Abstract
Thermophilic proteins have great potential to be utilized as biocatalysts in biotechnology. Machine learning algorithms are gaining increasing use in identifying such enzymes, reducing or even eliminating the need for experimental studies. While most previously used machine learning methods were based on manually designed features, we developed BertThermo, a model using Bidirectional Encoder Representations from Transformers (BERT), as an automatic feature extraction tool. This method combines a variety of machine learning algorithms and feature engineering methods, while relying on single-feature encoding based on the protein sequence alone for model input. BertThermo achieved an accuracy of 96.97% and 97.51% in 5-fold cross-validation and in independent testing, respectively, identifying thermophilic proteins more reliably than any previously described predictive algorithm. Additionally, BertThermo was tested by a balanced dataset, an imbalanced dataset and a dataset with homology sequences, and the results show that BertThermo was with the best robustness as comparied with state-of-the-art methods. The source code of BertThermo is available.
1. Introduction
Enzymes derived from thermophilic organisms often have great practical utility as biocatalysts. While higher temperatures tend to increase catalytic efficiency, conventional enzymes denature and lose effectiveness in high-temperature environments [1]. However, thermophilic proteins to do not suffer from this limitation, making them attractive for use in biotechnology.
Historically, various predictive methods using machine learning have been proposed to identify thermophilic proteins based on their amino acid sequence. However, these algorithms cannot directly use the protein sequence information as training input, necessitating various feature extraction methods to construct a feature matrix, typically requiring extensive human interventions [2]. The features extracted in this process, including amino acid composition [2,3,4,5,6,7,8,9,10,11,12], composition transition and distribution [2,7,11,12,13], and dipeptide deviation from the expected means [10,14,15], all have clear physicochemical relevance. Several machine learning classification schemes were proposed based on one or more of these physical and chemical features. Attempts to improve the accuracy of prediction relied on refining the aspects of feature extraction [16,17], feature selection [18,19,20,21], and better classifier selection [22,23]. Feng C. et al. [9] re-encoded amino acid sequences with RAAC [24], extracted physicochemical characteristics [25,26], utilized auto-cross covariance [27], and reduced the number of dipeptides, resulting in an algorithm with an accuracy of 0.982 in 10-fold cross-validation testing. In terms of feature selection, Guo Z. et al. [2] introduced MRMD2.0 as a feature selection algorithm to reduce the feature matrix to 119 dimensions, achieving a final accuracy of 0.9602. To improve classifier use, Ahmed Z et al. [10] adopted multi-layer perceptron as a machine learning algorithm, obtaining an accuracy of 0.9626 in independent tests. Of these approaches, re-encoding the amino acid sequence with RAAC was the most promising, achieving an accuracy of 0.982. Furthermore, as in these tests only 500 of the 915 randomly selected thermophilic proteins and 500 of the 793 non-thermophilic proteins were used for training purposes from a pre-existing dataset, these results could be theoretically improved further. In 2023, a model named DeepTP [12] was constructed using six artificial design features as inputs, combined with attention mechanism network and multilayer perceptron as classifiers. DeepTP achieved an accuracy of 87.1% for 10-fold cross-validation with the new benchmark dataset; DeepTP was also with better independently tested results than those of reportd methods for testing in various scenario, including a balanced dataset, a non-balanced dataset and homologous data.
Recent rapid developments in the field of natural language processing (NLP) made it possible to directly extract information from proteins using self-supervised learning algorithms [28,29,30,31]. These models treat protein sequences as natural language, interpreting amino acids as words in a sentence [32] and extracting meaningful information directly from the sequence [33]. Several deep representation learning methods have been used for such protein sequence analysis, including BERT [32,34], UniRep [35], Word2Vec (W2V) [36], and SSA [37]. These algorithms were utilized to predict protein interaction sites [38], and to identify peptides with bitter [39] or umami taste [40].
The work presented here extends this use of deep representation learning to extract features of amino acid sequences suitable to predict thermophilic characteristics in proteins [41]. This work is the first description of using sequence-based bidirectional representations from transformer (BERT) embedding features for this purpose. We named the method as BertThermo, which embedded amino acid via pre-trained BERT model and then used logistic regression as classifiers. BertThermo’s accuracy is 96.97% and 97.51% in 5-fold cross-validation and in independent testing, respectively, which is better than the state-of-the-art methods using the same benchmark dataset. It is believed that BertThermo would be a useful toolkit for researcher in the area of thermophilic proteins.
2. Materials and Methods
The framework of BertThermo is illustrated in Figure 1. We introduced a dataset containing 1443 non-thermophilic and 1366 thermophilic proteins to train the model. Each protein sequence was taken as text and feature-extracted by the BERT-bfd model. After tokenization, encoding, and pooling, BertThermo converted the sequences into 1024-dimensional feature vectors consisting of 1443 negative and 1366 positive samples. We then introduced the SMOTE to synthesize new positive samples to address the imbalance between positive and negative samples. After obtaining a balanced dataset, the feature selection method LGBM was used to overcome overfitting, converting the 1024-dimensional vectors into shorter ones. Then, Logistic Regression, which had the highest accuracy among the seven classifiers in this task, was used to classify these feature vectors. Finally, we upload the BertThermo on github for researchers in this area. The source code of BertThermo is available at https://github.com/zhibinlv/BertThermo (accessed on 27 January 2022).
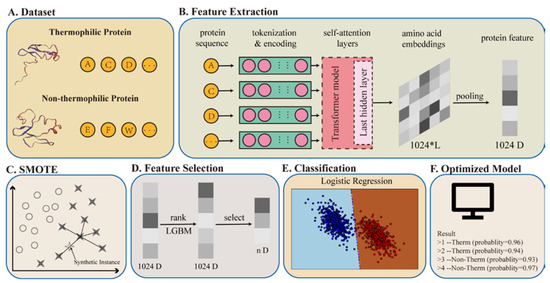
Figure 1.
Technology roadmap. (A) is the database of thermophilic and non-thermophilic protein sequences as input for feature extraction. (B) is the feature extraction part using the BERT-bfd to convert the sequence information into a 1024-dimensional feature vector. (C) is to balance the dataset using SMOTE. (D) is the feature selection part, which uses the LGBM to sort features in the 1024-dimensional feature vector, and selects the top n features as the input of the classifier. (E) naïve is the classifier using the Logistic Regression to train the model. (F) is a source code package we build which can predict the thermophilic characteristic of the sequences on user’ computer.
2.1. The Benchmark Dataset
We used a dataset extracted from the universal protein resource by Ahmed et al. [10]. Generally, thermophilic proteins are synthesized by heat tolerant microbes, characterized by a higher optimal growth temperature (OGT). As proposed by Lin et al. [5], proteins synthesized by microorganisms with OGT > 60 °C are considered thermophilic while proteins synthesized by microorganisms with OGT < 30 °C are considered non-thermophilic. Ahmed et al. improved the reliability of their curated dataset through the following steps: First, they removed proteins that have not been manually checked, eliminated ‘fuzzy fragments’ that could be part of other proteins, and discarded ‘proteins’ obtained solely via prediction or homology analysis. Next, the CD-HIT program with a cutoff of 30% was used to remove sequences with high similarity [42]. This processing resulted in a dataset containing 1443 non-thermophilic and 1366 thermophilic proteins. Then, the dataset [10] was divided into 80:20 ratios. The 80% dataset and the 20% dataset were used as the training dataset and the independent dataset in this work, respectively. In real practical application, the number of non-thermophilic proteins is much more than that of thermophilic proteins. Therefore, in order to evaluate the ability of the model in this regard, we redeployed our model using a new benchmark training dataset and three independent test sets for testing, which mentioned in DeepTP [12]. For more details about the datasets, please see reference of DeepTP thermophilic proteins [12].
2.2. Feature Extraction
Before classification, in a process referred to as feature extraction, protein sequences need to be converted into feature vectors [43]. In contrast to previous work, which relied on extracted features with physical and chemical significance, we used deep representation learning to extract features. In the development of our model we tested four feature extraction models: BERT [32,34], UniRep [35], SSA [37], and W2V [36]. All of these convert amino acid sequences of different lengths into fixed-length vectors that can be combined with downstream machine learning models for supervised learning and to complete various downstream tasks.
2.2.1. BERT
The training of the BERT model can be divided into two steps: pre-training and fine-tuning. Pre-training is performed on unlabeled datasets to obtain initial parameters, while fine-tuning is achieved by reassessing the initial parameters through labeled downstream tasks. Originally developed to process natural language, BERT can be trained to treat the amino acid sequences of proteins as sentences. In this context, the concept of a ‘word’ can be either a single amino acid, several adjacent amino acids [44], or a functional unit [45]. In this paper, we used the assumption that a single amino acid in protein was a ‘word’ in a sentence.
There are several distinct versions of the BERT that differ in their pre-training methods. In order to select the most suitable version for thermophilic protein identification, we tested BERT, BERT-bfd, and TAPE-BERT. During pre-training, BERT uses UniRef100 [46], BERT-bfd uses the Big Fantastic Database (BFD) [47], and TAPE-BERT uses the protein families database (Pfam) [48]. Of these, BFD is the most extensive collection, containing 2122 million proteins, while UniRef100 contains 216 million, and Pfam 31 million sequences.
BERT, BERT-bfd, and TAPE-BERT have similar structures. As an example, BERT-bfd is shown in Figure 1B. As illustrated, BERT-bfd first encodes and tokenizes an amino acid sequence of length L into a matrix. This process generates the input of a transformer function that produces a hidden state of the last self-attention layer as the output. This output is a matrix for BERT-bfd and BERT, and a matrix for TAPE-BERT. In the final step, this matrix is transformed into a feature vector through mean-pooling, to facilitate downstream machine learning. The final feature vector is 1024-dimensional for BERT and BERT-bfd, and 768-dimensional for TAPE-BERT.
2.2.2. UniRep
UniRep conducts unsupervised learning based on the UniRef50 database [46]. After the removal of proteins containing more than 5000 amino acids and sequences with noncanonical amino-acid symbols (X, B, Z, J), this consists of 24 million proteins. UniRep uses mLSTM for representation learning.
This approach first encodes amino acid sequences of length L to generate a amino acid feature matrix, forming the input of mLSTM. At this point the last hidden layer, a matrix, is created as the output. Finally, this matrix is transformed into a 1900-dimensional feature vector through mean-pooling.
2.2.3. SSA
The SSA algorithm can be utilized to predict structural similarity between two protein sequences. SSA is divided into two components: its pre-training language model is based on a multi-layer bidirectional LSTM (biLTSM) that encodes input protein sequences into feature vectors of fixed length (121D). The second component conducts structural similarity calculations based on Soft Symmetric Alignment. Since in the context of our work structure similarity was irrelevant, we only used the first part of this algorithm, the pre-training language model, using the Pfam database [48] for unsupervised learning.
2.2.4. W2V
W2V was trained on 524,529 untagged sequences from the UniProt database [49]. Unlike previously described characteristics-based representation models, W2V treats k-mers as words. After embedding the sequence into the doc2vec model, the algorithm is trained by predicting the middle k-mer based on context k-mer sequences.
2.3. The Synthetic Minority over Sampling Technique (SMOTE)
The SMOTE is a common oversampling technique used to address the imbalance between positive and negative samples in datasets. This approach uses an algorithm to synthesize new minority samples artificially. For example, if there are fewer positives and an excess number of negatives in a dataset, the SMOTE synthesizes new positive samples as illustrated in Figure 1C. First, for each positive sample, the Euclidean distance is taken as the standard to find its K-nearest neighbor. Next, a sampling ratio of n is set according to the proportion of sample imbalance. Random positive samples are selected from the K-nearest neighbors of a positive sample. Finally, each is connected with the original sample and a random point is selected on each line and used as a synthesized sample.
2.4. Feature Selection Method
Overfitting is a significant issue in machine learning [50] and feature selection is often necessary to avoid this problem caused by using a high-dimensional input with the intention of making full use of the implicit information [51]. In the work presented here input features and labels were sorted from most important to least important, using the Lighting Gradient Boosting Machine (LGBM) algorithm, as shown in Figure 1D. Using this approach, we selected the top n classifiers used in the next steps.
2.5. Classifiers
To identify thermophilic proteins by using the feature vectors extracted in the previous steps, we tested seven machine learning classifiers, which are briefly introduced below.
2.5.1. Logistic Regression (LR)
This paper primarily used LR, a widely used classifier in machine learning. The formula describing LR is similar to that of the linear regression model, in the form of . The difference is that linear regression uses directly as the dependent variable, while LR maps to a discrete logical output via the Sigmoid function [52].
2.5.2. Naïveive Bayes (NB)
NB is a classification method based on Bayes’ theorem [53]. Let the input space be an N-dimensional vector space , and the output space be the label set . The corresponding training set is:
where xi is the eigenvector of the instance. We used Bayes’ formula to calculate . This way the probability of the instance corresponding to Label can be obtained. Once the probabilities of all labels are calculated, the one with the highest probability is selected, and its corresponding is considered to be the Label corresponding to .
2.5.3. Linear Discriminant Analysis (LDA)
LDA is a classical linear discriminant analysis method. On a given training sample set, LDA attempts to project instances onto a straight line, rendering the projection points of similar instances as close as possible while keeping the projection points of different examples at the maximum distance [54]. When classifying a new sample, it is projected onto the line, and its category is determined according to the position of the projected point.
2.5.4. K-Nearest Neighbor (KNN)
KNN is a classification algorithm based on the assumption that a closer position indicates a higher degree of similarity [55]. KNN places the sample that is to be classified into input space , where is the n-dimensional real vector space , to generate a point. KNN finds the nearest k samples to , checks these neighbors, and forms a judgment according to the ‘minority is subordinate to the majority, every point corresponds to one vote’ principle. In this analysis the label with the most points becomes the label of x.
2.5.5. Random Forest (RF)
RF is an ensemble learning algorithm based on a decision tree [56]. RF obtains multiple estimators through training and combines the results of multiple estimators into a final result. RF incorporates multiple decision trees, with the dataset and features of each decision tree being randomly selected and returned [57]. RF uses these models to make predictions and takes an average or a plurality of categories to obtain the final forecast results.
2.5.6. Supporting Vector Machine (SVM)
The principle of SVM is to find a hyperplane in the input space to separate different samples [58]. This hyperplane should maximize the sum of the distances between each sample within a set [59]. This way, when new samples appear, they can be classified on this optimized hyperplane.
2.5.7. The Lighting Gradient Boosting Machine (LGBM)
The LGBM is an evolutionary version of the GBDT model [60]. The central concept of the LGBM is to use a weak classifier (decision tree) to train the model to obtain the optimal classifier in an iterative process. The growth mode of the LGBM tree is vertical, while other algorithms represent horizontal growth. Thus, the LGBM grows the leaves of the tree, while other algorithms grow the level. Consequently, growing the same leaves, this algorithm reduces information loss compared to level-wise algorithms.
2.6. Evaluation
We used the following five metrics to measure model performance [61]: ACC, Sn, Sp, Recall, Precision, MCC, AUC and AP, representing accuracy, sensitivity, specificity (recall reate), precision rate, Matthews’s correlation coefficient, area under receiver operating characteristic (ROC) curve, and average of precision, respectively.
where TP, FP, FN and TN mean number of samples for true positive, false positive, false negative and true negeative.
ROC curve is drawn according to probalibity threshold, with the true positive rate (sensitivity) as the vertical coordinate and the false positive rate (1-specificity) as the horizontal coordinate. AUC is defined as the area under the ROC curve. The value of AUC can range from 0.5 to 1. The closer the AUC is to 1.0, the higher the accuracy of the detection method. In contrast, a value of 0.5 indicates a random prediction.
For AP metrics, it is a more effective metric for an imbalanced dataset test. These metrics are calculated by and at the nth probability threshold. Generally, AUC is enough to evaluate the model test by a balanced dataset, while for an imbalanced dataset test, it is better to use AP as scoring.
3. Results
3.1. Performance of Models Using Various BERT Embedding Features
In order to select the most suitable BERT features for the classification of thermophilic proteins, BERT, BERT-bfd, and TAPE-BERT were used to extract features. The seven classifiers described in Materials and methods were used for training and prediction, and performance of the models, using different implementations of BERT was compared. As shown in Figure 2, models using BERT and BERT-bfd showed broadly comparable performance, while TAPE-BERT was clearly inferior. As indicated in Figure 2F, the average accuracy of the seven classifiers using the BERT, BERT-bfd, and TAPE-BERT models was 0.9502, 0.9513, and 0.8746, respectively. It was apparent from these data that BERT-bfd was the best performing model overall. Furthermore, all individual metrics, apart from Sn, reached their best values when using the BERT-bfd model.
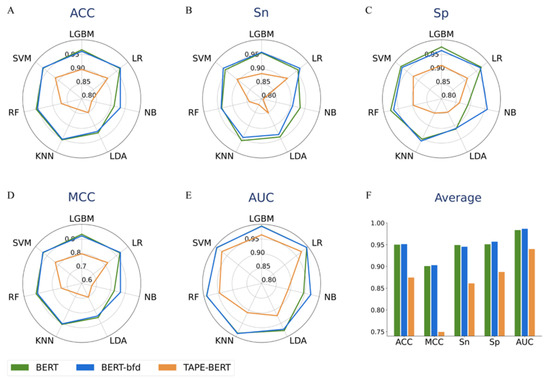
Figure 2.
The 5-fold cross-validation performance of models using BERT, BERT-bfd, and TAPE-BERT features, with seven different classifiers. (A) is the radar map of ACC. (B) is the radar map of Sn. (C) is the radar map of Sp. (D) is the radar map of the MCC. (E) is the radar map of AUC. (F) is the histogram of average performance, which is the average performance obtained after the average of seven different classifiers.
Since our goal was to build the most efficient classifier, in addition to the average performance, we also evaluated the optimal performance of individual components in each of the three models. Using different classifiers, the highest accuracies achieved were 0.9671 with BERT using the SVM classifier, 0.9684 for BERT-bfd with SVM, and 0.9145 for TAPE-BERT with SVM. Since the model using BERT-bfd achieved the highest accuracy, this was used for the purposes of further comparisons. Compared with Bert feature pre-trained by using UniRef50 and TAPE-BERT feature pre-trained using the Pfam database, the Bert-bdf feature was pre-trained by using larger protein data BFD, so the Bert-bdf feature contains more abundant protein sequence information than the other two [32].
3.2. Performance of Models Using BERT-Bfd and Other Deep Representation Learning Features
Next, we run the three other deep representation learning models, UniRep, SSA, and W2V, using all seven classifiers, to extract features and compare their performance with BERT-bfd. The results of these comparisons are summarized in Figure 3. The data clearly indicated that BERT-bfd outperformed UniRep, SSA, and W2V in feature extraction, irrespective of what classifier was used for training and prediction. The best accuracy achievable with BERT-bfd was 0.9513, compared to 0.7673, 0.8611, and 0.8107 achieved with UniRep, SSA, and W2V, respectively. Furthermore, the lowest accuracy of models using BERT-bfd was over 0.92, while the ACC value of the next best option, SSA with the LGBM classifier, was 0.9021. BERT-bfd appeared to be the most suitable model for the prediction and classification of thermophilic proteins. Additionally, the best model is the one with BERT-bfd feature, which is superior to others, which may be because BERT-bfd is more novel and more efficient at capturing contextual information stored in protein sequences [32].
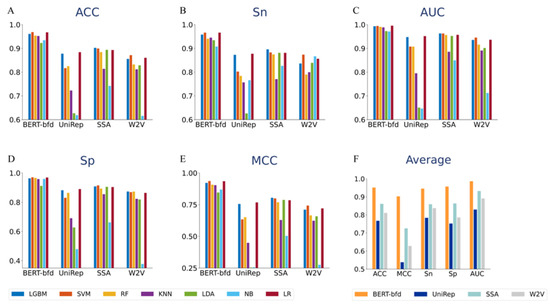
Figure 3.
Performance of models using BERT-bfd and other deep representation learning features, UniRep, SSA, and W2V, with seven different classifiers. (A) is the histogram of ACC. (B) is the histogram of Sn. (C) is the histogram of AUC. (D) is the histogram of Sp. (E) is the histogram of MCC. (F) is the histogram of average performance of seven different classifiers.
3.3. Performance of Models Using BERT-Bfd with Seven Different Classifiers
After demonstrating that BERT-bfd was the most suitable feature extraction model for the identification of thermophilic proteins, we needed to establish the optimal classifier that could be utilized. We compare the performance of models using BERT-bfd feature with seven classifiers. We tested the potential combinations, either with or without using the SMOTE, in both cross-validation experiments and independent tests. As shown in Table 1, the highest accuracy achievable in 5-fold cross-validation was 0.9697, obtained when LR was used as classifier while applying the SMOTE. The highest accuracy in the independent test was 0.9751, observed again using LR as the classifier but this time without the SMOTE. We also compared the average (independent test and cross-validation) performance of the models. Here, the highest average accuracy, 0.9713, was obtained by the LR classifier without the SMOTE, indicating that this model had the best overall performance in cross-validation and independent tests. Therefore, we intend to utilize this classifier in in the rest of the results section.

Table 1.
Performance of models using BERT-bfd feature with seven different classifiers.
3.4. Performance after Feature Selection
Our dataset consisted of 1368 thermophilic proteins and 1443 non-thermophilic proteins, and the feature matrix had 1024 dimensions. A dataset of this size and complexity almost invariably leads to the overfitting of AI models. In this respect, it is important to point out that in previous studies, features used for classification usually had a range of dimensions between 20 and 30. Thus, the 1024 dimensions extracted by BERT-bfd were likely to be redundant. Therefore, we used the LGBM to reduce the feature dimension generated by BERT-bfd. In these predictions, we tested the performance of the model using BERT-bfd and LR as a classifier in either cross-validation or independent tests, both with and without the SMOTE. The number of top features used in these trials was increased from 10 to 300 with increments of 10 additional features added in each iterative round. Table 2 shows the comparison between the performance of predictions using the selected top 90 features against the unselected complete feature set. These results clearly indicated that after the SMOTE, selecting the top 90 features resulted in the highest average accuracy of 0.9724. In addition, other performance metrics were also higher compared to running the model without feature selection. Therefore, the 1024-dimensional feature set extracted by BERT-bfd was indeed highly redundant, with feature selection resulting in improved model performance. Although we gave only the results of LR in this part, we had tested all the seven classifiers on the reduced set of features, which were listed in Supporting Information Table S1. Additionally, it could be found that the LR model had the best average ACC. The unabridged results have been uploaded on the github for consistency of results.

Table 2.
Before or after feature selection and with or without the SMOTE, performance of the model on the independent test and cross-validation.
3.5. Feature Analysis Using Dimension Reduction
To provide a visual representation of the performance of the various models using deep representation learning, we used a dimension-reduction method, uniform manifold approximation and projection (UMAP). In this, we reduced the feature vector into a 2-dimensional space using UAMP and displayed the vectors as a series of points on this plane, as shown in Figure 4. Red points represent thermophilic proteins (positive samples), while blue points indicate non-thermophilic proteins (negative samples). The decision function line of LR were also drawn in Figure 4. As shown in Figure 4A–C, there was considerable overlap between positive and negative samples when protein prediction was carried out using UniRep, SSA, or W2V. Consequently, classifying samples accurately using these features is almost impossible in a 2-dimensional space. However, as shown in Figure 4D, most positive and negative samples projected in the 2D representation of BERT-bfd vectors concentrated into two distinct areas, with positive samples mostly in the yellow area and negative samples in the cyan area in Figure 4D. This finding visually indicated that LR classified most of the samples correctly in this 2-dimensional space, indicating the superiority of BERT-bfd-based feature extraction.
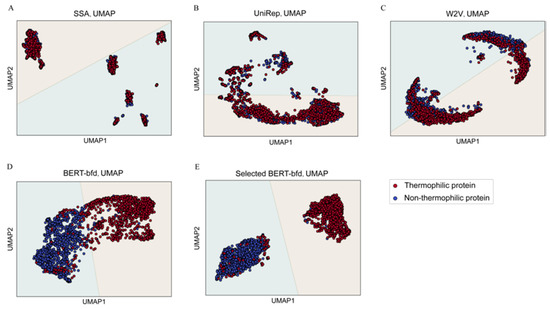
Figure 4.
Dimension reduction visualization of BERT-bfd and other deep representation learning features. Red dots represent thermophilic proteins and blue dots represent non-thermophilic proteins. Horizontal axis and vertical axis are the first feature and the second feature of the reduced 2-dimensional vectors, respectively. These dots are classified by LR (yellow area for positive samples, while cyan area for negative samples). (A) shows the SSA feature reduced by UMAP. (B) shows the UniRep feature reduced by UMAP. (C) shows the W2V feature reduced by UMAP. (D) shows the BERT-bfd feature reduced by UMAP. (E) shows the selected BERT-bfd (top 90) feature reduced by UMAP.
As described in Section 3.4, feature selection could further improve the performance of the BertThermo model. Therefore, we also introduced dimension reduction using the top 90 BERT-bfd features and displayed the results as a 2D figure, shown in Figure 4E. The distribution of positive and negative samples identified under these conditions was concentrated into smaller areas. However, there are still considerable overlap between positive and negative samples. Therefore, better feature selection methods or feature extraction methods were still needed for better identification in the future. Additionally, as with the improvement of computational resources, for training, interpreting and predicting, a better performance model will be developed.
3.6. Comparison with Previous Models
Previously reported methods for the prediction of thermophilic proteins used physicochemical feature extraction methods. Although deep learning had already been used to identify thermophilic proteins by previous works, such as DeepTP [12], these models did not avoid using physicochemical features such as AAC, DC, and CTD. In a striking departure from this approach, our work is the first demonstration of the utility of deep representation learning algorithms for identifying thermophilic enzymes. We compared the performance of our BertThermo model with the previously proposed prediction algorithms. Since most previous models reported outcomes based cross validation experiments, while others showed the outcome of independent tests, we summarized these comparisons in two tables. Table 3 illustrates the findings of cross validation tests, while Table 4 illustrates the outcomes of independent tests using the same test set as us.

Table 3.
Comparison of our model and previous models on cross-validation.

Table 4.
Comparison of our model and previous models on independent test with a iThermo benchmark dataset.
Our model showed the highest accuracy in cross validation tests, reaching an Sn and Sp of 0.9697. These values are significantly better than those obtained with previous models. In terms independent tests, the best accuracy achieved by our model was 0.9751, with other metrics being also better than those produced by the previous model, iThermo. Since our BertThermo algorithm was tested on the same dataset as iThermo [10], these results represent direct comparisons and clearly demonstrate the improved performance of the new approach. From another perspective, compared with the complex classifiers and higher computational complexity chosen by other groups, such as SVM [2,5,6,7,8] and MLP [10], our model uses LR as a classifier, resulting in reduced computational complexity. This also means that BertThermo is easier to train and is more stable. In more general terms, the impressive performance of BertThermo shows the promise of NLP-based algorithms in predicting thermophilic proteins and suggests the utility of NLP-based approaches in studying other features of protein function and behavior in the future.
3.7. Comparison with a Different Benchmark Dataset Used by DeepTP
In order to further evaluate the performance of BertThermo, the benchmark training set of DeepTP [12] was used to retrain the model, and the three benchmark independent test sets used by DeepTP were used to score the model. The scoring results are listed in Table 5 and Table 6. First, compared with DeepTP, BertThermo shows better values for the 10-fold cross-validation with the same training set (see Table 5). Second, for independent testing of balanced data, the metrics of BertThemo, except for Sp, are greater than the other state-of-the-art methods as far as we know. In fact, the number of non-thermophilic proteins is far more than that of thermophilic proteins. This means that data imbalance is a common situation. From the independent test results of an imbalanced dataset in Table 6, it can see that BertThermo has better generalization performance in the case of data imbalance. At the same time, in practical application, proteins with homology may show different thermophilicity. In Table 6, the results of testing 100 thermophilic proteins and 100 non-thermophilic proteins with a similarity of more than 40% showed that BertThermo also had the best performance.

Table 5.
Ten-fold cross-validation metrics for BertTherm reimplemented by a dataset from DeepTP.

Table 6.
Comparison with state-of-the-art methods with various independent datasets.
4. Conclusions
The work presented here compares the performance of models using three different implementations of BERT in identifying thermophilic proteins. We found that the use of BERT-bfd is the most suitable approach for this task, significantly outperforming three other deep representation learning models, UniRep, SSA, and W2V. We refined the algorithm further, showing that out of seven tested classifiers, LR results in the highest accuracy in both cross-validation and independent tests. In addition, the use of feature engineering methods, such as the SMOTE and the LGBM, could increase the performance of the model even further. In final testing, the optimized model achieved an accuracy of 96.97% in 5-fold cross-validation and 97.51% in independent tests. Comparing the performance of the BertThermo algorithm reported here with previous models proves that BertThermo is currently the best available model for identifying thermophilic proteins. This model provides the scientific community with a promising new tool for the detection of enzymes that are stable at higher temperatures, and may provide valuable information for use in biotechnology.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app13052858/s1, Table S1. The evaluation metrics score of models with various machine learning method.
Author Contributions
H.P. performed the data analyses and wrote the manuscript; J.L., J.J. and S.M. performed the data collection; M.L. helped write the source code; Q.Z. contributed significantly to constructive discussions; Z.L. supervised the work. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (No. 62001090, No. 62250028, No. 62131004), the Sichuan Provincial Science Fund for Distinguished Young Scholars (2021JDJQ0025), the Municipal Government of Quzhou (No. 2022D040) and Fundamental Research Funds for the Central Universities of Sichuan University (No. YJ2021104).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset for model training and testing is available at https://github.com/zhibinlv/BertThermo (accessed on 27 January 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ahmed, Z.; Zulfiqar, H.; Tang, L.; Lin, H. A Statistical Analysis of the Sequence and Structure of Thermophilic and Non-Thermophilic Proteins. Int. J. Mol. Sci. 2022, 23, 10116. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, P.; Liu, Z.; Zhao, Y. Discrimination of Thermophilic Proteins and Non-thermophilic Proteins Using Feature Dimension Reduction. Front. Bioeng. Biotechnol. 2020, 8, 584807. [Google Scholar] [CrossRef]
- Bhasin, M.; Raghava, G.P.S. Classification of nuclear receptors based on amino acid composition and dipeptide composition. J. Biol. Chem. 2004, 279, 23262–23266. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Suresh, M.X. Discrimination of mesophilic and thermophilic proteins using machine learning algorithms. Proteins 2008, 70, 1274–1279. [Google Scholar] [CrossRef]
- Lin, H.; Chen, W. Prediction of thermophilic proteins using feature selection technique. J. Microbiol. Methods 2011, 84, 67–70. [Google Scholar] [CrossRef]
- Nakariyakul, S.; Liu, Z.-P.; Chen, L. Detecting thermophilic proteins through selecting amino acid and dipeptide composition features. Amino Acids 2012, 42, 1947–1953. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Fu, Z.; Xia, J. Prediction of thermophilic protein with pseudo amino Acid composition: An approach from combined feature selection and reduction. Protein Pept. Lett. 2011, 18, 684–689. [Google Scholar] [CrossRef]
- Fan, G.-L.; Liu, Y.-L.; Wang, H. Identification of thermophilic proteins by incorporating evolutionary and acid dissociation information into Chou’s general pseudo amino acid composition. J. Theor. Biol. 2016, 407, 138–142. [Google Scholar] [CrossRef]
- Feng, C.; Ma, Z.; Yang, D.; Li, X.; Zhang, J.; Li, Y. A Method for Prediction of Thermophilic Protein Based on Reduced Amino Acids and Mixed Features. Front. Bioeng. Biotechnol. 2020, 8, 285. [Google Scholar] [CrossRef]
- Ahmed, Z.; Zulfiqar, H.; Khan, A.A.; Gul, I.; Dao, F.-Y.; Zhang, Z.-Y.; Yu, X.L.; Tang, L. iThermo: A Sequence-Based Model for Identifying Thermophilic Proteins Using a Multi-Feature Fusion Strategy. Front. Microbiol. 2022, 13, 790063. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Schaduangrat, N.; Moni, M.A.; Lio, P.; Manavalan, B.; Shoombuatong, W. SAPPHIRE: A stacking-based ensemble learning framework for accurate prediction of thermophilic proteins. Comput. Biol. Med. 2022, 146, 105704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yan, W.; Yang, Y. DeepTP: A Deep Learning Model for Thermophilic Protein Prediction. Int. J. Mol. Sci. 2023, 24, 2217. [Google Scholar] [CrossRef]
- Dubchak, I.; Muchnik, I.; Mayor, C.; Dralyuk, I.; Kim, S.H. Recognition of a protein fold in the context of the Structural Classification of Proteins (SCOP) classification. Proteins 1999, 35, 401–407. [Google Scholar] [CrossRef]
- Saravanan, V.; Gautham, N. Harnessing Computational Biology for Exact Linear B-Cell Epitope Prediction: A Novel Amino Acid Composition-Based Feature Descriptor. Omics 2015, 19, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, P.; Zou, Q. Prediction of Thermophilic Proteins Using Voting Algorithm. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 8–10 May 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 195–203. [Google Scholar]
- Zhao, W.; Xu, G.; Yu, Z.; Li, J.; Liu, J. Identification of nut protein-derived peptides against SARS-CoV-2 spike protein and main protease. Comput. Biol. Med. 2021, 138, 104937. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Xu, C.; Luo, M.; Wang, P.; Xu, Z.; Xue, G.; Jin, X.; Huang, Y.; Li, Y.; Nie, H.; et al. MutCov: A pipeline for evaluating the effect of mutations in spike protein on infectivity and antigenicity of SARS-CoV-2. Comput. Biol. Med. 2022, 145, 105509. [Google Scholar] [CrossRef]
- Cao, C.; Kossinna, P.; Kwok, D.; Li, Q.; He, J.; Su, L.; Guo, X.; Zhang, Q.; Long, Q. Disentangling genetic feature selection and aggregation in transcriptome-wide association studies. Genetics 2022, 220, 34849857. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Kwok, D.; Edie, S.; Li, Q.; Ding, B.; Kossinna, P.; Campbell, S.; Wu, J.; Greenberg, M.; Long, Q. kTWAS: Integrating kernel machine with transcriptome-wide association studies improves statistical power and reveals novel genes. Brief. Bioinform. 2021, 22, bbaa270. [Google Scholar] [CrossRef]
- Cao, C.; Wang, J.; Kwok, D.; Cui, F.; Zhang, Z.; Zhao, D.; Li, M.J.; Zou, Q. webTWAS: A resource for disease candidate susceptibility genes identified by transcriptome-wide association study. Nucleic Acids Res. 2022, 50, D1123–D1130. [Google Scholar] [CrossRef]
- Canzhuang, S.; Yonge, F. Identification of Disordered Regions of Intrinsically Disordered Proteins by Multi-features Fusion. Curr. Bioinform. 2021, 16, 1126–1132. [Google Scholar] [CrossRef]
- Iraji, M.S.; Tanha, J.; Habibinejad, M. Druggable protein prediction using a multi-canal deep convolutional neural network based on autocovariance method. Comput. Biol. Med. 2022, 151 Pt A, 106276. [Google Scholar] [CrossRef]
- Jian, G.L.; Chen, H.B. A Path-based Method for Identification of Protein Phenotypic Annotations. Curr. Bioinform. 2021, 16, 1214–1222. [Google Scholar]
- Zheng, L.; Huang, S.; Mu, N.; Zhang, H.; Zhang, J.; Chang, Y.; Yang, L.; Zuo, Y. RAACBook: A web server of reduced amino acid alphabet for sequence-dependent inference by using Chou’s five-step rule. Database 2019, 2019, baz131. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Wei, L.; Yu, J.; Wang, C. Identifying Plant Pentatricopeptide Repeat Coding Gene/Protein Using Mixed Feature Extraction Methods. Front. Plant Sci. 2018, 9, 1961. [Google Scholar] [CrossRef]
- Cai, C.Z.; Han, L.Y.; Ji, Z.L.; Chen, X.; Chen, Y.Z. SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003, 31, 3692–3697. [Google Scholar] [CrossRef]
- Liu, B.; Wang, S.; Dong, Q.; Li, S.; Liu, X. Identification of DNA-binding proteins by combining auto-cross covariance transformation and ensemble learning. IEEE Trans. Nanobiosci. 2016, 15, 328–334. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Xia, W.; Zheng, L.; Fang, J.; Li, F.; Zhou, Y.; Zeng, Z.; Zhang, B.; Li, Z.; Li, H.; Zhu, F. PFmulDL: A novel strategy enabling multi-class and multi-label protein function annotation by integrating diverse deep learning methods. Comput. Biol. Med. 2022, 145, 105465. [Google Scholar] [CrossRef]
- Long, H.; Sun, Z.; Li, M.; Fu, H.; Lin, M. Predicting Protein Phosphorylation Sites Based on Deep Learning. Curr. Bioinform. 2020, 15, 300–308. [Google Scholar] [CrossRef]
- Ao, C.; Jiao, S.; Wang, Y.; Yu, L.; Zou, Q. Biological Sequence Classification: A Review on Data and General Methods. Research 2022, 2022, 11. [Google Scholar] [CrossRef]
- Elnaggar, A.; Heinzinger, M.; Dallago, C.; Rehawi, G.; Wang, Y.; Jones, L.; Gibbs, T.; Feher, T.; Angerer, C.; Steinegger, M.; et al. ProtTrans: Toward Understanding the Language of Life Through Self-Supervised Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 7112–7127. [Google Scholar] [PubMed]
- Detlefsen, N.S.; Hauberg, S.; Boomsma, W. Learning meaningful representations of protein sequences. Nat. Commun. 2022, 13, 1914. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Bhattacharya, N.; Thomas, N.; Duan, Y.; Chen, X.; Canny, J.; Abbeel, P.; Song, Y. Evaluating Protein Transfer Learning with TAPE. arXiv 2019, arXiv:1906.08230. [Google Scholar]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef]
- Yang, K.K.; Wu, Z.; Bedbrook, C.N.; Arnold, F.H. Learned protein embeddings for machine learning. Bioinformatics 2018, 34, 2642–2648. [Google Scholar] [CrossRef]
- Bepler, T.; Berger, B. Learning protein sequence embeddings using information from structure. arXiv 2019, arXiv:1902.08661. [Google Scholar]
- Hosseini, S.; Ilie, L. PITHIA: Protein Interaction Site Prediction Using Multiple Sequence Alignments and Attention. Int. J. Mol. Sci. 2022, 23, 12814. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Lin, X.; Jiang, Y.; Jiang, L.; Lv, Z. Identify Bitter Peptides by Using Deep Representation Learning Features. Int. J. Mol. Sci. 2022, 23, 7877. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, J.; Wang, X.; Zhang, Y.; Zheng, B.; Liu, S.; Zhang, Y.; Liu, C.; Wan, Y.; Xiang, D.; et al. IUP-BERT: Identification of Umami Peptides Based on BERT Features. Foods 2022, 11, 3742. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yu, L. EPSOL: Sequence-based protein solubility prediction using multidimensional embedding. Bioinformatics 2021, 37, btab463. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zou, Q.; Tang, F.; Yu, L. WMSA: A novel method for multiple sequence alignment of DNA sequences. Bioinformatics 2022, 38, 5019–5025. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F. Predicting Thermophilic Proteins by Machine Learning. Curr. Bioinform. 2020, 15, 493–502. [Google Scholar]
- Asgari, E.; McHardy, A.C.; Mofrad, M.R.K. Probabilistic variable-length segmentation of protein sequences for discriminative motif discovery (DiMotif) and sequence embedding (ProtVecX). Sci. Rep. 2019, 9, 3577. [Google Scholar] [CrossRef] [PubMed]
- Coin, L.; Bateman, A.; Durbin, R. Enhanced protein domain discovery by using language modeling techniques from speech recognition. Proc. Natl. Acad. Sci. USA 2003, 100, 4516–4520. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Steinegger, M.; Mirdita, M.; Söding, J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 2019, 16, 603–606. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, D.; Ding, H.; Zhong, B.; Xu, L. Escherichia coli DNA N-4-Methycytosine Site Prediction Accuracy Improved by Light Gradient Boosting Machine Feature Selection Technology. IEEE Access 2020, 8, 14851–14859. [Google Scholar] [CrossRef]
- Tang, Y.-J.; Pang, Y.-H.; Liu, B. IDP-Seq2Seq: Identification of intrinsically disordered regions based on sequence to sequence learning. Bioinformatics 2021, 36, 5177–5186. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, J.C. Logistic regression: A brief primer. Acad. Emerg. Med. 2011, 18, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xuan, Z.; Feng, X.; Zou, Q.; Wang, L. A novel collaborative filtering model for LncRNA-disease association prediction based on the Naïve Bayesian classifier. BMC Bioinform. 2019, 20, 396. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Meng, Q.; Chen, Y.; Wu, P. Subcellular location prediction of apoptosis proteins using two novel feature extraction methods based on evolutionary information and LDA. BMC Bioinform. 2020, 21, 212. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient kNN Classification with Different Numbers of Nearest Neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1774–1785. [Google Scholar] [CrossRef]
- Lv, Z.; Jin, S.; Ding, H.; Zou, Q. A Random Forest Sub-Golgi Protein Classifier Optimized via Dipeptide and Amino Acid Composition Features. Front. Bioeng. Biotechnol. 2019, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, K. iPromoter-2L2.0: Identifying Promoters and Their Types by Combining Smoothing Cutting Window Algorithm and Sequence-Based Features. Mol. Nucleic Acids 2019, 18, 80–87. [Google Scholar] [CrossRef]
- Huo, Y.; Xin, L.; Kang, C.; Wang, M.; Ma, Q.; Yu, B. SGL-SVM: A novel method for tumor classification via support vector machine with sparse group Lasso. J. Theor. Biol. 2020, 486, 110098. [Google Scholar] [CrossRef]
- Tan, J.X.; Li, S.H.; Zhang, Z.M.; Chen, C.X.; Chen, W.; Tang, H.; Lin, H. Identification of hormone binding proteins based on machine learning methods. Math. Biosci. Eng. 2019, 16, 2466–2480. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, S.; Xie, R.; Li, J.; Leier, A.; Marquez-Lago, T.T.; Akutsu, T.; Smith, A.I.; Ge, Z.; Wang, J.; et al. PeNGaRoo, a combined gradient boosting and ensemble learning framework for predicting non-classical secreted proteins. Bioinformatics 2020, 36, 704–712. [Google Scholar] [CrossRef]
- Yu, L.; Wang, M.; Yang, Y.; Xu, F.; Zhang, X.; Xie, F.; Gao, L.; Li, X. Predicting therapeutic drugs for hepatocellular carcinoma based on tissue-specific pathways. PLoS Comput. Biol. 2021, 17, e1008696. [Google Scholar] [CrossRef]
- Meng, C.; Ju, Y.; Shi, H. TMPpred: A support vector machine-based thermophilic protein identifier. Anal. Biochem. 2022, 645, 114625. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Chotpatiwetchkul, W.; Lee, V.S.; Nantasenamat, C.; Shoombuatong, W. A novel sequence-based predictor for identifying and characterizing thermophilic proteins using estimated propensity scores of dipeptides. Sci. Rep. 2021, 11, 23782. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).