An Explainable Brain Tumor Detection Framework for MRI Analysis
Abstract
1. Introduction
2. Related Work
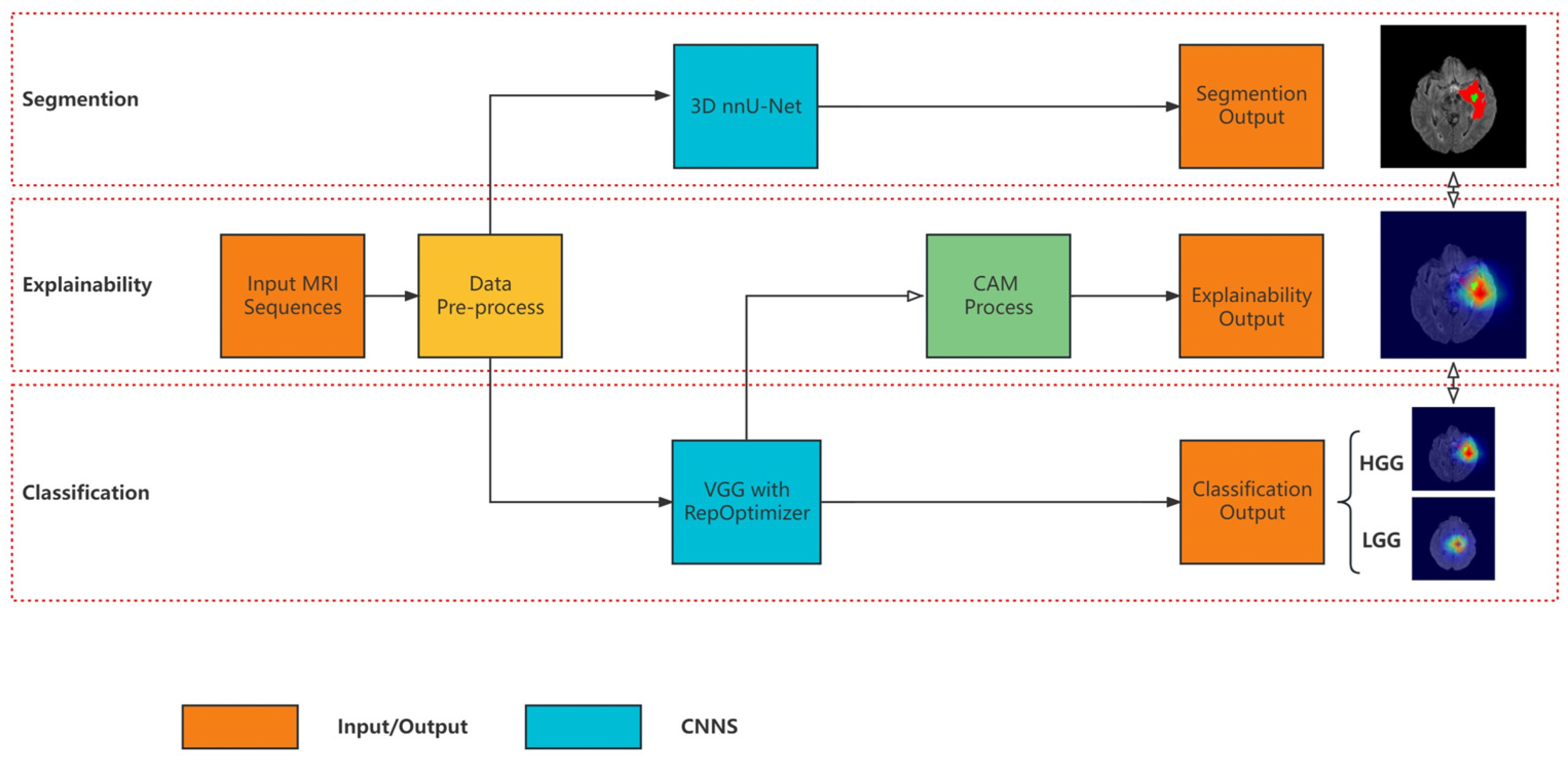
3. Materials and Methods
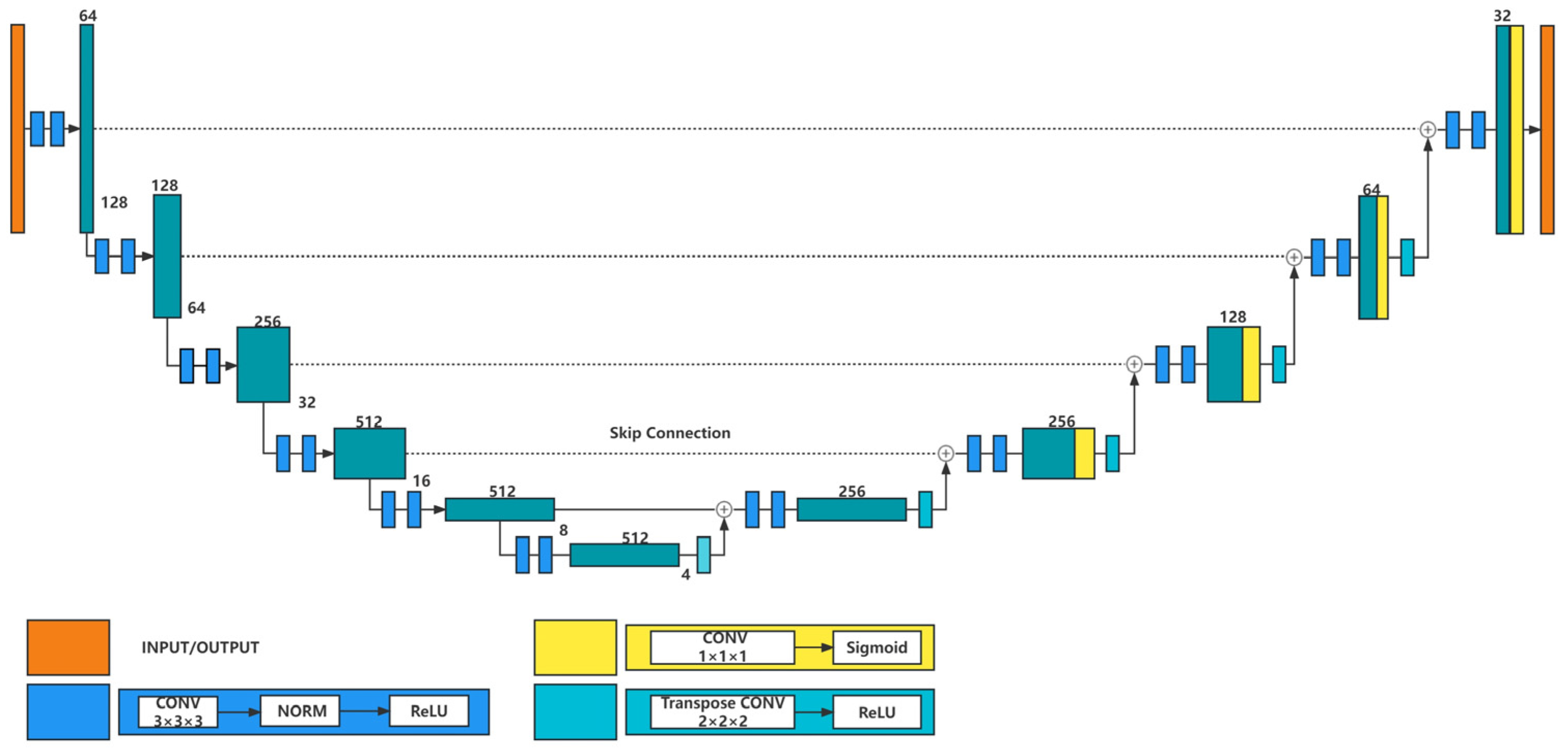
3.1. Segmentation Model
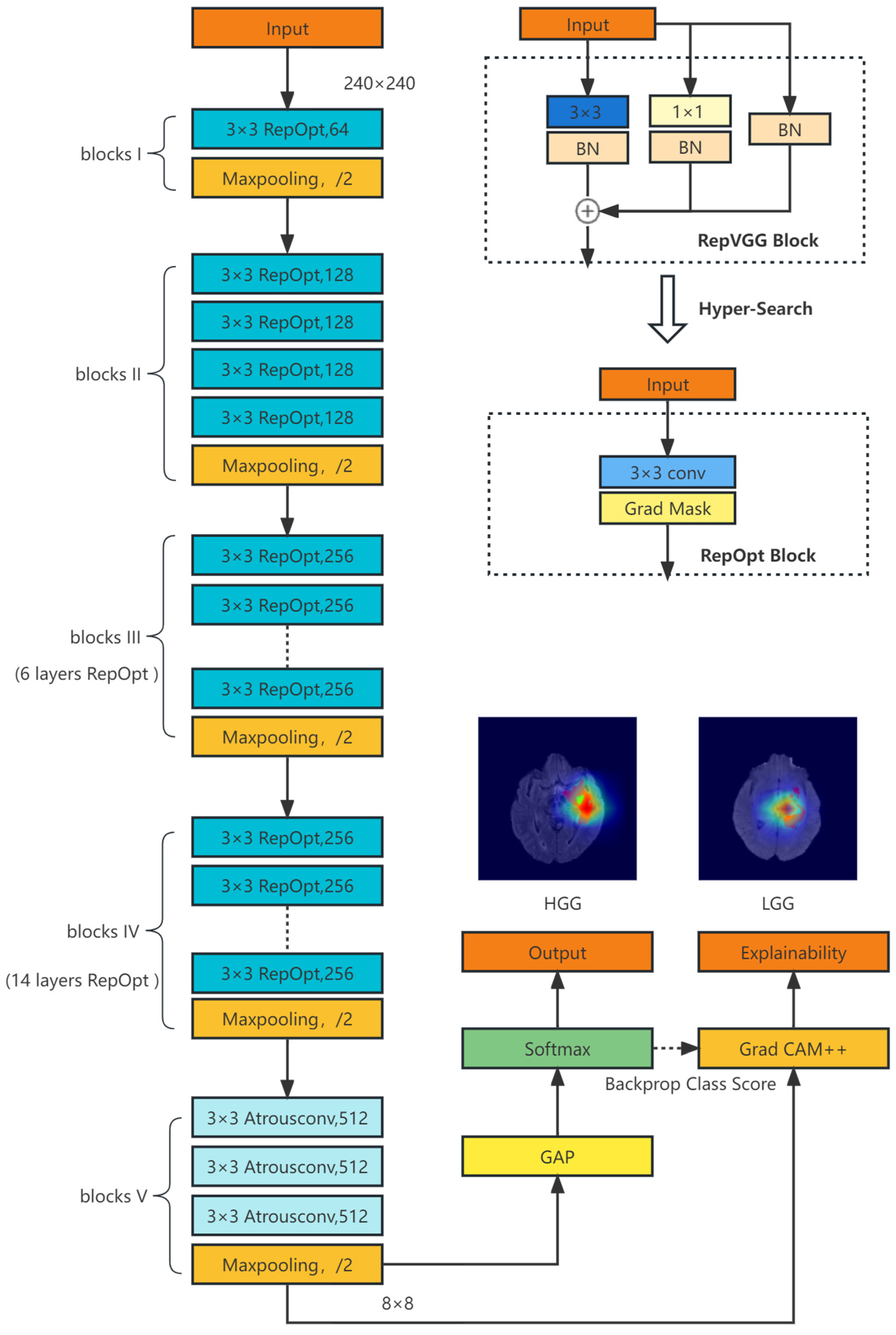
3.2. Classification Model Based on Re-Parameterization
3.3. Explainability
4. Results and Discussion
4.1. Dataset and Pre-Processing
4.2. Experimental Setting
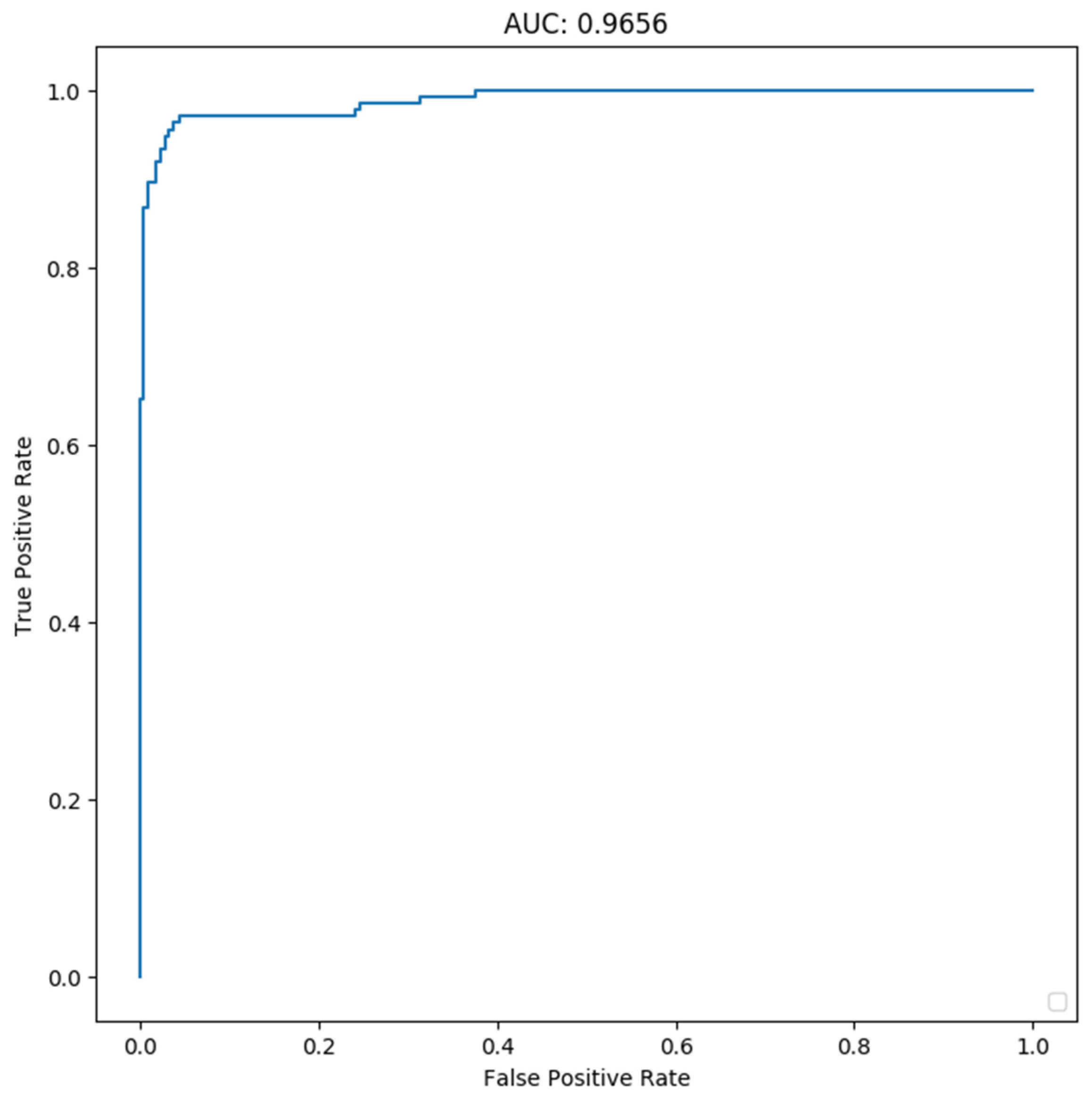
4.3. Results and Analysis
4.3.1. Segmentation Results
4.3.2. Classification Results
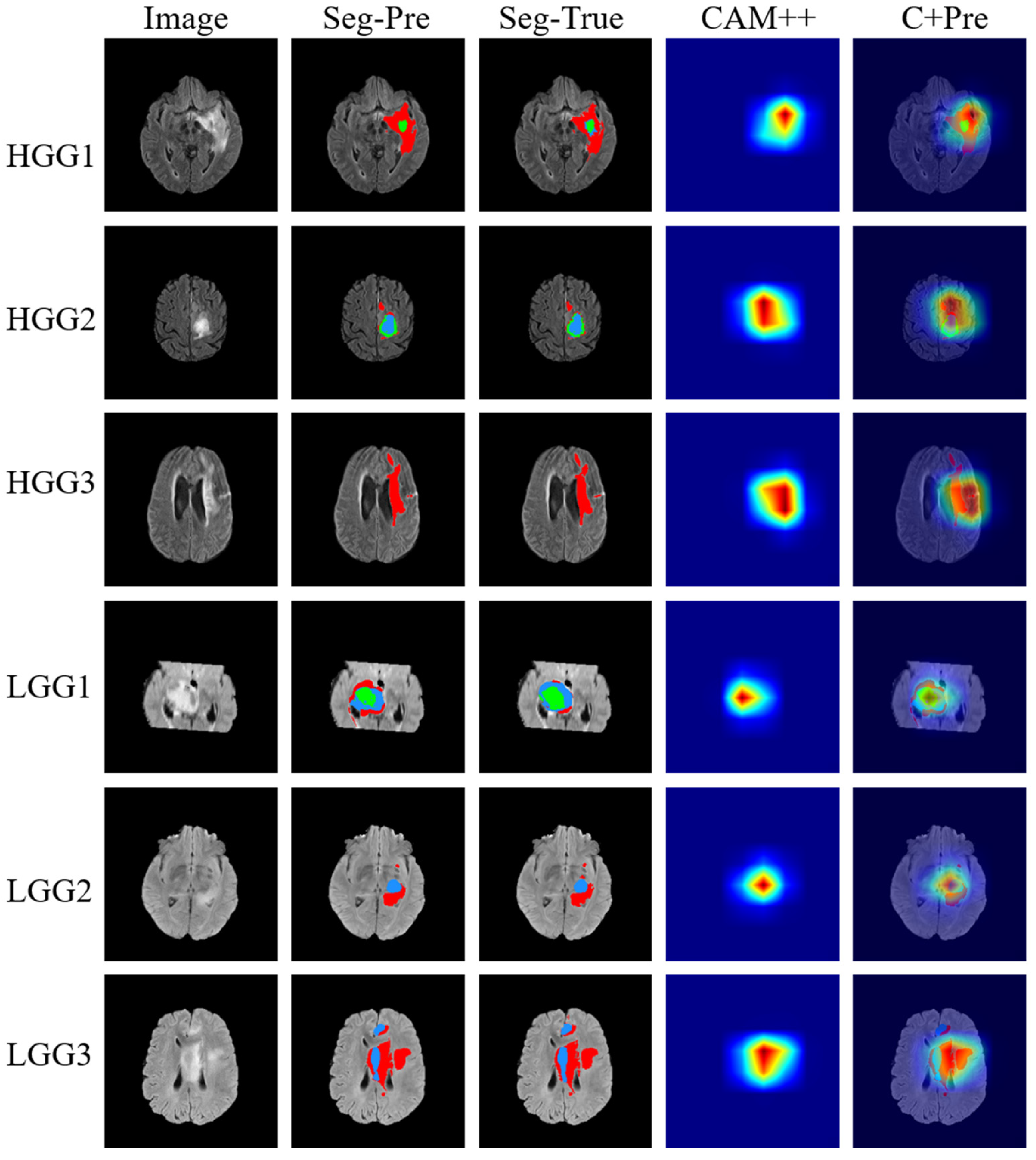
4.3.3. Comparison of Segmentation and Explainability
4.4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jemal, A.; Thomas, A.; Murray, T.; Thun, M. Cancer statistics. Ca-Cancer J. Clin. 2002, 52, 23–47. [Google Scholar]
- Miner, R.C. Image-guided neurosurgery. J. Med. Imaging Radiat. Sci. 2017, 48, 328–335. [Google Scholar] [CrossRef]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2017; pp. 287–297. [Google Scholar]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Adadi, A.; Berrada, M. Peeking inside the black-box: A survey on explainable artificial intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.-Z. XAI—Explainable artificial intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef] [PubMed]
- Doshi-Velez, F.; Kim, B. Towards a rigorous science of interpretable machine learning. arXiv 2017, arXiv:1702.08608. [Google Scholar]
- Tonekaboni, S.; Joshi, S.; McCradden, M.D.; Goldenberg, A. What clinicians want: Contextualizing explainable machine learning for clinical end use. In Proceedings of the Machine Learning for Healthcare Conference, Ann Arbor, MI, USA, 8–10 August 2019; pp. 359–380. [Google Scholar]
- Messina, P.; Pino, P.; Parra, D.; Soto, A.; Besa, C.; Uribe, S.; Andía, M.; Tejos, C.; Prieto, C.; Capurro, D. A survey on deep learning and explainability for automatic report generation from medical images. ACM Comput. Surv. 2022, 54, 1–40. [Google Scholar] [CrossRef]
- Temme, M. Algorithms and transparency in view of the new general data protection regulation. Eur. Data Prot. Law Rev. 2017, 3, 473–485. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 3319–3328. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Tian, J.; Li, C.; Shi, Z.; Xu, F. A diagnostic report generator from CT volumes on liver tumor with semi-supervised attention mechanism. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 702–710. [Google Scholar]
- Han, Z.; Wei, B.; Leung, S.; Chung, J.; Li, S. Towards automatic report generation in spine radiology using weakly supervised framework. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 185–193. [Google Scholar]
- Teixeira, L.O.; Pereira, R.M.; Bertolini, D.; Oliveira, L.S.; Nanni, L.; Cavalcanti, G.D.; Costa, Y.M. Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images. Sensors 2021, 21, 7116. [Google Scholar] [CrossRef]
- Ramzan, F.; Khan, M.U.G.; Iqbal, S.; Saba, T.; Rehman, A. Volumetric segmentation of brain regions from MRI scans using 3D convolutional neural networks. IEEE Access 2020, 8, 103697–103709. [Google Scholar] [CrossRef]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Rangarajan, A.; Ranka, S. Visual explanations from deep 3D convolutional neural networks for Alzheimer’s disease classification. In Proceedings of the AMIA Annual Symposium Proceedings, San Francisco, CA, USA, 3–7 November 2018; pp. 1571–1580. [Google Scholar]
- Wickstrøm, K.; Kampffmeyer, M.; Jenssen, R. Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps. Med. Image Anal. 2020, 60, 101619. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, M.; Vettukattil, R.; Banitalebi, H.; Krogh, N.R.; Geitung, J.T. Explainable artificial intelligence for human-machine interaction in brain tumor localization. J. Pers. Med. 2021, 11, 1213. [Google Scholar] [CrossRef]
- Saleem, H.; Shahid, A.R.; Raza, B. Visual interpretability in 3D brain tumor segmentation network. Comput. Biol. Med. 2021, 133, 104410. [Google Scholar] [CrossRef]
- Natekar, P.; Kori, A.; Krishnamurthi, G. Demystifying brain tumor segmentation networks: Interpretability and uncertainty analysis. Front. Comput. Neurosci. 2020, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, J.; Gilmer, J.; Muelly, M.; Goodfellow, I.; Hardt, M.; Kim, B. Sanity checks for saliency maps. Adv. Neural Inf. Process. Syst. 2018, 31, 9505–9515. [Google Scholar]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 106–114. [Google Scholar]
- Narayanan, B.N.; De Silva, M.S.; Hardie, R.C.; Kueterman, N.K.; Ali, R. Understanding deep neural network predictions for medical imaging applications. arXiv 2019, arXiv:1912.09621. [Google Scholar]
- Isensee, F.; Jäger, P.; Full, P.; Vollmuth, P.; Maier-Hein, K. nnU-Net for Brain Tumor Segmentation in Brainlesion: Glioma. In Proceedings of the Multiple Sclerosis, Stroke and Traumatic Brain Injuries-6th International Workshop, BrainLes, Lima, Peru, 4 October 2020. [Google Scholar]
- Yan, F.; Wang, Z.; Qi, S.; Xiao, R. A Saliency Prediction Model Based on Re-Parameterization and Channel Attention Mechanism. Electronics 2022, 11, 1180. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, X.; Ma, N.; Han, J.; Ding, G.; Sun, J. Repvgg: Making vgg-style convnets great again. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 19–20 June 2021; pp. 13733–13742. [Google Scholar]
- Ding, X.; Chen, H.; Zhang, X.; Huang, K.; Han, J.; Ding, G. Re-parameterizing Your Optimizers rather than Architectures. arXiv 2022, arXiv:2205.15242. [Google Scholar]
- Krizhevsky, A.; Hinton, G. Learning Multiple Layers of Features from Tiny Images. Master’s Thesis, University of Tront, Toronto, ON, Canada, 2009. [Google Scholar]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 5894–5897. [Google Scholar]
- Rehman, A.; Khan, M.A.; Saba, T.; Mehmood, Z.; Tariq, U.; Ayesha, N. Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture. Microsc. Res. Tech. 2021, 84, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.; Nanda, A. An improved whale optimization algorithm-based radial neural network for multi-grade brain tumor classification. Vis. Comput. 2022, 38, 3525–3540. [Google Scholar] [CrossRef]


| Method | ET | TC | WT |
|---|---|---|---|
| U-Net | 0.8250 | 0.8473 | 0.9005 |
| VAE U-Net | 0.8145 | 0.8041 | 0.9042 |
| nnU-Net | 0.7945 | 0.8524 | 0.9119 |
| Model | Top-1 Accuracy | Speed | Params (M) | FLOPs (B) |
|---|---|---|---|---|
| ResNet-50 | 76.31% | 719 | 25.53 | 3.9 |
| ResNet-101 | 77.21% | 430 | 44.49 | 7.6 |
| VGG-16 | 72.21% | 415 | 138.35 | 15.5 |
| RepVGG-B1 | 78.42% | 685 | 51.82 | 11.8 |
| RepOpt-B1 | 78.48% | 1254 | 51.82 | 11.9 |
| Model | Method | DataSet | Accuracy |
|---|---|---|---|
| Ge et al., 2018 [42] | 2D CNNS | BraTS 2017 | 90.87% |
| Khan et al., 2020 [26] | VGG and EML | BraTS 2018 | 92.5% |
| Rehman et al., 2021 [43] | 3D CNNS | BraTS 2018 | 92.67% |
| Dixit et al., 2022 [44] | FCM-IWOA-RBNN | BraTS 2018 | 96% |
| Our Model | RepOpt | BraTS 2018 | 95.46% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, F.; Chen, Y.; Xia, Y.; Wang, Z.; Xiao, R. An Explainable Brain Tumor Detection Framework for MRI Analysis. Appl. Sci. 2023, 13, 3438. https://doi.org/10.3390/app13063438
Yan F, Chen Y, Xia Y, Wang Z, Xiao R. An Explainable Brain Tumor Detection Framework for MRI Analysis. Applied Sciences. 2023; 13(6):3438. https://doi.org/10.3390/app13063438
Chicago/Turabian StyleYan, Fei, Yunqing Chen, Yiwen Xia, Zhiliang Wang, and Ruoxiu Xiao. 2023. "An Explainable Brain Tumor Detection Framework for MRI Analysis" Applied Sciences 13, no. 6: 3438. https://doi.org/10.3390/app13063438
APA StyleYan, F., Chen, Y., Xia, Y., Wang, Z., & Xiao, R. (2023). An Explainable Brain Tumor Detection Framework for MRI Analysis. Applied Sciences, 13(6), 3438. https://doi.org/10.3390/app13063438






