Abstract
A growing societal awareness is calling upon scientists to reconsider the use of animals in research, which stimulates the development of translational in vitro models. The physiological and architectural interactions between different cell types within an organ present a challenge to these models, particularly for a complex organ such as the brain. Thus far, in vitro brain models mostly consist of a single cell type and demonstrate little predictive value. Here, we present a co-culture of an epileptic human neocortical biopsy on a layer of human induced pluripotent stem cell (hiPSC)-derived cortical neurons. The activity of the cortical neurons was recorded by a 120-electrode multi-electrode array. Recordings were obtained at 0, 3, and 6 days after assembly and compared to those obtained from cortical neurons without a biopsy. On all three recording days, the hybrid model displayed a firing rate, burst behavior, number of isolated spikes, inter-spike interval, and network bursting pattern that aligns with the characteristics of an epileptic network as reported by others. Thus, this novel model may be a non-animal, translational alternative for testing new therapies up to six days after resection.
1. Introduction
Whereas the burden of brain diseases is ever increasing, the treatment of these disorders often falls short. In search of new treatments, animal models were instrumental in unravelling disease mechanisms and yielded potential drug targets, yet up to 96% of human trials fail to reproduce drug effects that are eminent in animal models [1]. As animal models often partly mimic human pathophysiology, they show limited external validity [2]. To replace animal-based modelling, there is a growing interest in the development of alternative, clinically relevant disease models [3,4]; for example, in epilepsy, which is a devastating disease affecting nearly 50 million people worldwide [5] with about one in three patients whom suffer from drug-refractory seizures [6]. As a last drastic resort, 10–15% of these drug-refractory patients may undergo resective brain surgery, a procedure whereby the epileptic focus is surgically removed. In addition to health gain for the patient, the hereby obtained biopsy is a unique source for research on the disease pathology.
Traditionally, the neuronal activity of brain biopsies is evaluated in organotypic slices. Although this technique is commonly used in rodent studies [7], it is less often applied on human brain biopsies. Interestingly, organotypic slice cultures of human brain biopsies received more attention since just 10 years ago. In these slices, some studies reported localized (single-electrode) epileptiform activity [8,9,10], yet the culture duration of the slices was limited up to 20 h. To evaluate the activity of an epileptic network, it is necessary to record a number of neurons over a longer period. This may be obtained from a multi-electrode array (MEA), yet there are challenges, such as loss of contact between electrode and biopsy and sufficient perfusion require attention. For example, Hsiao et al. applied a MEA to a temporal lobe slice from an epilepsy patient [11]. Although they reported artefacts due to a slice shift on the MEA, they were able to record 4-aminopyridine-evoked epileptiform activity for up to 11 h. In addition, Dossi et al. evaluated MEA recordings from human epileptic cortical tissue and reported repetitive epileptic activity over a period of 24 h [12]. The most comprehensive study on long-term recordings of epileptic activity in organotypic cultures was achieved by Le Duigou et al. (2018) [13]. Yet, these recordings were obtained by patch clamp, and consequently, hardly represent network activity, as cultures are on porous cell culture inserts. The latter are not compatible with MEA chips and setup. They successfully demonstrate single-neuron recordings by patch clamping for 6 weeks in culture. This can only be achieved by a highly specialized setup, including viral vector techniques with promoters for transgene expression, a customized culture medium, and sophisticated hardware. Indeed, it was shown that dedicated medium, such as human cerebrospinal fluid (hCSF), promotes longer-term neuronal viability and network function in human organotypic brain slice cultures, when compared with commercially available ones [14]. Yet, hCSF is not widely accessible, as it requires ethical approval, infrastructure, and only a limited amount can be collected per patient (making the experiments expensive and it is not readily available).
The introduction of the human induced pluripotent stem cell (hiPSC) technology, pioneered by Takahashi et al. [15], and advancements in 2D and 3D brain models [16,17] greatly expanded the toolkit for human disease modelling and personalized treatment [18]. Regardless of the sophistication of these brain models (a.k.a. brain organoids), a major drawback of this approach is a lack of a complete set of cell types and the histoarchitecture of the brain. This is not trivial when discerning neurocircuitries and their role in pathology. Still, the morphology and functionality of neurons derived from differentiated hiPSCs can meet that of neurons in the human brain and thus be studied as models for brain diseases [19,20]. Epileptic network activity was studied by using a MEA, mainly on animal-derived tissues [21], but also on hiPSCs from a patient [22]. A major drawback of hiPSCs is that rather immature electrophysiological profiles, more closely mimicking neurons of the developing embryonic brain, are expressed [17].
In this feasibility study, we mounted a freshly obtained, patient-derived temporal lobe brain biopsy onto a layer of hiPSCs-derived cortical neurons that was cultured on a MEA, with the aim to obtain a clinically relevant electrophysiological model for epilepsy. This “hybrid model” was cultured to evaluate the development of network activity over time and to investigate whether transferring the signal from the biopsy plug to the electrodes via a layer of differentiated hiPSC-derived neurons potentiates long-term measurements (days). The benefit of having a layer of neurons derived from hiPSCs in our model is that it acts as a kind of “antenna” for signals coming from the biopsy; as in, biopsy-derived neuronal activity can be read from the cell layer for multiple days and this information is not available when no layer of hiPSC-derived neurons is present. Epileptiform activity was defined by quantifying electrophysiological characteristics that align with neurophysiology [23]. This model may thus present a new, animal-free, and clinically relevant manner to investigate (personalized) treatments without the need to use dedicated medium (such as hCSF) or slicing.
2. Materials and Methods
2.1. Concept of the Model
The model setup consisted of hiPSC-derived cortical neurons (product ax0015; Axol Bio science, Cambridge, UK) that were cultured on a 120 electrode MEA (120MEA100/30iR-ITO-pr; Multi Channel Systems GmbH, Reutlingen, Germany). A brain biopsy from a patient with temporal lobe epilepsy was, immediately upon resection, sampled and then mounted onto the layer of cortical neurons (Figure 1A,B). The layer of hiPSC-derived cortical neurons served as a transducer of electrical activity in the biopsy to the MEA. Three conditions were compared: (1) the biopsy on the MEA, recorded at 1 h after resection, (2) the hiPSC-derived cortical cells on the MEA, recorded at culture on day 16 (days in vitro = DIV), and (3) the biopsy mounted on DIV16 hiPSC-derived cortical neurons, recorded at 1 h, 3 days and 6 days after resection (Figure 1C–E).
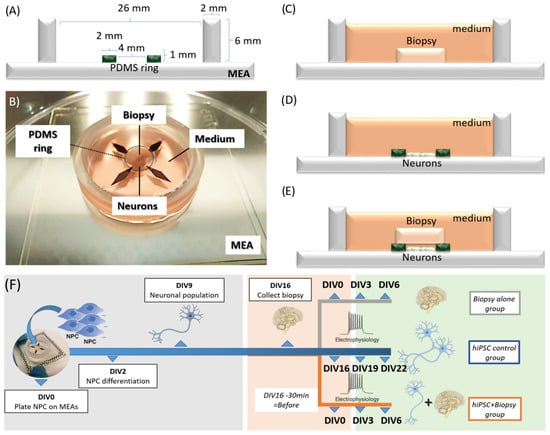
Figure 1.
Design of the hybrid brain biopsy plug-hiPSC-derived neuronal cell monolayer model. (A) Schematic cross section showing dimensions of the setup, multi-electrode array (MEA) and the PDMS ring. (B) Picture of the biopsy plug placed directly on the hiPSC-seeded MEA device. Schematics of the three culturing conditions: (C) only the biopsy plug; (D) only the hiPSC-derived cortical neuronal cell monolayer; and (E) the hybrid model with the biopsy in close contact with the hiPSC-derived neurons. (F) Time line of the experiments showing the days in vitro (DIV) for the hiPSC-derived neuronal cultures before the biopsy collection (grey), when the biopsy is collected and assembled (DIV16 neuronal culture/DIV0 biopsy culture, pink) and for the experiment (DIV19 to 22 neuronal culture/DIV3 to 6 biopsy culture, green). Data points stating “Before” represent the day of the biopsy assembly (DIV16) minus 30 min. Measurements were taken “before” to characterize both the hiPSC neuronal culture alone and the biopsy plug alone before the biopsy was mounted on the neuronal cell layer.
2.2. Preparation of Human Induced Pluripotent Stem Cell (hiPSC)-Derived Neuron Cultures
A polydimethylsiloxane (PDMS, Sylgrad 184, Dow Corning, Midland, MI, USA) ring insert (1 mm high, inner diameter 4 mm, and outer diameter 8 mm) was prepared in-house and mounted on a MEA plate to form a microwell on top of the electrodes (Figure 1A). Subsequently, the MEAs were coated with ReadySet (Axol Bioscience, Cambridge, UK) at a volume of 250 µL/cm2 for 1 h at 37 °C, followed by washing with distilled water and coating with Surebond FX (Axol Bioscience) at a volume of 200 µL/cm2 for 1 h at 37 °C in 5% CO2. Cells were thawed quickly at 37 °C, centrifuged at 200× g for 5 min, and resuspended in 1 mL of plating medium (Axol Bioscience). The excess of the coating solution was aspirated from the MEA and washed three times in phosphate-buffered saline (PBS, Sigma-Aldrich) for 5 min and replaced with plating medium prior to cell seeding (and kept inside an incubator). Cells (85,000 cells/cm2) were seeded inside the PDMS ring in plating medium (DIV0) and differentiation was initiated according to the manufacturer’s instructions (synchronous differentiation, system B delivered with dedicated plating, differentiation and maintenance media from Axol Bioscience). Plating medium was refreshed after 1 h of seeding and changed to maintenance medium (Axol Bioscience) on DIV1 and switched to differentiation medium on DIV2 and kept until DIV9, as specified by the manufacturer (Figure 1F). All incubations were at 37 °C in a 5% CO2 incubator. Following 7 days of differentiation, the medium was changed back to maintenance medium for the remainder of the experiment, with media refreshing every 3 days until DIV22, the end of the experiment. On DIV16, three experimental groups were generated: biopsy alone, hiPSC controls, and hiPSC with biopsy (Figure 1F grey, blue and orange, respectively). All media were supplemented with 1% penicillin and 1% streptomycin. For additional histology analysis, polystyrene 12-well plates (Corning® multi-well cell culture plates, article no. 29442-040) were coated with ReadySet and Surebond FX, and cells seeded therein also at 85,000 cells/cm2, as described above for the MEAs.
2.3. Biopsy Collection and Assembly of Hybrid Model
Following Maastricht University Medical Center+’s medical ethical committee protocol #2018-0945, a pre-operatively signed informed consent was obtained. Patients underwent a unilateral temporal lobe resection including an amygdalohippocampectomy as treatment of drug-refractory temporal lobe epilepsy [24]. This type of surgery commonly includes resection of the most anterior 4 cm approximately part of the temporal lobe. Part of the biopsies was used for routine histopathological analysis; the remainder was used for the current study. Histopathological evaluation of resected neocortical and hippocampal tissue typically showed hippocampal sclerosis. Immediately following resection of the brain tissue, the biopsy was transported from the operation theatre to the laboratory (approximately 10 min) in maintenance medium at room temperature and without oxygenation. In the laboratory, the collection tube was opened in a laminar flow hood, and the biopsy was placed in a sterile tissue culture dish and cut into 3D-tissue plugs using a 5 mm inner diameter biopsy puncher (Biopunch® Ted Pella, Inc., Redding, CA, USA). The plug dimensions were 5 mm in diameter and 3 mm in height. For the hybrid model, three plugs were transferred onto a neuronal layer grown on three separate MEAs (DIV16), covered with maintenance medium, and incubated for 30 min at 37 °C and 5% CO2 prior to the first recording (Figure 1B,F). From this point onwards (only maintenance), CSF-free medium was used (and special CSF-like media was not needed). All MEA devices were carefully transferred from the incubator to a preheated MEA head stage and allowed 5 min of rest before recordings started. Cultures were maximally 15 min outside the incubator. As a control, a 2D layer of hiPSC-derived cortical neurons was kept in culture on three MEAs without the biopsy plug. A second control consisted of three biopsy plugs that were directly placed on a MEA, i.e., without the neuronal cell layer as interface.
2.4. Data Collection by Multi-Electrode Array (MEA) Recording
A MEA2100 system (Multi Channel Systems, Reutlingen, Germany) was used to record electrophysiological activity on 120MEA100/30iR-ITO-pr plates (Multi Channel Systems) with the heating stage set at 37 °C. MEA chips have titanium nitride (TiN) electrodes and a silicon nitride (SiN) isolator with contact pads and tracks transparent (Indium tin oxide, ITO). It contains four internal reference electrodes alongside the 120 recording electrodes (inter-electrode space = 100 µm, electrode diameter = 30 µm). Because the electrodes are bigger than neurons, the recorded signals are considered to represent multi-unit activity. Multiple recordings of 2 min at 20 kHz were taken 30 min after mounting the biopsy (DIV0) and after 3 and 6 days in culture (DIV3 and DIV6). Three representative recordings of 2 min each were analyzed for each condition and DIV point. Electric activity was recorded using the Multi Channel Systems experimenter software set at a 200 Hz Butterworth high pass filter and 50 Hz notch filter. Recorded raw data (.mcs) were converted to a HDF5 format using Multi Channel manager software. To run the analysis on the large amount of recorded data, we used a custom-made MEA-ToolBox (for settings see Table 1) [25]. This permitted the extraction of eight electrophysiological properties (for definitions see Supplementary Table S1) that align with earlier reported parameters [26,27,28].

Table 1.
Parameters for the post processing calculation of the selected endpoints.
2.5. Immunohistochemistry
HiPSC-derived cortical neuron cultures from the same batch of thawed cells used in our hybrid model were also cultured on polystyrene tissue culture plates in parallel to the cultures on MEA to assess their histological architecture by staining. The hiPSC-derived cortical neurons were fixed at DIV7 and DIV16 in 4% paraformaldehyde for 1 h and permeabilized with 1% Triton X-100 (Sigma-Aldrich, St. Louis, MI, USA) in PBS for 20 min, followed by three wash steps in PBS for 5 min each. Cells were then incubated with 1% bovine serum albumin (BSA; no 9048468, Sigma-Aldrich) for 30 min and again washed in PBS three times for 5 min each. As first antibody, MAP-2 (mouse monoclonal ab254143, Abcam), Beta tubulin III (mouse monoclonal ab70878, Abcam), and synaptophysin (rabbit monoclonal ab52636, Abcam), were diluted 1:200 in 1% BSA in dPBS (i.e., PBS without calcium and magnesium, Sigma-Aldrich) and incubated for 1.5 h at room temperature, followed by three wash steps in PBS for 5 min each. As secondary, antibody, either Cy5 (goat anti mouse CY5 ab6563, Abcam) or Alexa Fluor® 488 (goat anti rabbit ab0077, Abcam) was diluted 1:500 in 1% BSA in dPBS and incubated for 1.5 h at room temperature in the dark. Nuclear staining was performed using two drops of NucBlue (Life Technologies, Carlsbad, CA, USA) for 20 min. Next, samples were washed again three times in PBS for 5 min each. Fluorescent images were acquired using a Keyence BZ-810 microscope system (Osaka, Japan).
3. Results
3.1. Immunohistochemical Characterization of hiPSC-Derived Neurons
Figure 1 depicts how the hybrid model is assembled and the experimental procedure timeline. Prior to mounting the 3D tissue plugs, optical inspection of the hiPSC-derived cortical neuronal culture showed a carpet-like morphology with network formation from DIV7 and MAP2 positive (Figure 2A). In addition, on DIV16, immunohistochemical-stained cultures confirmed that these cells expressed neuronal markers beta tubulin III and synaptophysin (Figure 2B). Bright-field visual inspection of DIV16 neurons that were cultured on a MEA showed neurons that looked similar to the ones that were used for immunohistochemistry (Figure 2C).
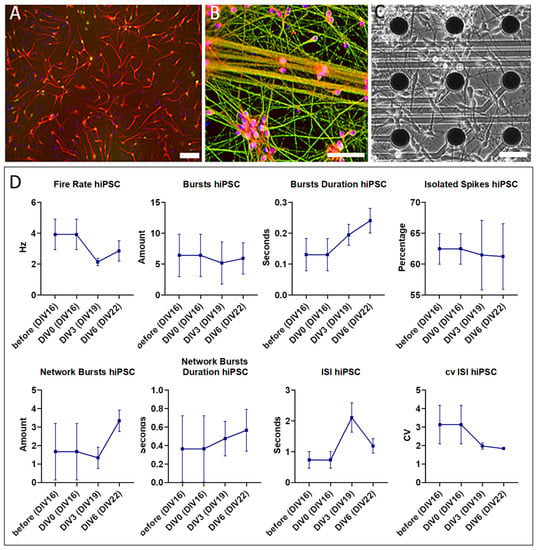
Figure 2.
Image of hiPSC-derived cortical neurons cultured on (control) polystyrene (A,B) and the MEA (C) showing eight endpoints measured in parallel as controls (D); (A) after 7 days of differentiation, cells were (immuno)histochemically stained for neuronal marker MAP2 (red), indicating a successful induction of a neuronal lineage (Magnification 10×); (B) after 16 days (i.e., 7 days of differentiation and maintained for an additional 9 days in culture; DIV16), cells were (immuno) histochemically stained with markers for more mature neurons, including synaptophysin (red) and Beta tubulin III (green), and with nuclear marker DAPI (blue), indicating that the neurons matured and started to develop a network (Magnification 20×); (C) bright field image of hiPSC-derived cortical neurons that were cultured for 16 days on a MEA, representing the stage as used for assembly with the biopsy (Magnification 20×). All scale bars = 50 µm; (D) electrophysiological characterization of the control hiPSC-derived neuronal culture, measured at DIV16 (=before the assembly with the biopsy) and in parallel during the week of experiment (DIV0 to DIV6 neuronal cell + biopsy co-culture equivalent to DIV16 to DIV22 neuronal cell culture). Error bars represent mean with standard error of the mean (SEM) of three 2 min recordings per condition.
3.2. Electrophysiological Phenotype of the Hybrid Model
Electrical activity recordings from the control construct, i.e., the biopsy mounted on the MEA without a neuronal cell layer as an interface, did not surpass the signal-to-noise ratio. Hence, these recordings were not further analyzed and are displayed in Supplementary Figure S1. The second control construct, i.e., DIV16 cortical neurons cultured on the MEA, did show electrophysiological activity above the signal-to-noise ratio. Consequently, its electrophysiological phenotype was used as baseline to compare with that of the construct with the subsequently added biopsy (Figure 2D). At DIV16, hiPSC-derived neuronal cultures exhibited a firing rate of ±3 Hz and 5–10 burst events per minute with a burst duration of 0.1–0.3 s. In addition, these cells displayed isolated spikes in 60% of the recording time, 0–3 network bursts per minute with a duration 0.4–0.8 s per burst, an inter spike interval (ISI) up to 2 s, and a coefficient of variation between 2 and 4. Addition of the biopsy plug on the neuronal cell layer changed the baseline phenotype into hyperactive network bursting activity (example 1 h after the assembly of the biopsy plug shown in Figure 3A). The raw single-channel network burst signal that was recorded across the network with the threshold for spike detection is also shown (Figure 3B). The identified bursts and single spikes from the analysis are also presented (Figure 3C).
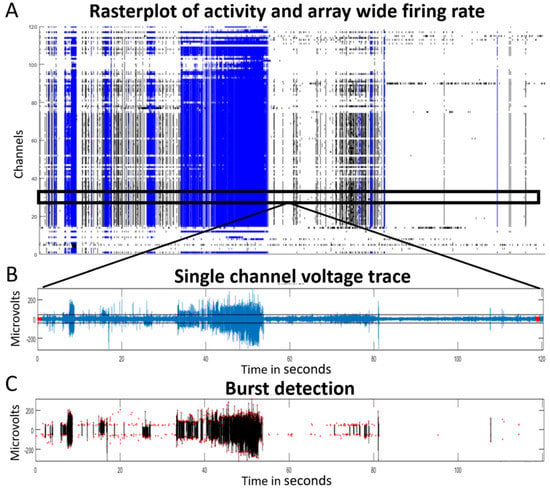
Figure 3.
Samples of electrophysiological activity in the hybrid model recorded by a 120-electrode MEA device one hour after assembly (DIV0). (A) Temporo-spatial distribution of neuronal activity (array wide raster plot) of a 2 min recording. Network burst activity (blue) suggests an epileptiform phenotype; (B) example of a single-channel full trace with detection threshold (7 RMS), and (C) the corresponding detected network bursts (black lines represent the blue lines in A and show the actual network bursting events). Red dots in (C) indicate detected single spikes.
This seizure-like activity lasted for 6 days (Figure 4A–C). Quantitative analysis of MEA recordings comparing the hybrid model with hiPSC alone revealed an increased firing rate, burst rate, burst duration, coefficient of variation in ISI and network burst durations, and a decreased number of isolated spikes and ISI (Figure 4D). Prior to the biopsy plug addition, electrophysiological MEA characterization at DIV16 demonstrated stable firing (around 3 Hz, Figure 2D), which is considered normal for such 2D hiPSC-derived neuronal cultures [29]. Some network bursts were already present in the neuronal culture (average of two network bursts detected, Figure 2D), prior to biopsy plug assembly, with shorter durations (up to 0.8 s). Figure 5 compares the electrophysiological phenotype of the hybrid model (orange, triangles) with the control hiPSC neuronal cultures (blue, squares). It shows that the firing rate increased considerably upon addition of the biopsy plug up to 25 Hz on DIV0 and then gradually decreased to 10 Hz at DIV6. Single-channel bursts increased on average from 3 to 10 bursts per minute up to 18 to 36 bursts per minute when the biopsy plug was assembled, with recorded network bursts duration increasing from 0.7 to 15 s (DIV0) and 28 s (DIV3) and dropping down again to 5 s on DIV6, indicating that the model was no longer capable of recording strong epileptic/seizurogenic signatures after this time point. In addition, isolated spikes in the hybrid model showed a considerable decrease, with typically 60 to 70% isolated spikes recorded from the hiPSC culture alone compared to 6 to 20% in the assembled biopsy-hiPSC co-culture for the first 3 days, and increasing up to 50% at day 6. Similarly, the inter-spike interval values were lower in the assembled biopsy-hiPSC co-culture for the whole duration of the experiment (lower than 0.5 s), while the coefficient of variation in the inter-spike interval was considered elevated, especially during the first day of recording, when the value was above 5. Such types of firing patterns (Figure 5, Biopsy + hiPSC, orange) were not observed when we cultured the biopsy or the iPSC-derived neuronal layer separately on the MEA (Figure 5, hiPSC, blue).
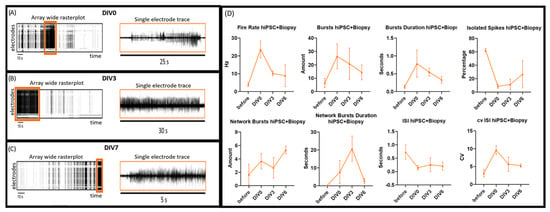
Figure 4.
Electrophysiological recordings of the hybrid model one hour after assembly (DIV0) (A); at DIV3 (B) and at DIV6 (C). The left of each panel shows an array-wide rasterplot (120 electrodes on the Y-axis vs. time in seconds (s) on the X-axis), from which some epileptiform phenotypes were extracted (orange rectangles). The right of each panel represents a single-channel trace from the corresponding timeframe at a higher temporal resolution, showing voltage activity. This suggests that burst activity of 25−30 s can be detected for up to 3 days after resection of the biopsy and diminishes to bursts of 5 s by culture day 6; (D) Electrophysiological characterization (fire rate, burst rate and durations, isolated spikes, network bursts and durations, as well as ISI and cv of ISI) of the hybrid model, with hiPSC neuronal derived cultures (DIV16) measured before and after biopsy assembly for 6 days (hiPSC + Biopsy, orange). Error bars represent mean with SEM of three 2 min recordings per condition.
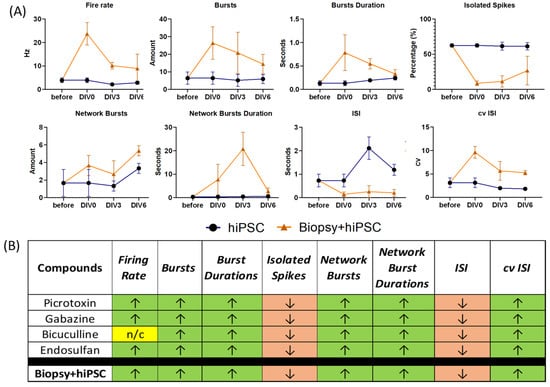
Figure 5.
Electrophysiological characterization. (A) Fire rate, bursts and burst durations, isolated spikes, network bursts and durations, as well as ISI and cv of ISI of the hybrid model (hiPSC + Biopsy, orange), with hiPSC neuronal derived cultures (DIV16 = DIVO on graph) measured before and after biopsy assembly for 6 days and compared with the control hiPSC neuronal-derived cultures (DIV16) (hiPSC, blue) during the same 6-day period. Error bars represent mean with SEM of three 2 min recordings per condition. (B) The hybrid model displayed a higher firing rate, number of bursts, longer burst durations, network burst and durations, and a higher CV of ISI than the biopsy or neuronal monolayer alone. The hybrid model was also characterized by a decreased percentage of isolated spikes and a reduced inter-spike interval. Taken together, the electrophysiological output signature of the hybrid model corresponds to the seizurogenic profile of hiPSC cultures treated with picrotoxin, gabazine, bicuculline, or endosulfan, as reported by Bradley et al. (2018) [30].; “n/c” = no change. Adapted with permission from [30]. Copyright 2018, Oxford University Press.
To gain insight into the reproducibility of the model, a biopsy from another epilepsy patient was submitted to the same procedures. In this second case, the recorded electrical activity was similar to that of the first case, with the exception of ISI, which showed an inconsistent and less pronounced increase over days in culture (Supplementary Figure S2).
4. Discussion
This study describes the assembly of a hybrid brain model, combining a neocortical biopsy from a patient with epilepsy with a layer of hiPSC-derived cortical neurons cultured on a MEA. This setup allowed recordings of spontaneous electrophysiological activity for 6 days, with the main advantage being the possibility to study patient-derived biopsy postoperatively with commercially available consumables and a standardized readout system. During the first week of co-culturing of the hybrid model, we confirmed that the network activity of the neuronal cell layer can serve as an interface between the patient-derived 3D tissue plug and the MEA electrodes. Gradually, the co-culture became less responsive as of day 6, which suggests its degradation. Various read-outs (fire rate, burst rate, inter-spike interval, etc.; see Supplementary Table S1) were extracted from the electrophysiological signal. Based on literature [26,27,28], eight read-outs (endpoints) were selected to characterize the network’s behaviour of our hybrid model and revealed that it was reminiscent of pro-convulsive and epileptiform electrophysiological signatures [30,31,32,33,34]. The hereby obtained epileptiform phenotype was not observed in setups where either the biopsy alone or the hiPSC-derived cortical neuron layer alone were cultured on a MEA. Here we will discuss the simplicity of our approach and the results obtained with our hybrid system, but also the challenges remaining.
Recordings from the stand-alone biopsy in 37 °C maintenance medium hardly surpassed the signal-to-noise ratio, i.e., single electrodes showed some local field potentials in the first few minutes. There was no network activity observed and all signals were lost within the hour (Supplementary Figure S1). Technically, without a dedicated chip (i.e., cortical neurons grown onto a MEA) and suction ports [35] to keep the biopsy in place, an alternative is to place the MEA in the recording apparatus and load the biopsy atop the electrodes directly and measure for as long as possible (up to a few hours) before discarding it. Moving the MEA chip back and forth between the recorder and the incubator can displace the biopsy, thereby increasing the risk that electrical activity is not always obtained from the same location. Therefore, we added a PDMS ring that kept the biopsy in place. However, for the stand-alone biopsy, adding a PDMS ring to fix the biopsy did not improve the electrophysiological signal, as the neurons of the biopsy plugs were not in close enough contact with the electrodes of the MEA chip. However, loading the 3D biopsy plug onto a PDMS ring containing a layer of cortical neurons at the bottom resulted in a good signal-to-noise ratio of spontaneous electrophysiological activity on all electrodes for up to 6 days. When we measured activity from the layer of hiPSC-derived cortical neurons prior to assembly of the biopsy plug, we retrieved an expected spike activity and bursting behaviour (Figure 2D), which is commonly seen for such cultures [25], but no seizure-like discharge patterns. DIV16 was chosen to assemble the biopsy plugs with the neuronal culture because morphological (i.e., immunohistochemical positivity of neuronal markers beta tubulin III, MAP2 and synaptophysin) and electrophysiological (i.e., a firing rate of ~3 Hz and the presence of network bursts) read-outs indicated that the hiPSC-derived neuronal cell layer reached a stage that permitted their functioning as a transducer between the biopsy and the MEA.
When comparing the electrophysiological recordings of the three conditions (i.e., the hiPSC-derived neuronal layer alone (Figure 1D), the hybrid setup (Figure 1E) and the biopsy alone (Figure S1)), in the hybrid model we identified not only an increase in firing and bursting rates, but also an increase in burst duration for both single-channel and network bursts, as well as an increase in inter-spike interval coefficient of variation. Increases coincided with a decrease in isolated spikes and inter-spike intervals. Independent confirmation from an experiment with a second brain biopsy of another patient with epilepsy, in which we were able to record electrical activity for up to 6 days after assembly of the biopsy plug, again revealed seizures, albeit less pronounced (Supplementary Figure S2). It must remain unexplained at this stage of research whether variations in the MEA recordings are due to differences in the experimental condition; for example, the hiPSC-derived neurons connected with the electrodes of the MEA alone versus the biopsy plug atop. Whereas the latter could simply be a source of additional chemicals triggering seizures in the cultured cortical cell layer. On the other hand, there is a likelihood that pathology of the patients, and/or variation in tissue environment of which the plug originated from, could be the reason. Our findings here at least demonstrate that such an approach to ex vivo modelling is feasible.
Notably, such electrophysiological recordings were reported as a pro-convulsive response from neurons to picrotoxin, gabazine, bicuculline, and endosulfan in epileptic models (Figure 5B) [30]. In that study, cortical neurons cultured on MEAs were exposed to known drugs, and the response was categorized in terms of electrophysiological endpoints, including firing and bursting rates (at both the single-channel and the network level), burst duration and inter-spike interval coefficient of variation, isolated spikes, and inter-spike interval. Depending on these parameters increasing or decreasing, the target drugs could be categorized as pro-convulsive, toxic, or non-responsive. Our findings matched that of a pro-convulsive response, indicating that the seizure-like electrical signature of the biopsy could be recorded by the interfacing neuronal layer. This potentially offers an approach for the development of models to directly assess human diseased brain tissue and allow for a prolonged time the monitoring of physiological responses to, for example, drug treatment.
Due to the limited access to human neocortical biopsies (surgically collected during brain surgery), it was not possible to culture a non-diseased biopsy in our model. Hence, we had to compare our findings with those of “control” (hiPSC neurons alone) conditions, where the only difference was the assembly of the biopsy. In addition, our previous 3D culture study, using a bioreactor on top of a MEA containing the same hiPSC-derived neurons, also did not demonstrate any epileptiform signals [36]. Thus, we are ensured that the recorded response introduced into the system was caused by the assembly of the biopsy to the hiPSC-derived cortical neurons. Baseline measurements prior to the assembly of the biopsy showed normal electrical behaviour, and as previously mentioned, the biopsy plug alone did not result in measurable network activity. In this respect, it is of interest that Osaki et al. (2020) [37] showed in their 3D brain model that neurons from the organoid can produce outgrowths and physically connect and innervate muscle cells. In our hybrid model, there is no firm evidence that outgrowing and connecting neurons are involved in changing the read-out layer network topology because the signal is only present for 6 days. It is possible that the biopsy plug simply secretes substances, such as neurotransmitters, onto the hiPSC-derived cortical neuron cell layer, which could result in network activity in the read-out layer capturing the pro-convulsive electrophysiological phenotype. It was previously shown that a freshly operated brain biopsy may slowly release compounds, such as microRNAs [38], which can leak onto the hiPSC-derived neuronal cell layer and change its response over time. Future studies on elaborating the origin of the electrophysiologic activity may address the connectivity between the neuronal cell layer and the biopsy.
One challenge is that the biopsy tissue also contains healthy material that surrounds diseased tissue when the latter is surgically removed. This means that some brain biopsy plugs may not contain diseased material, or just very little. Therefore, we aimed to retrieve the biopsy plugs from the center of the surgically removed biopsy. Human brain biopsies are hard to obtain, and typically, brain slices generated from them can only be measured for 6 to 12 h [39]. Recording long-term network activity via multiple electrodes from a 3D biopsy or tissue construct was shown to be even more challenging [40]. In this respect, it is remarkable that we were able to record electrical activity from our hybrid model for 6 consecutive days within a relatively simple to use set-up and minimal culturing efforts. Four major advances of our new 3D model are, (1) extended time of measuring electrical activity from an intact brain biopsy tissue architecture ex vivo including all extracellular matrix compounds and the full plethora of cells to be found in human brain tissues, (2) good network functionality in a practical approach to assemble such biopsies on MEA that allows for the extracting of disease-relevant electrophysiological fingerprints by means of a stable and reproducible hiPSC-derived neuron layer for taking reliable readouts, and (3) sufficient tissue volume to test various (non-) pharmacological interventions (e.g., neuromodulation, radiotherapy) and (4) capable of testing biopsy plugs punched along a gradient from healthy to diseased tissue that form the edge of the biopsy tissue towards the center, if sufficient material was retrieved during surgery, which has no further use in the conventional investigations of the pathologist’s laboratory. These advantages of our model offer a unique opportunity to study functional aspects of epilepsy in far greater detail than before. Finally, our approach to construct a hybrid ex vivo model may also be combined with 3D MEA technology, for example, a spike electrode array [41], to obtain information from measuring within the brain biopsy. As rodent tissue is more readily available than human biopsy material, future experiments targeted at a better understanding of the interplay between the biopsy and the cell layer could use, for instance, mouse biopsies of specific regions of the brain in a hybrid model with mouse iPSC-derived neurons.
5. Conclusions
By assembling a human epileptic brain biopsy onto a layer of cortical neurons that was grown on a 120-electrode chip, we were able to record spontaneous electrophysiological activity for 6 days. The activity was characterized as epileptiform based on firing rate, amount and duration of bursts, isolated spikes, inter-spike interval, coefficient of variation in interspike interval, number of network bursts, and network burst durations. As a proof-of-principle, this approach demonstrates the potential as a new, animal-free, and clinically relevant model of epilepsy to evaluate novel (personalized) treatments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app13031432/s1, Figure S1: Electrophysiological characterization of the biopsy alone; Figure S2: Electrophysiological characteristics of the second hybrid model conditions. Table S1: Selected electrophysiological endpoint parameters and definitions.
Author Contributions
R.L., G.H. and J.-P.F. conceived the project and the main conceptual ideas. J.-P.F. developed and performed the experiments. M.H.Y.H. developed the MEA data analysis toolbox and assisted in analysing MEA recordings. K.R., O.E.M.G.S. and J.T.A.D. performed the surgery and provided access to the biopsy tissue. A.M.J.M.v.d.M. commented on the manuscript and co-supervized the work on the MEA data analysis toolbox and MEA recordings. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Netherlands Organ-on-Chip Initiative, an NWO Gravitation project (024.003.001) funded by the Ministry of Education, Culture and Science of the government of the Netherlands, the Health Holland LSH-TKI Brain@home (114025101). This work was further supported by ZonMW OffRoad grant 40-08125-98-160 and by the European Union’s 580 Horizon 2020 Research And Innovation Programme H2020- 581 FETPROACT-2018-01 under Grant Agreement No. 824070.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee of Maastricht University Medical Centre (protocol code METC 2018-0945, date of approval 07-05-2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data available on request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Akhtar, A. The Flaws and Human Harms of Animal Experimentation. Camb. Q. Healthc. Ethics 2015, 24, 407–419. [Google Scholar] [CrossRef]
- Sayed, N.; Liu, C.; Wu, J.C. Translation of Human-Induced Pluripotent Stem Cells from Clinical Trial in a Dish to Precision Medicine. J. Am. Coll. Cardiol. 2016, 67, 2161–2176. [Google Scholar] [CrossRef]
- Hubrecht, R.C.; Carter, E. The 3Rs and Humane Experimental Technique: Implementing Change. Animals 2019, 9, 754. [Google Scholar] [CrossRef]
- Hampshire, V.A.; Gilbert, S.H. Refinement, Reduction, and Replacement (3R) Strategies in Preclinical Testing of Medical Devices. Toxicol. Pathol. 2019, 47, 329–338. [Google Scholar] [CrossRef]
- Beghi, E.; Giussani, G.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; Abraha, H.N.; Adib, M.G.; Agrawal, S.; Alahdab, F.; Awasthi, A.; et al. Global, Regional, and National Burden of Epilepsy, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 357–375. [Google Scholar] [CrossRef]
- Dalic, L.; Cook, M.J. Managing Drug-Resistant Epilepsy: Challenges and Solutions. Neuropsychiatr. Dis. Treat. 2016, 12, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Humpel, C. Neuroscience Forefront Review Organotypic Brain Slice Cultures: A Review. Neuroscience 2015, 305, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Eugène, E.; Cluzeaud, F.; Cifuentes-Diaz, C.; Fricker, D.; Le Duigou, C.; Clemenceau, S.; Baulac, M.; Poncer, J.C.; Miles, R. An Organotypic Brain Slice Preparation from Adult Patients with Temporal Lobe Epilepsy. J. Neurosci. Methods 2014, 235, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.S.G.; da Silva, A.B.; Whittaker, R.G.; Woodhall, G.L.; Cunningham, M.O. Human Brain Slices for Epilepsy Research: Pitfalls, Solutions and Future Challenges. J. Neurosci. Methods 2016, 260, 221–232. [Google Scholar] [CrossRef]
- Valente, C.A.; Meda, F.J.; Carvalho, M.; Sebastião, A.M. A Model of Epileptogenesis in Rhinal Cortex-Hippocampus Organotypic Slice Cultures. J. Vis. Exp. 2021, 169, e61330. [Google Scholar] [CrossRef]
- Hsiao, M.C.; Yu, P.N.; Song, D.; Liu, C.Y.; Heck, C.N.; Millett, D.; Berger, T.W. An in VitroSeizure Model from Human Hippocampal Slices Using Multi-Electrode Arrays. J. Neurosci. Methods 2015, 244, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Dossi, E.; Blauwblomme, T.; Nabbout, R.; Huberfeld, G.; Rouach, N. Multi-electrode Array Recordings of Human Epileptic Postoperative Cortical Tissue. J. Vis. Exp. 2014, 92, e51870. [Google Scholar] [CrossRef] [PubMed]
- Le Duigou, C.; Savary, E.; Morin-Brureau, M.; Gomez-Dominguez, D.; Sobczyk, A.; Chali, F.; Milior, G.; Kraus, L.; Meier, J.C.; Kullmann, D.M.; et al. Imaging pathological activities of human brain tissue in organotypic culture. J. Neur. Meth. 2018, 298, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, N.; Hedrich, U.B.S.; Schwarz, H.; Harshad, P.A.; Dammeier, N.; Auffenberg, E.; Bedogni, F.; Honegger, J.B.; Lerche, H.; Wuttke, T.V.; et al. Human Cerebrospinal fluid promotes long-term neuronal viability and network function in human neocortical organotypic brain slice cultures. Sci. Rep. 2017, 7, 12249. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Logan, S.; Arzua, T.; Canfield, S.G.; Seminary, E.R.; Sison, S.L.; Ebert, A.D.; Bai, X. Studying Human Neurological Disorders Using Induced Pluripotent Stem Cells: From 2D Monolayer to 3D Organoid and Blood Brain Barrier Models. Compr. Physiol. 2019, 9, 565–611. [Google Scholar] [CrossRef]
- Mateos-Aparicio, P.; Bello, S.A.; Rodríguez-Moreno, A. Challenges in Physiological Phenotyping of HiPSC-Derived Neurons: From 2D Cultures to 3D Brain Organoids. Front. Cell Dev. Biol. 2020, 8, 797. [Google Scholar] [CrossRef]
- Jalink, P.; Caiazzo, M. Brain Organoids: Filling the Need for a Human Model of Neurological Disorder. Biology 2021, 10, 740. [Google Scholar] [CrossRef]
- Farkhondeh, A.; Li, R.; Gorshkov, K.; Chen, K.G.; Might, M.; Rodems, S.; Lo, D.C.; Zheng, W. Induced Pluripotent Stem Cells for Neural Drug Discovery. Drug Discov. Today 2019, 24, 992–999. [Google Scholar] [CrossRef]
- Nikolakopoulou, P.; Rauti, R.; Voulgaris, D.; Shlomy, I.; Maoz, B.M.; Herland, A. Recent Progress in Translational Engineered in Vitro Models of the Central Nervous System. Brain 2021, 143, 3181–3213. [Google Scholar] [CrossRef]
- Tóth, E.; Fabó, D.; Entz, L.; Ulbert, I.; Eross, L. Intracranial Neuronal Ensemble Recordings and Analysis in Epilepsy. J. Neurosci. Methods 2016, 260, 261–269. [Google Scholar] [CrossRef]
- Du, X.; Parent, J.M. Using Patient-Derived Induced Pluripotent Stem Cells to Model and Treat Epilepsies. Curr. Neurol. Neurosci. Rep. 2015, 15, 1–9. [Google Scholar] [CrossRef]
- Dulla, C.G.; Janigro, D.; Jiruska, P.; Raimondo, J.V.; Ikeda, A.; Lin, C.C.K.; Goodkin, H.P.; Galanopoulou, A.S.; Bernard, C.; de Curtis, M. How Do We Use in Vitro Models to Understand Epileptiform and Ictal Activity? A Report of the TASK1-WG4 Group of the ILAE/AES Joint Translational Task Force. Epilepsia Open 2018, 3, 460–473. [Google Scholar] [CrossRef]
- Smeets, J.S.J.; Horstman, A.M.H.; Schijns, O.E.M.G.; Dings, J.T.A.; Hoogland, G.; Gijsen, A.P.; Goessens, J.P.B.; Bouwman, F.G.; Wodzig, W.K.W.H.; Mariman, E.C.; et al. Brain Tissue Plasticity: Protein Synthesis Rates of the Human Brain. Brain 2018, 141, 1122–1129. [Google Scholar] [CrossRef]
- Hu, H.; Frega, M.; Tolner, E.A.; van den Maagdenberg, A.M.J.M.; Frimat, J.P.; le Feber, J. MEA-ToolBox: An Open Source Toolbox for Standardized Analysis of Multi-Electrode Array Data. Neuroinformatics 2022, 20, 1077–1092. [Google Scholar] [CrossRef]
- Wagenaar, D.; Demarse, T.B.; Potter, S.M. MeaBench: A Toolset for Multi-Electrode Data Acquisition and on-Line Analysis. In Proceedings of the 2nd International IEEE EMBS Conference on Neural Engineering, Arlington, VA, USA, 16–19 March 2005; pp. 518–521. [Google Scholar] [CrossRef]
- Cotterill, E.; Charlesworth, P.; Thomas, C.W.; Paulsen, O.; Eglen, S.J. A Comparison of Computational Methods for Detecting Bursts in Neuronal Spike Trains and Their Application to Human Stem Cell-Derived Neuronal Networks. J. Neurophysiol. 2016, 116, 306–321. [Google Scholar] [CrossRef]
- Mendis, G.D.C.; Morrisroe, E.; Petrou, S.; Halgamuge, S.K. Use of Adaptive Network Burst Detection Methods for Multielectrode Array Data and the Generation of Artificial Spike Patterns for Method Evaluation. J. Neural Eng. 2016, 13, 026009. [Google Scholar] [CrossRef]
- Mossink, B.; Verboven, A.H.A.; van Hugte, E.J.H.; Klein Gunnewiek, T.M.; Parodi, G.; Linda, K.; Schoenmaker, C.; Kleefstra, T.; Kozicz, T.; van Bokhoven, H.; et al. Human Neuronal Networks on Micro-Electrode Arrays Are a Highly Robust Tool to Study Disease-Specific Genotype-Phenotype Correlations in Vitro. Stem Cell Rep. 2021, 16, 2182–2196. [Google Scholar] [CrossRef]
- Bradley, J.A.; Luithardt, H.H.; Metea, M.R.; Strock, C.J. In Vitro Screening for Seizure Liability Using Microelectrode Array Technology. Toxicol. Sci. 2018, 163, 240–253. [Google Scholar] [CrossRef]
- Ishii, M.N.; Yamamoto, K.; Shoji, M.; Asami, A.; Kawamata, Y. Human Induced Pluripotent Stem Cell (HiPSC)-Derived Neurons Respond to Convulsant Drugs When Co-Cultured with HiPSC-Derived Astrocytes. Toxicology 2017, 389, 130–138. [Google Scholar] [CrossRef]
- Tukker, A.M.; Wijnolts, F.M.J.; de Groot, A.; Westerink, R.H.S. Human IPSC-Derived Neuronal Models for in Vitro Neurotoxicity Assessment. Neurotoxicology 2018, 67, 215–225. [Google Scholar] [CrossRef]
- Odawara, A.; Saitoh, Y.; Alhebshi, A.H.; Gotoh, M.; Suzuki, I. Long-Term Electrophysiological Activity and Pharmacological Response of a Human Induced Pluripotent Stem Cell-Derived Neuron and Astrocyte. Biochem. Biophys. Res. Commun. 2014, 443, 1176–1181. [Google Scholar] [CrossRef]
- Odawara, A.; Katoh, H.; Matsuda, N.; Suzuki, I. Physiological Maturation and Drug Responses of Human Induced Pluripotent Stem Cell-Derived Cortical Neuronal Networks in Long-Term Culture. Sci. Rep. 2016, 6, 26181. [Google Scholar] [CrossRef]
- Tang, Y.T.; Kim, J.; López-Valdés, H.E.; Brennan, K.C.; Ju, Y.S. Development and Characterization of a Microfluidic Chamber Incorporating Fluid Ports with Active Suction for Localized Chemical Stimulation of Brain Slices. Lab Chip 2011, 11, 2247–2254. [Google Scholar] [CrossRef]
- Yalcin, Y.D.; Bastiaens, A.J.; Frimat, J.P.; Luttge, R. Long-term brain-on-chip: Multielectrode array recordings in 3D neural cell cultures. J. Vac. Sci. Technol. B 2021, 39, 064004. [Google Scholar] [CrossRef]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. On-Chip 3D Neuromuscular Model for Drug Screening and Precision Medicine in Neuromuscular Disease. Nat. Protoc. 2020, 15, 421–449. [Google Scholar] [CrossRef]
- Kubaczkova, V.; Sedlarikova, L.; Bollova, B.; Sandecka, V.; Stork, M.; Pour, L.; Sevcikova, S. Liquid Biopsies—The Clinics and the Molecules. Klin. Onkol. 2017, 30, 2S13–2S20. [Google Scholar] [CrossRef]
- Buskila, Y.; Breen, P.P.; Tapson, J.; Van Schaik, A.; Barton, M.; Morley, J.W. Extending the Viability of Acute Brain Slices. Sci. Rep. 2014, 4, 5309. [Google Scholar] [CrossRef]
- Didier, C.; Kundu, A.; Rajaraman, S. Capabilities and Limitations of 3D Printed Microserpentines and Integrated 3D Electrodes for Stretchable and Conformable Biosensor Applications. Microsyst. Nanoeng. 2020, 6, 15. [Google Scholar] [CrossRef]
- Liu, M.G.; Chen, X.F.; He, T.; Li, Z.; Chen, J. Use of Multi-Electrode Array Recordings in Studies of Network Synaptic Plasticity in Both Time and Space. Neurosci. Bull. 2012, 28, 409–422. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).