Isolation, Specificity, and Application of β-N-Acetylhexosaminidases from Penicillium crustosum
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
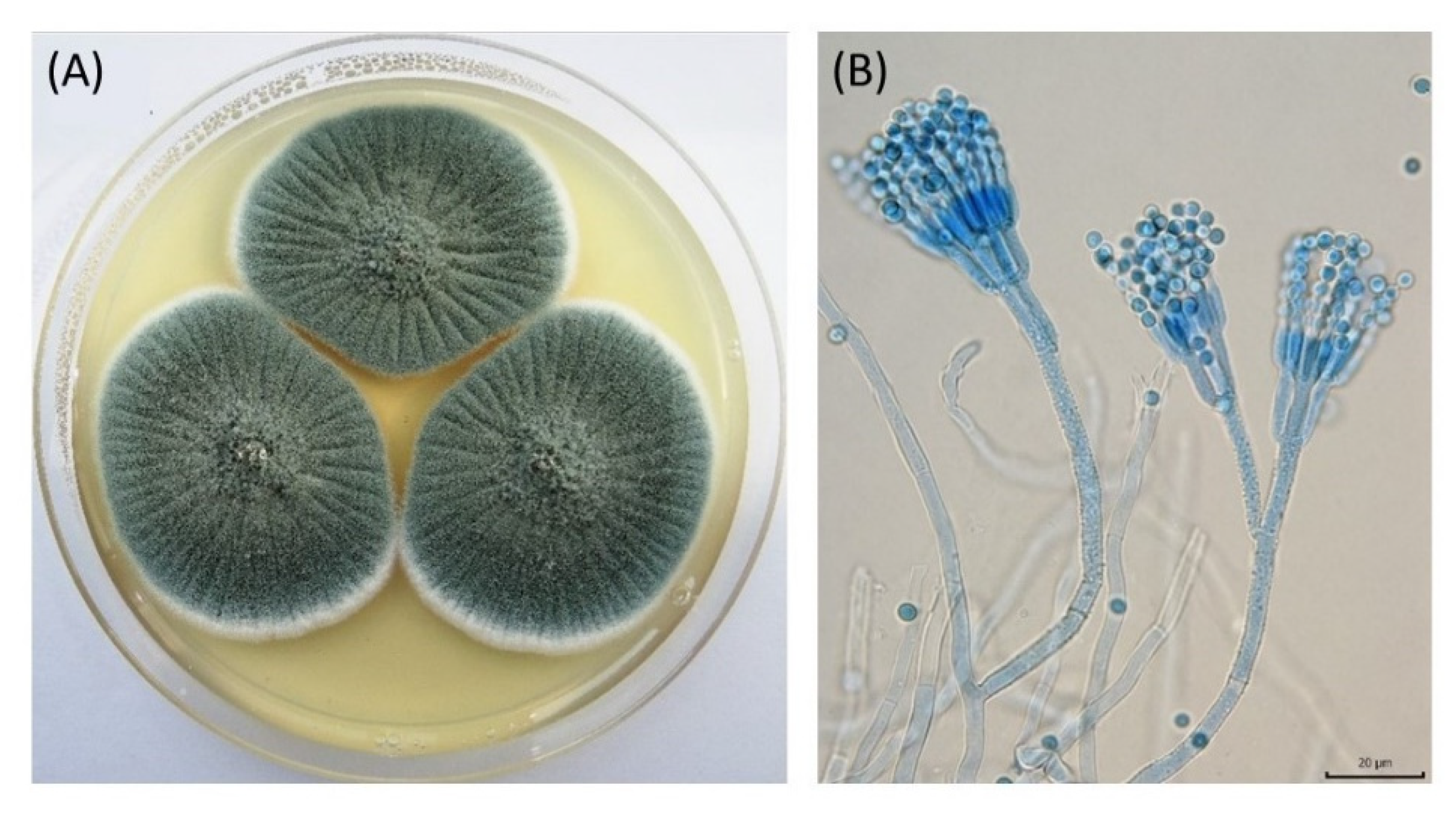
2.2. Microorganisms
2.3. Cell Cultivation and Purification of ß-N-Acetylhexosaminidase
2.4. Enzyme Assay
2.5. Resolution of the Anomeric Mixture of 4MU-β-GalNAc by Enzyme Hydrolysis
3. Results and Discussion
3.1. Production, Purification, and Characterization of ß-N-Acetylhexosaminidases from P. crustosum Strains
3.2. Effect of pH and Temperature on the Activity of ß-N-Acetylhexosaminidases
3.3. Substrate Specificity toward 4NP-ß-Glc/GalNAc and 2NP-ß-Glc/GalNAc
3.4. Substrate Specificity toward Nonconventional Substrates
3.5. Separation of an Anomeric Mixture of 4MU-α/β-GalNAc by Selective Hydrolysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayer, C.; Vocadlo, D.J.; Mah, M.; Rupitz, K.; Stoll, D.; Warren, R.A.J.; Withers, S.G. Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from Cellulomonas fimi. FEBS J. 2006, 273, 2929–2941. [Google Scholar] [CrossRef]
- Ogata, M.; Zeng, X.; Usui, T.; Uzawa, H. Substrate specificity of N-acetylhexosaminidase from Aspergillus oryzae to artificial glycosyl acceptors having various substituents at the reducing ends. Carbohydr. Res. 2007, 342, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Slámová, K.; Bojarová, P.; Petrásková, L.; Křen, V. β-N-Acetylhexosaminidase: What’s in a name…? Biotechnol. Adv. 2010, 28, 682–693. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Hamlyn, P.F.; Wales, D.; Sagar, B.F. Extracellular Enzymes of Penicillium. In Penicillium and Acremonium; Peberdy, J.F., Ed.; Springer: Boston, MA, USA, 1987; pp. 245–284. [Google Scholar] [CrossRef]
- Rigo, E.; Ninow, J.L.; Tsai, S.M.; Durrer, A.; Foltran, L.L.; Remonatto, D.; Sychoski, M.; Vardanega, R.; de Oliveira, D.; Treichel, H.; et al. Preliminary Characterization of Novel Extra-cellular Lipase from Penicillium crustosum Under Solid-State Fermentation and its Potential Application for Triglycerides Hydrolysis. Food Bioprocess Technol. 2012, 5, 1592–1600. [Google Scholar] [CrossRef]
- Sutivisedsak, N.; Leathers, T.; Bischoff, K.; Nunnally, M.; Peterson, S.W. Novel sources of β-glucanase for the enzymatic degradation of schizophyllan. Enzym. Microb. Technol. 2013, 52, 203–210. [Google Scholar] [CrossRef]
- Sonjak, S.; Frisvad, J.; Gunde-Cimerman, N. Comparison of secondary metabolite production by Penicillium crustosum strains, isolated from Arctic and other various ecological niches. FEMS Microbiol. Ecol. 2005, 53, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Burtseva, I.V.; Sova, V.; Pivkin, M.; Anastiuk, S.; Gorbach, V.; Zviagintseva, T.N. Distribution of O-glycosylases in marine fungi of the Sea of Japan and the Sea of Okhotsk: Characterization of exocellular N-acetyl-beta-D-glucosaminidase of the marine fungus Penicillium canescens. Prikl. Biokhim. Mikrobiol. 2010, 46, 700–708. [Google Scholar] [PubMed]
- Wang, S.Y.; Laborda, P.; Lu, A.M.; Wang, M.; Duan, X.C.; Liu, L.; Voglmeir, J. Chemo-enzymatic approach to access diastereopure α-substituted GlcNAc derivatives. J. Carbohydr. Chem. 2016, 35, 423–434. [Google Scholar] [CrossRef]
- Hronská, H.; Štefuca, V.; Ondrejková, E.; Bláhová, M.; Višňovský, J.; Rosenberg, M. Chemo-Enzymatic Production of 4-Nitrophenyl-2-acetamido-2-deoxy-α-D-galactopyranoside Using Immobilized β-N-Acetylhexosaminidase. Catalysts 2022, 12, 474. [Google Scholar] [CrossRef]
- Slámová, K.; Bojarová, P.; Gerstorferová, D.; Fliedrová, B.; Hofmeisterová, J.; Fiala, M.; Pompach, P.; Křen, V. Sequencing, cloning and high-yield expression of a fungal β-N-acetylhexosaminidase in Pichia pastoris. Protein Expr. Purif. 2012, 82, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Plíhal, O.; Sklenář, J.; Hofbauerová, K.; Novák, P.; Man, P.; Pompach, P.; Kavan, D.; Ryšlavá, H.; Weignerová, L.; Charvátová-Pišvejcová, A.; et al. Large propeptides of fungal beta-N-acetylhexosaminidases are novel enzyme regulators that must be intracellularly processed to control activity, dimerization, and secretion into the extracellular environment. Biochemistry 2007, 46, 2719–2734. [Google Scholar] [CrossRef]
- Ryšlavá, H.; Kalendová, A.; Doubnerová, V.; Skočdopol, P.; Kumar, V.; Kukačka, Z.; Pompach, P.; Vaněk, O.; Slámová, K.; Bojarová, P.; et al. Enzymatic characterization and molecular modeling of an evolutionarily interesting fungal β-N-acetylhexosaminidase: β-N-Acetylhexosaminidase from Penicillium oxalicum. FEBS J. 2011, 278, 2469–2484. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Kulik, N.; Slámová, K.; Hubálek, M.; Kotik, M.; Cvačka, J.; Pelantová, H.; Křen, V. Selective β-N-acetylhexosaminidase from Aspergillus versicolor—A tool for producing bioactive carbohydrates. Appl. Microbiol. Biotechnol. 2019, 103, 1737–1753. [Google Scholar] [CrossRef]
- Scigelova, M.; Crout, D.H.G. Microbial β-N-acetylhexosaminidases and their biotechnological applications. Enzym. Microb. Technol. 1999, 25, 3–14. [Google Scholar] [CrossRef]
- Fialová, P.; Weignerová, L.; Rauvolfová, J.; Přikrylová, V.; Pišvejcová, A.; Ettrich, R.; Kuzma, M.; Sedmera, P.; Křen, V. Hydrolytic and transglycosylation reactions of N-acyl modified substrates catalysed by β-N-acetylhexosaminidases. Tetrahedron 2004, 60, 693–701. [Google Scholar] [CrossRef]
- Pócsi, I.; Pusztahelyi, T.; Bogáti, M.; Szentirmai, A. The formation of N-acetyl-β-D-hexosaminidase is repressed by glucose inPenicillium chrysogenum. J Basic Microbiol. 1993, 33, 259–267. [Google Scholar] [CrossRef]
- Weignerová, L.; Vavrušková, P.; Pišvejcová, A.; Thiem, J.; Křen, V. Fungal β-N-acetylhexosaminidases with high β-N-acetylgalactosaminidase activity and their use for synthesis of β-GalNAc-containing oligosaccharides. Carbohydr. Res. 2003, 338, 1003–1008. [Google Scholar] [CrossRef]
- Yamamoto, K.; Lee, K.; Kumagai, H.; Tochdcura, T. Purification and Characterization of β-N -Acetylhexosaminidase from Penicillium oxalicum. Agric. Biol. Chem. 1985, 49, 611–619. [Google Scholar] [CrossRef]
- Orenga, S.; James, A.; Manafi, M.; Perry, J.; Pincus, D.H. Enzymatic substrates in microbiology. J. Microbiol. Methods 2009, 79, 139–155. [Google Scholar] [CrossRef]
- Profeta, G.S.; Pereira, J.A.; Costa, S.G.; Azambuja, P.; Garcia, E.S.; Moraes, C.D.S.; Genta, F.A. Standardization of a Continuous Assay for Glycosidases and Its Use for Screening Insect Gut Samples at Individual and Populational Levels. Front. Physiol. 2017, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Van Tilbeurgh, H.; Loontiens, F.; De Bruyne, C.; Claeyssens, M. Fluorogenic and chromogenic glycosides as substrates and ligands of carbohydrases. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1988; Volume 160, pp. 45–59. [Google Scholar] [CrossRef]
- Konkol, N.; McNamara, C.; Mitchell, R. Fluorometric detection and estimation of fungal biomass on cultural heritage materials. J. Microbiol. Methods 2010, 80, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Matthews, B.; Huber, R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance: LacZ β-Galactosidase. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.M.D.; Jaramillo, O.B.; Teixeira, R.; Espindola, F.S. Salivary Surrogates of Plasma Nitrite and Catecholamines during a 21-Week Training Season in Swimmers. PLoS ONE 2013, 8, e64043. [Google Scholar] [CrossRef]
- Yumasdhika, F.; Suharsini, M.; Indiarti, I.S.; Anggraeni, H.D. Correlation between FLACC Pain Score and Salivary Alpha-Amylase Level (A Review on Children with Down Syndrome). J. Int. Dent. Med. Res. 2017, 10, 529–532. [Google Scholar]
- Ferreira, C.R.; Gahl, W.A. Lysosomal storage diseases. TRD 2017, 2, 1–71. [Google Scholar] [CrossRef] [PubMed]






| Substrate | Substrate Concentration | Reaction Volume | Enzyme Activity | Spectrophotometric Determination (Wavelength) | HPLC Determination | ||
|---|---|---|---|---|---|---|---|
| Column | Mobile Phase | Flow Rate | |||||
| 4MU-β-GalNAc | 4 mM | 2 mL | 0.2–0.3 U | 347 nm | Nucleodur C18ec, 250 × 4 mm, 30 °C | 0.5% o-phosphoric acid (A), acetonitrile (B); gradient: 0–5 min, held at 21% B; 5–10 min, 21–40% B; 10–13 min, held at 40% B; 13–55 min, 40–21% B. | 1 mL/min |
| X-ß-GalNAc | 0.5 mM | 2 mL | 0.1–0.7 U | - | Nucleodur C18ec, 250 × 4 mm, 30 °C | Water (A), acetonitrile (B); gradient: 0–10 min, 40–80% B; 10–15 min, 80–40% B; 15–20 min, held at 40% B. | 0.7 mL/min |
| 2Cl-4NP-ß-Gal | 1 mM | 2 mL | 0.2–0.3 U | 420 nm | Nucleodur C18ec, 250 × 4 mm, 30 °C | Water (55%), acetonitrile (45%). | 0.7 mL/min |
| Specific Activity (U/mg Protein) | ||||||
|---|---|---|---|---|---|---|
| 4NPGalNAcase | 4NPGlcNAcase | Ratio 4NPGalNAcase/ GlcNAcase | 2NPGalNAcase | 2NPGlcNAcase | Ratio 2NPGalNAcase/ GlcNAcase | |
| PcHex1 | 17.40 ± 0.52 | 16.61 ± 0.33 | 1.0 | 23.94 ± 1.13 | 20.05 ± 0.89 | 1.2 |
| PcHex3437 | 15.31 ± 0.44 | 11.89 ± 0.23 | 1.3 | 16.40 ± 0.61 | 12.54 ± 0.43 | 1.3 |
| PcHex3210 | 14.46 ± 0.24 | 18.76 ± 0.52 | 0.8 | 26.76 ± 0.83 | 21.52 ± 0.64 | 1.3 |
| PcHex62558 | 15.81 ± 0.49 | 15.96 ± 0.51 | 1.0 | 20.32 ± 0.79 | 24.48 ± 1.09 | 0.8 |
| Specific Activity (U/mg Protein) | |||
|---|---|---|---|
| 4MU-β-GalNAc | X-β-GalNAc | 2Cl-4NP-β-Gal | |
| PcHex1 | 15.4 ± 1.2 | 0.181 ± 0.004 | 3.32 ± 0.29 |
| PcHex3437 | 5.67 ± 0.51 | 0.171 ± 0.025 | 3.97 ± 0.30 |
| PcHex3210 | 5.05 ± 0.47 | 0.221 ± 0.019 | 2.082 ± 0.009 |
| PcHex62558 | 5.89 ± 0.42 | 0.287 ± 0.025 | 2.50 ± 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ondrejková, E.; Hronská, H.; Štefuca, V.; Bláhová, M.; Rosenberg, M. Isolation, Specificity, and Application of β-N-Acetylhexosaminidases from Penicillium crustosum. Appl. Sci. 2023, 13, 3399. https://doi.org/10.3390/app13063399
Ondrejková E, Hronská H, Štefuca V, Bláhová M, Rosenberg M. Isolation, Specificity, and Application of β-N-Acetylhexosaminidases from Penicillium crustosum. Applied Sciences. 2023; 13(6):3399. https://doi.org/10.3390/app13063399
Chicago/Turabian StyleOndrejková, Ema, Helena Hronská, Vladimír Štefuca, Mária Bláhová, and Michal Rosenberg. 2023. "Isolation, Specificity, and Application of β-N-Acetylhexosaminidases from Penicillium crustosum" Applied Sciences 13, no. 6: 3399. https://doi.org/10.3390/app13063399
APA StyleOndrejková, E., Hronská, H., Štefuca, V., Bláhová, M., & Rosenberg, M. (2023). Isolation, Specificity, and Application of β-N-Acetylhexosaminidases from Penicillium crustosum. Applied Sciences, 13(6), 3399. https://doi.org/10.3390/app13063399







