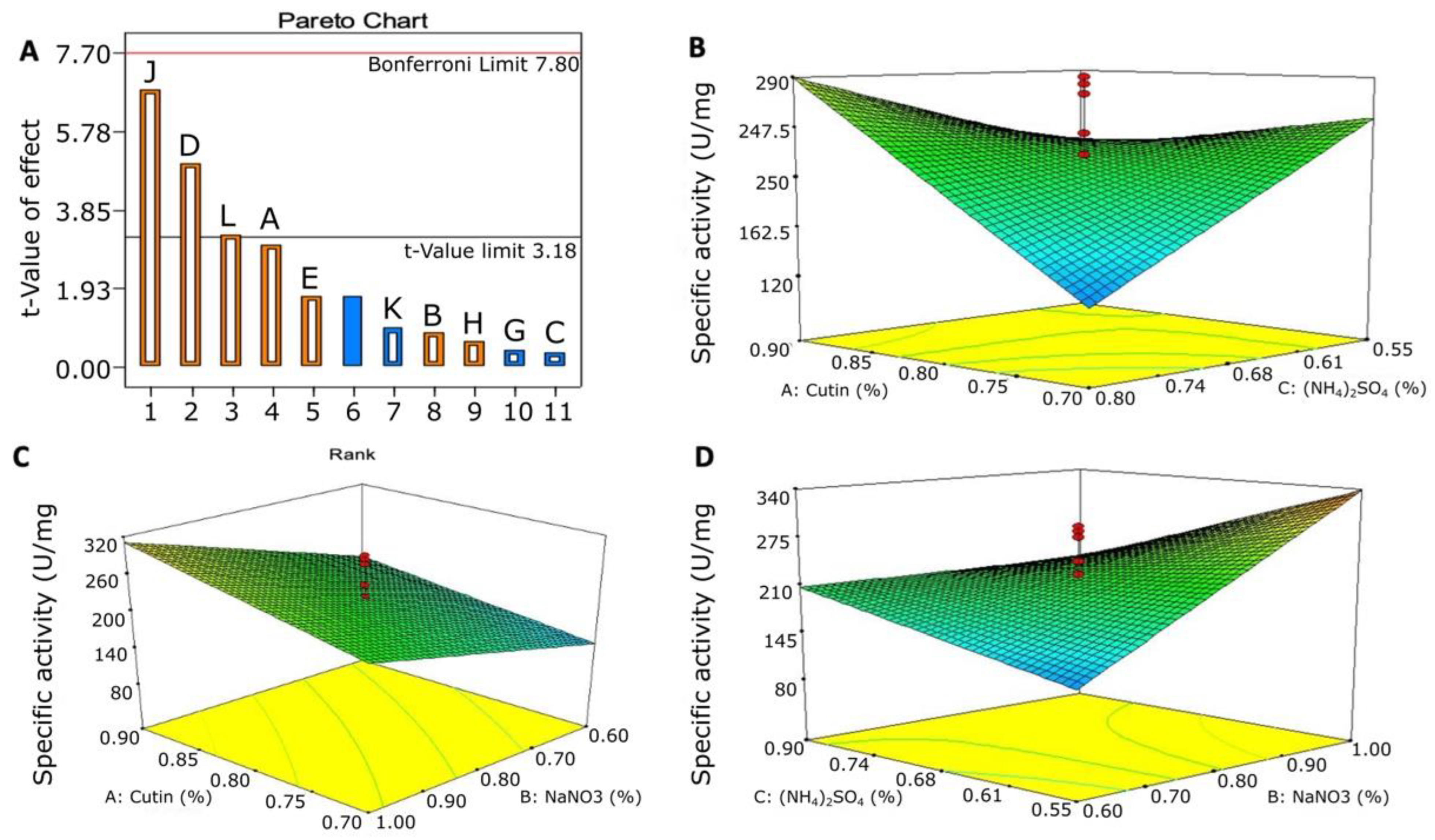
3.5.1. Plackett–Burman Design for Optimizing the Nutritional Condition
3.5.2. Optimization of the Significant Variable Using a Central Composite Design
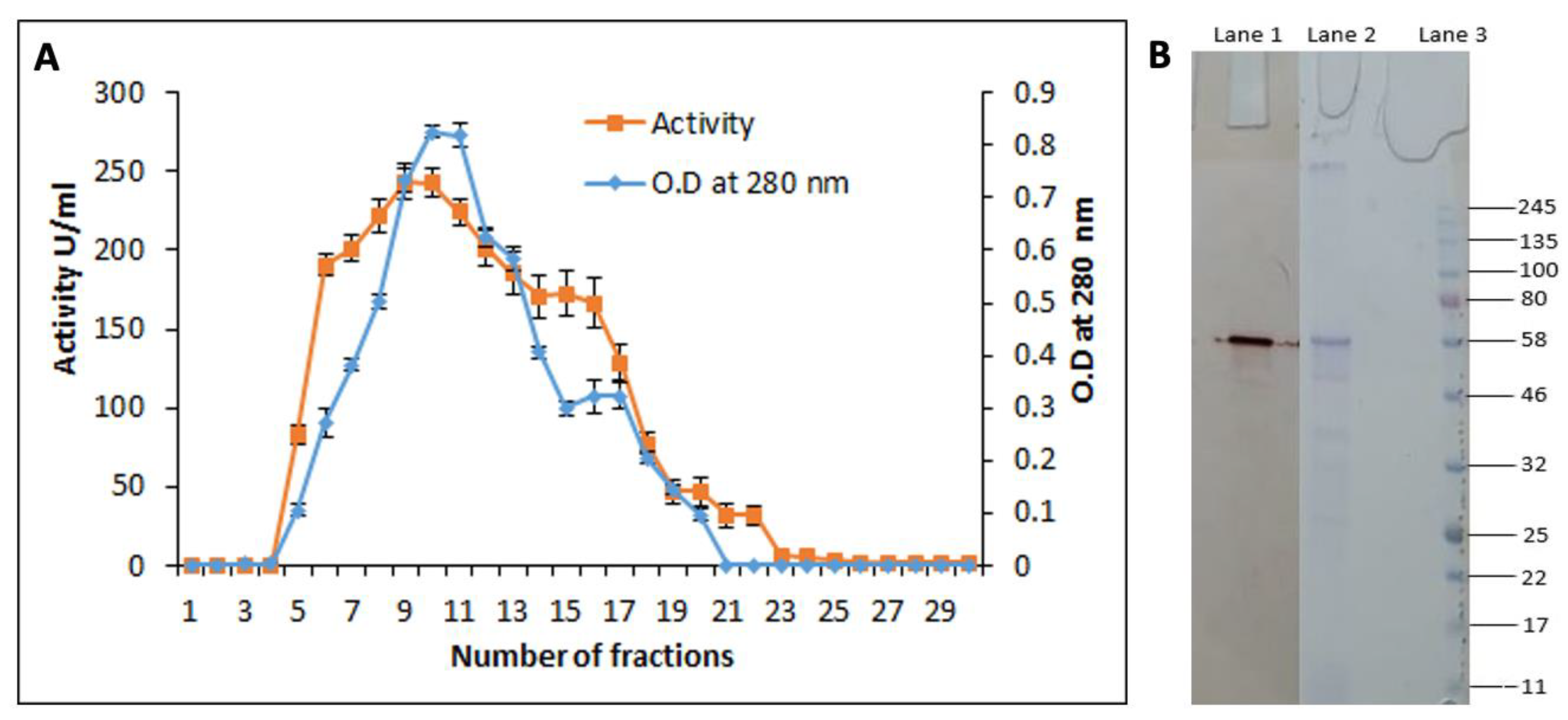
3.6. Purification of the Cutinase-like Enzyme from S. maltophilia PRS8
3.7. Characterization of Purified Cutinase
3.7.1. Effect of Temperature and pH on the Activity and Stability
3.7.2. Effect of Metals Ions
3.7.3. Effect of the Organic Solvent and Surfactants
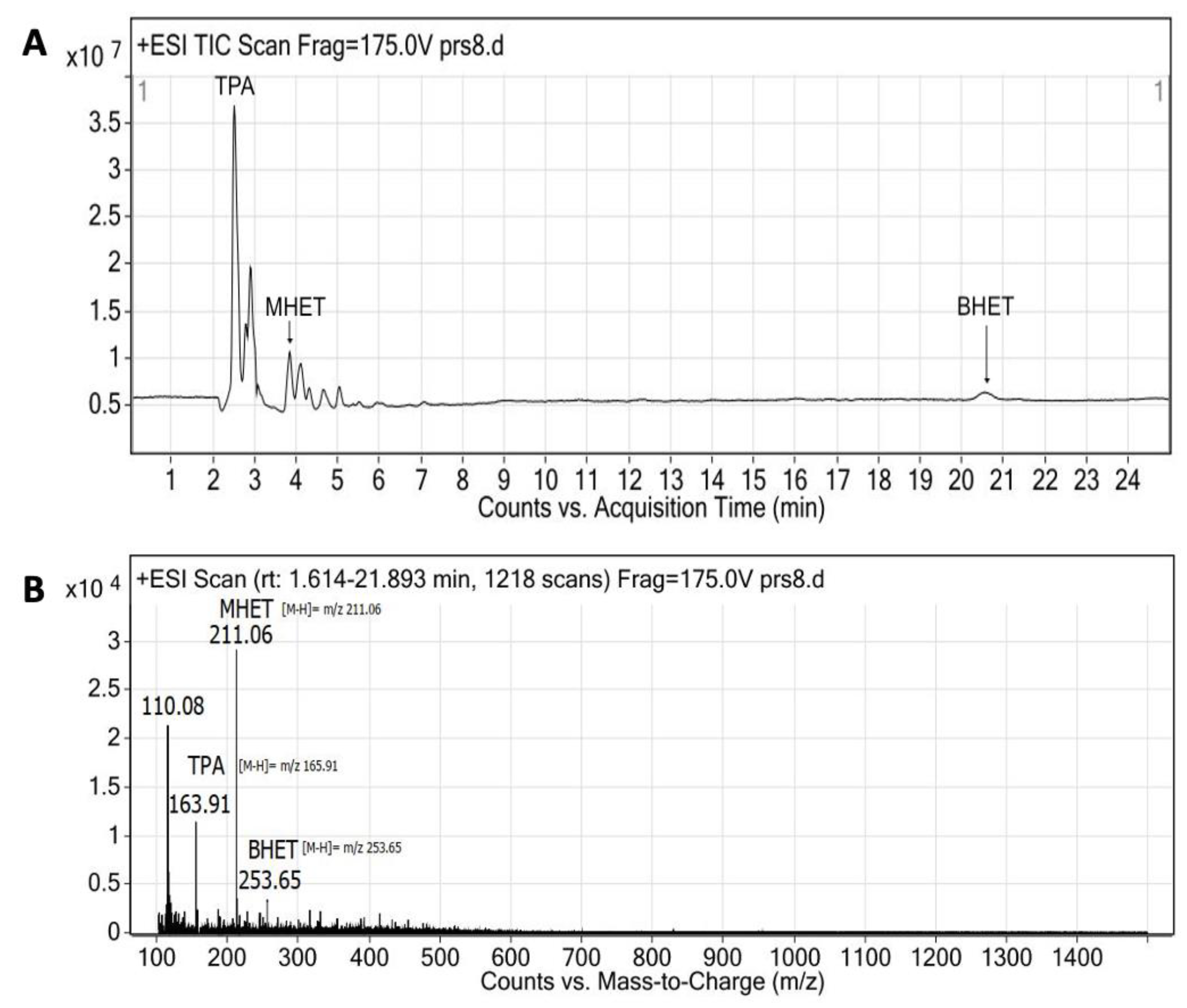
3.8. PET Bottle Flakes Degradation Products
3.9. Activity on the PET Powder
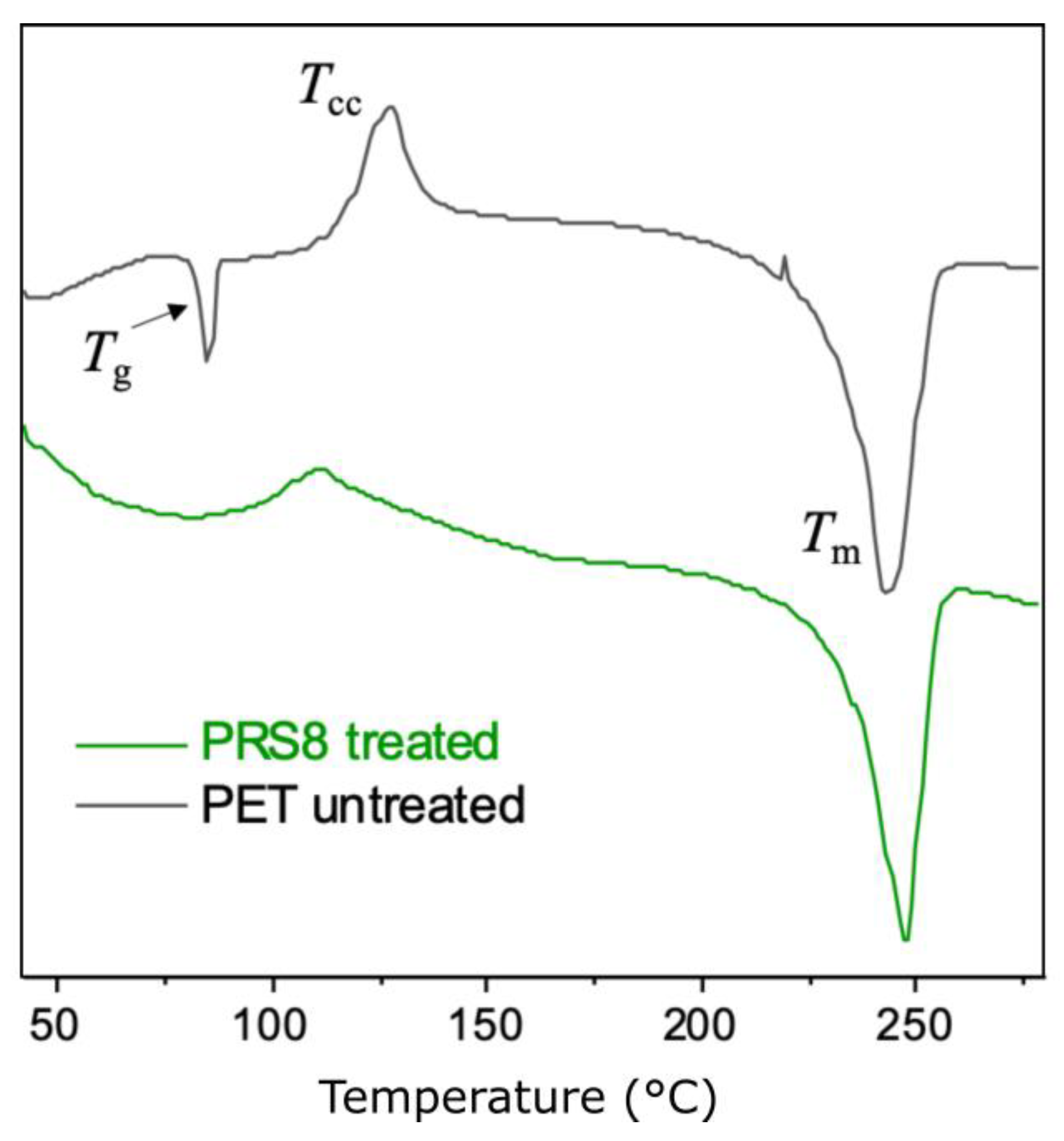
Thermal Properties and the Crystallinity of the PET Powders
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Müller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- PlasticsEurope, E. Plastics—The Facts 2019. An Analysis of European Plastics Production, Demand and Waste Data, PlasticEurope. 2019. Available online: https://www.plasticseurope.org/en/resources/publications/1804-plastics-facts-2019 (accessed on 17 November 2022).
- Nielsen, T.D.; Hasselbalch, J.; Holmberg, K.; Stripple, J. Politics and the plastic crisis: A review throughout the plastic life cycle. Wiley Interdiscip. Rev. Energy Environ. 2020, 9, e360. [Google Scholar] [CrossRef]
- Prata, J.C. Microplastics in wastewater: State of the knowledge on sources, fate and solutions. Mar. Pollut. Bull. 2018, 129, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Human consumption of microplastics. Environ. Sci. Technol. 2019, 53, 7068–7074. [Google Scholar] [CrossRef]
- Pinter, E.; Welle, F.; Mayrhofer, E.; Pechhacker, A.; Motloch, L.; Lahme, V.; Grant, A.; Tacker, M. Circularity study on PET bottle-to-bottle recycling. Sustainability 2021, 13, 7370. [Google Scholar] [CrossRef]
- Kaabel, S.; Arciszewski, J.; Borchers, T.H.; Therien, J.D.; Friščić, T.; Auclair, K. Solid-State Enzymatic Hydrolysis of Mixed PET/Cotton Textiles. ChemSusChem 2023, 16, e202201613. [Google Scholar] [CrossRef]
- MacLeod, M.; Arp, H.P.H.; Tekman, M.B.; Jahnke, A. The global threat from plastic pollution. Science 2021, 373, 61–65. [Google Scholar] [CrossRef]
- Barboza, L.G.A.; Lopes, C.; Oliveira, P.; Bessa, F.; Otero, V.; Henriques, B.; Raimundo, J.; Caetano, M.; Vale, C.; Guilhermino, L. Microplastics in wild fish from North East Atlantic Ocean and its potential for causing neurotoxic effects, lipid oxidative damage, and human health risks associated with ingestion exposure. Sci. Total Environ. 2020, 717, 134625. [Google Scholar] [CrossRef]
- Zrimec, J.; Kokina, M.; Jonasson, S.; Zorrilla, F.; Zelezniak, A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. mBio 2021, 12, e0215521. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, S.; Cai, D.; Shao, C.; Zhang, C.; Zhou, J.; Cui, Z.; He, T.; Chen, C.; Chen, B. Depolymerization of post-consumer PET bottles with engineered cutinase 1 from Thermobifida cellulosilytica. Green Chem. 2022, 24, 5998–6007. [Google Scholar] [CrossRef]
- Leitão, A.L.; Enguita, F.J. Structural insights into carboxylic polyester-degrading enzymes and their functional depolymerizing neighbors. Int. J. Mol. Sci. 2021, 22, 2332. [Google Scholar] [CrossRef]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Egea, P.; Tosi, V.; Wang, P.; Grey, C.; Zhang, B.; Linares-Pastén, J.A. Assessment of Is PETase-Assisted Depolymerization of Terephthalate Aromatic Polyesters and the Effect of the Thioredoxin Fusion Domain. Appl. Sci. 2021, 11, 8315. [Google Scholar] [CrossRef]
- Brooke, J.S. Advances in the microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e00030-19. [Google Scholar] [CrossRef] [PubMed]
- Arulazhagan, P.; Al-Shekri, K.; Huda, Q.; Godon, J.-J.; Basahi, J.M.; Jeyakumar, D. Biodegradation of polycyclic aromatic hydrocarbons by an acidophilic Stenotrophomonas maltophilia strain AJH1 isolated from a mineral mining site in Saudi Arabia. Extremophiles 2017, 21, 163–174. [Google Scholar] [CrossRef]
- Alvarado-Gutiérrez, M.L.; Ruiz-Ordaz, N.; Galíndez-Mayer, J.; Curiel-Quesada, E.; Santoyo-Tepole, F. Degradation kinetics of carbendazim by Klebsiella oxytoca, Flavobacterium johnsoniae, and Stenotrophomonas maltophilia strains. Environ. Sci. Pollut. Res. 2020, 27, 28518–28526. [Google Scholar] [CrossRef]
- Coleman, N.V.; Mattes, T.E.; Gossett, J.M.; Spain, J.C. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl. Environ. Microbiol. 2002, 68, 2726–2730. [Google Scholar] [CrossRef]
- Satti, S.M.; Shah, A.A.; Auras, R.; Marsh, T.L. Isolation and characterization of bacteria capable of degrading poly (lactic acid) at ambient temperature. Polym. Degrad. Stab. 2017, 144, 392–400. [Google Scholar] [CrossRef]
- Pompilio, A.; Piccolomini, R.; Picciani, C.; D’Antonio, D.; Savini, V.; Di Bonaventura, G. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: The role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 2008, 287, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Adıgüzel, A.O.; Tunçer, M. Purification and characterization of cutinase from Bacillus sp. KY0701 isolated from plastic wastes. Prep. Biochem. Biotechnol. 2017, 47, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, B.M.; Champakalakshmi, R.; Sakharkar, M.K.; Lim, C.S.; Sakharkar, K.R. Biodegradation of low density polythene (LDPE) by Pseudomonas species. Indian J. Microbiol. 2012, 52, 411–419. [Google Scholar] [CrossRef]
- Drobota, M.; Persin, Z.; Zemljic, L.F.; Mohan, T.; Stana-Kleinschek, K.; Doliska, A.; Bracic, M.; Ribitsch, V.; Harabagiu, V.; Coseri, S. Chemical modification and characterization of poly (ethylene terephthalate) surfaces for collagen immobilization. Cent. Eur. J. Chem. 2013, 11, 1786–1798. [Google Scholar] [CrossRef]
- Ioakeimidis, C.; Fotopoulou, K.; Karapanagioti, H.; Geraga, M.; Zeri, C.; Papathanassiou, E.; Galgani, F.; Papatheodorou, G. The degradation potential of PET bottles in the marine environment: An ATR-FTIR based approach. Sci. Rep. 2016, 6, 23501. [Google Scholar] [CrossRef]
- Chaudhari, S.A.; Singhal, R.S. Cutin from watermelon peels: A novel inducer for cutinase production and its physicochemical characterization. Int. J. Biol. Macromol. 2015, 79, 398–404. [Google Scholar] [CrossRef]
- Determann, H. Gel Chromatography Gel Filtration·Gel Permeation Molecular Sieves: A Laboratory Handbook; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Karpushova, A.; Brümmer, F.; Barth, S.; Lange, S.; Schmid, R. Cloning, recombinant expression and biochemical characterisation of novel esterases from Bacillus sp. associated with the marine sponge Aplysina aerophoba. Appl. Microbiol. Biotechnol. 2005, 67, 59–69. [Google Scholar] [CrossRef]
- Cheng, S.Z.; Pan, R.; Wunderlich, B. Thermal analysis of poly (butylene terephthalate) for heat capacity, rigid-amorphous content, and transition behavior. Die Makromol. Chem. Macromol. Chem. Phys. 1988, 189, 2443–2458. [Google Scholar] [CrossRef]
- Aristizábal-Lanza, L.; Mankar, S.V.; Tullberg, C.; Zhang, B.; Linares-Pastén, J.A. Comparison of the enzymatic depolymerization of polyethylene terephthalate and Akestra TM using Humicola insolens cutinase. Front. Chem. Eng. 2022, 4, 1048744. [Google Scholar] [CrossRef]
- Hadar, Y.; Sivan, A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl. Microbiol. Biotechnol. 2004, 65, 97–104. [Google Scholar]
- Carvalho, C.M.; Aires-Barros, M.R.; Cabral, J.M. Cutinase: From molecular level to bioprocess development. Biotechnol. Bioeng. 1999, 66, 17–34. [Google Scholar] [CrossRef]
- Liang, X.; Zou, H. Biotechnological Application of Cutinase: A Powerful Tool in Synthetic Biology. SynBio 2022, 1, 54–64. [Google Scholar] [CrossRef]
- Noor, H.; Satti, S.M.; ud Din, S.; Farman, M.; Hasan, F.; Khan, S.; Badshah, M.; Shah, A.A. Insight on esterase from Pseudomonas aeruginosa strain S3 that depolymerize poly (lactic acid)(PLA) at ambient temperature. Polym. Degrad. Stab. 2020, 174, 109096. [Google Scholar] [CrossRef]
- Li, M.; Yang, L.-R.; Xu, G.; Wu, J.-P. Cloning and characterization of a novel lipase from Stenotrophomonas maltophilia GS11: The first member of a new bacterial lipase family XVI. J. Biotechnol. 2016, 228, 30–36. [Google Scholar] [CrossRef]
- Parapouli, M.; Foukis, A.; Stergiou, P.-Y.; Koukouritaki, M.; Magklaras, P.; Gkini, O.A.; Papamichael, E.M.; Afendra, A.-S.; Hatziloukas, E. Molecular, biochemical and kinetic analysis of a novel, thermostable lipase (LipSm) from Stenotrophomonas maltophilia Psi-1, the first member of a new bacterial lipase family (XVIII). J. Biol. Res. -Thessalon. 2018, 25, 4. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Mao, X.; Lu, P.; Secundo, F.; Xue, C.; Sun, J. Cloning, expression, and characterization of a novel thermostable and alkaline-stable esterase from Stenotrophomonas maltophilia OUC_Est10 catalytically active in organic solvents. Catalysts 2019, 9, 401. [Google Scholar] [CrossRef]
- Çolak, A.; Şişik, D.; Saglam, N.; Güner, S.; Çanakçi, S.; Beldüz, A.O. Characterization of a thermoalkalophilic esterase from a novel thermophilic bacterium, Anoxybacillus gonensis G2. Bioresour. Technol. 2005, 96, 625–631. [Google Scholar] [CrossRef]
- Then, J.; Wei, R.; Oeser, T.; Barth, M.; Belisário-Ferrari, M.R.; Schmidt, J.; Zimmermann, W. Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnol. J. 2015, 10, 592–598. [Google Scholar] [CrossRef]
- Geyer, B.; Lorenz, G.; Kandelbauer, A. Recycling of poly (ethylene terephthalate)—A review focusing on chemical methods. Express Polym. Lett. 2016, 10, 559–586. [Google Scholar] [CrossRef]
- Degani, O. Production and purification of cutinase from fusarium oxysporum using modified growth media and a specific cutinase substrate. Adv. Biosci. Biotechnol. 2015, 6, 245. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Choi, G.-S.; Kim, S.-B.; Yoon, G.-S.; Kim, Y.-S.; Ryu, Y.-W. Screening and characterization of a novel esterase from a metagenomic library. Protein Expr. Purif. 2006, 45, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Clarke, D.J.; Dobson, A.D. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Front. Microbiol. 2020, 11, 571265. [Google Scholar] [CrossRef] [PubMed]
- Menzel, T.; Weigert, S.; Gagsteiger, A.; Eich, Y.; Sittl, S.; Papastavrou, G.; Ruckdäschel, H.; Altstädt, V.; Höcker, B. Impact of enzymatic degradation on the material properties of poly (ethylene terephthalate). Polymers 2021, 13, 3885. [Google Scholar] [CrossRef]
- Falkenstein, P.; Gräsing, D.; Bielytskyi, P.; Zimmermann, W.; Matysik, J.; Wei, R.; Song, C. UV pretreatment impairs the enzymatic degradation of polyethylene terephthalate. Front. Microbiol. 2020, 11, 689. [Google Scholar] [CrossRef] [PubMed]

| Purification Steps | Volume (mL) | Activity (U/mL) | Total Activity (Units) | Protein (mg/mL) | Specific Activity (U/mg) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|---|---|
| Crude Extract | 500 | 138.13 | 69,068.18 | 1.65 | 70.47 | 100 | 1 |
| (NH4)2SO4 | 200 | 260.40 | 52,081.82 | 1.18 | 153.42 | 75.40 | 2.17 |
| Sephadex (G-100) | 90 | 373.28 | 33,595.91 | 0.75 | 450.58 | 48.64 | 6.39 |
| Metals | Residual Activity (%) | |
|---|---|---|
| 15 mM | 2 mM | |
| Control | 100 ± 0.0 | 100 ± 0.0 |
| Ca2+ | 111.32 ± 0.6 | 102.34 ± 0.5 |
| Fe2+ | 98.82 ± 0.7 | 100.39 ± 0.6 |
| Cu2+ | 107.81 ± 1.0 | 100.78 ± 0.2 |
| Co2+ | 55.85 ± 0.2 | 58.98 ± 0.3 |
| Ni2+ | 100.39 ± 0.4 | 100 ± 0.1 |
| Zn2+ | 114.45 ± 0.5 | 107.42 ± 0.4 |
| K+ | 115.23 ± 0.5 | 111.71 ± 0.3 |
| Cd2+ | 1.95 ± 0.2 | 3.51 ± 0.3 |
| Na+ | 85.54 ± 0.1 | 91.79 ± 0.2 |
| Mg2+ | 107.42 ± 0.4 | 105.07 ± 0.3 |
| Ni2+ | 71.48 ± 0.3 | 66.01 ± 0.5 |
| Hg2+ | 0 ± 0.00 | 0 ± 0.00 |
| Ba2+ | 54.29 ± 0.1 | 55.46 ± 0.3 |
| Organic Solvents (10%) | Residual Activity % (Min) | |||
|---|---|---|---|---|
| 30 min | 60 min | 90 min | 120 min | |
| Control | 100 | 100 | 100 | 100 |
| Ethanol | 54.26 | 54.26 | 45.29 | 42.60 |
| Methanol | 104.03 | 103.13 | 98.65 | 91.03 |
| Acetonitrile | 93.27 | 90.13 | 91.03 | 83.40 |
| Acetone | 97.03 | 96.86 | 96.41 | 95.51 |
| Ethyl Acetate | 103.13 | 98.65 | 96.41 | 92.82 |
| Propanol | 105.82 | 101.79 | 101.34 | 100 |
| DMSO | 98.20 | 95.06 | 94.61 | 94.17 |
| Butanol | 65.08 | 50.67 | 49.32 | 28.25 |
| Formaldehyde | 94.61 | 89.68 | 85.65 | 85.20 |
| Surfactants | Residual Activity (%) | |
|---|---|---|
| 0.5% | 5% | |
| Control | 100 ± 0.00 | 100 ± 0.00 |
| Tween-20 | 109.41 ± 0.30 | 106.66 ± 0.53 |
| Tween-40 | 112.54 ± 0.72 | 116.47 ± 0.30 |
| Tween-60 | 110.98 ± 0.81 | 110.58 ± 0.90 |
| Tween-80 | 113.72 ± 0.70 | 105.09 ± 0.54 |
| CTAB | 11.76 ± 0.40 | 7.84 ± 0.31 |
| SDS | 0 ± 0.00 | 0 ± 0.00 |
| Triton X-100 | 109.80 ± 0.21 | 112.15 ± 0.12 |
| Enzyme | Ratio (E/PET) | T (°C) | pH | Degradation Products (mg/L) | Relative Production (%) | PET Depolymerization (%) | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|
| TPA | MHET | BHET | |||||||
| Extracellular medium | 0.005 * | 40 | 9 | 2246 | 38 | 60 | 2 | 22.5 | This work |
| IsPETase | 0.004 | 37 | 7.2 | 2304 | 24.2 | 73.7 | 2.1 | 23 | [17] |
| Trx-IsPETase | 0.005 | 37 | 7.2 | 3230 | 31.5 | 66.7 | 1.8 | 32 | [17] |
| HiCut (HiC) | 0.008 | 70 | 7.5 | 437 | 100 | n.d | n.d | 12.7 | [33] |
| Polymer | Tg (°C) | Tcc (°C) | Tm (°C) | ΔHcc (J/g) | ΔHm (J/g) | Crystallinity * (%) |
|---|---|---|---|---|---|---|
| PET (untreated) | 83 | 127 | 242 | 9.0 | 35.9 | 19.3 |
| PET (treated) | - | 110 | 247 | 2.9 | 32.4 | 21.1 |
| Enzyme | Strain | MW (kDa) | Sequence (GenBank) | Optimum Temperature (°C) | Optimum pH | Reference |
|---|---|---|---|---|---|---|
| Cutinase-like | PSR8 | 58 | Unknown | 40 | 8.0 | This work |
| Lipase (LipSM54) | GS11 | 52.8 | KX353755 | 35 | 8.0 | [38] |
| Lipase (LipSm) | Psi-1 | 40.7 | KC014616 | 64.5 | 8.0–9.0 | [39] |
| Esterase (est7) | OUC_Est10 | 76.6 | MH253883 | 45 | 9.0 | [41] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Din, S.U.; Kalsoom; Satti, S.M.; Uddin, S.; Mankar, S.V.; Ceylan, E.; Hasan, F.; Khan, S.; Badshah, M.; Beldüz, A.O.; et al. The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature. Appl. Sci. 2023, 13, 3686. https://doi.org/10.3390/app13063686
Din SU, Kalsoom, Satti SM, Uddin S, Mankar SV, Ceylan E, Hasan F, Khan S, Badshah M, Beldüz AO, et al. The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature. Applied Sciences. 2023; 13(6):3686. https://doi.org/10.3390/app13063686
Chicago/Turabian StyleDin, Salah Ud, Kalsoom, Sadia Mehmood Satti, Salah Uddin, Smita V. Mankar, Esma Ceylan, Fariha Hasan, Samiullah Khan, Malik Badshah, Ali Osman Beldüz, and et al. 2023. "The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature" Applied Sciences 13, no. 6: 3686. https://doi.org/10.3390/app13063686
APA StyleDin, S. U., Kalsoom, Satti, S. M., Uddin, S., Mankar, S. V., Ceylan, E., Hasan, F., Khan, S., Badshah, M., Beldüz, A. O., Çanakçi, S., Zhang, B., Linares-Pastén, J. A., & Shah, A. A. (2023). The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature. Applied Sciences, 13(6), 3686. https://doi.org/10.3390/app13063686








