Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data
Abstract
:1. Introduction
Aim of the Article
2. Materials
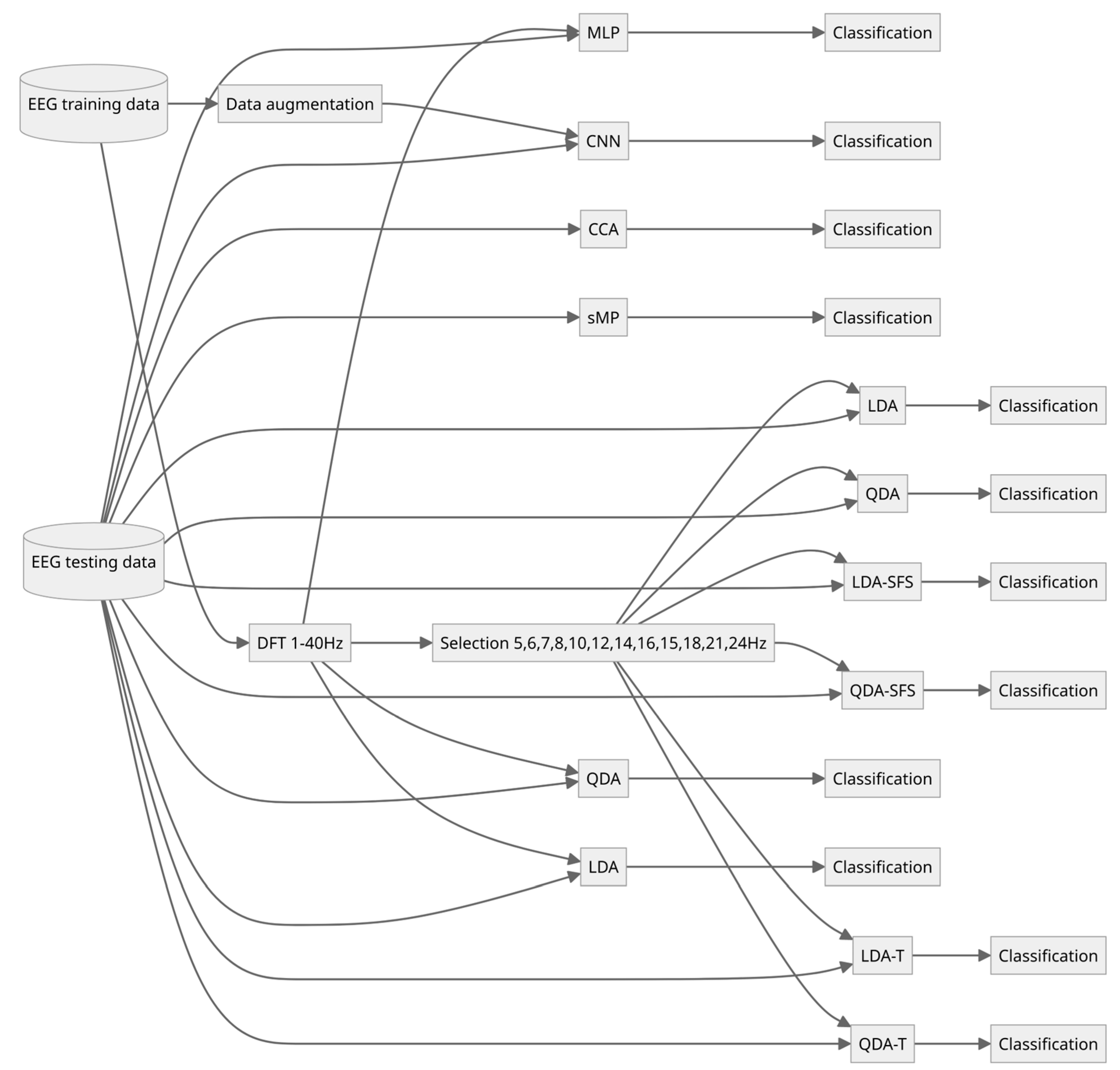
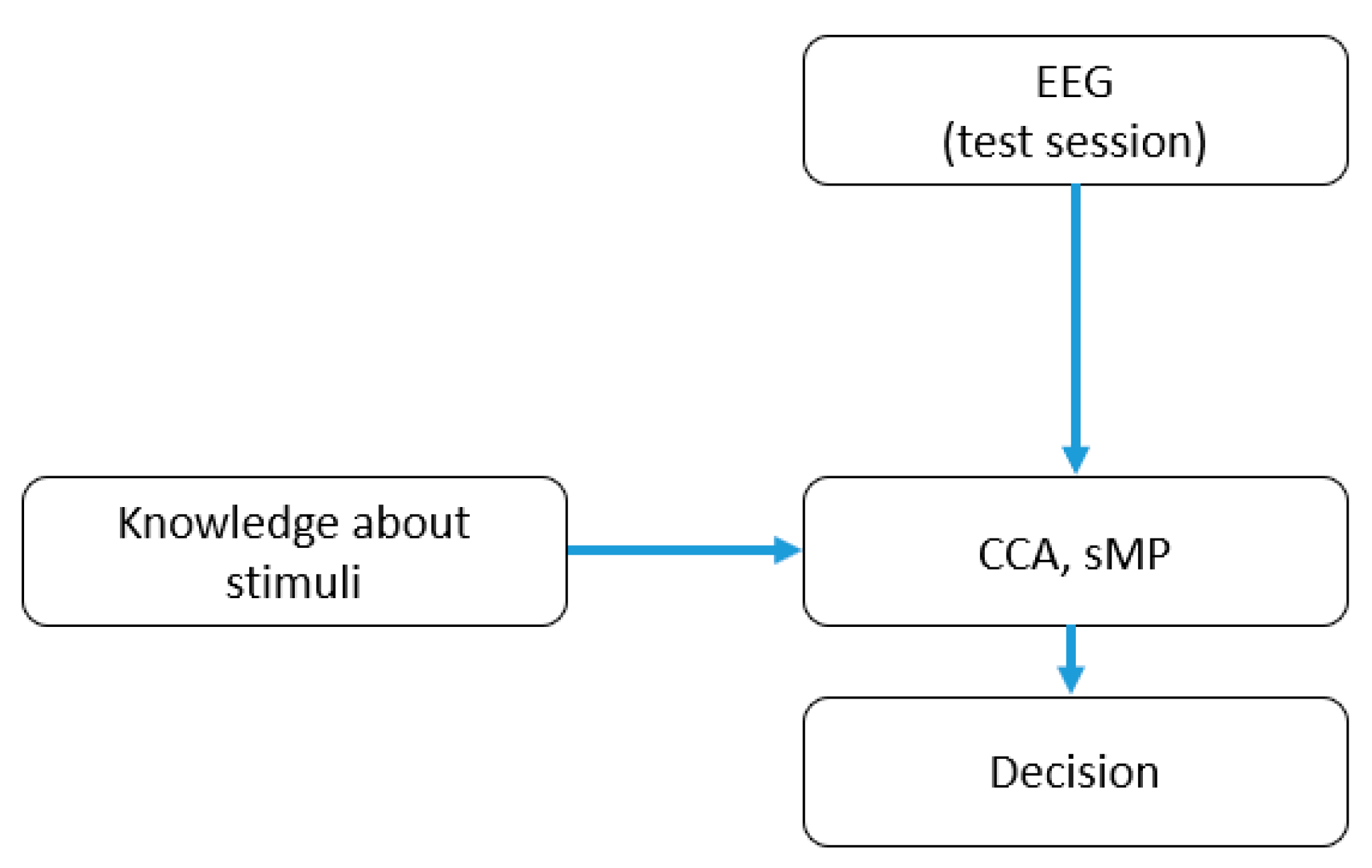
3. Methods
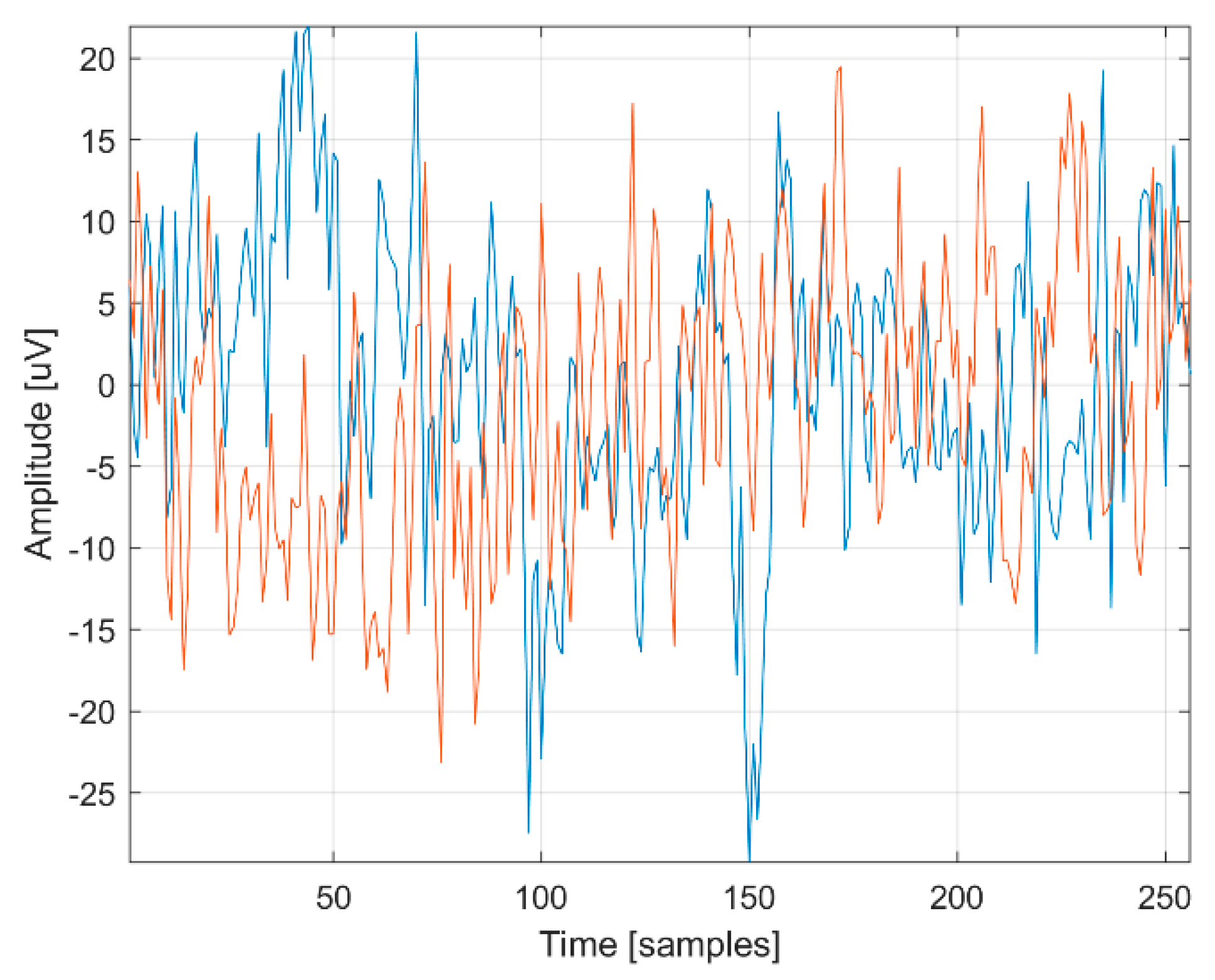
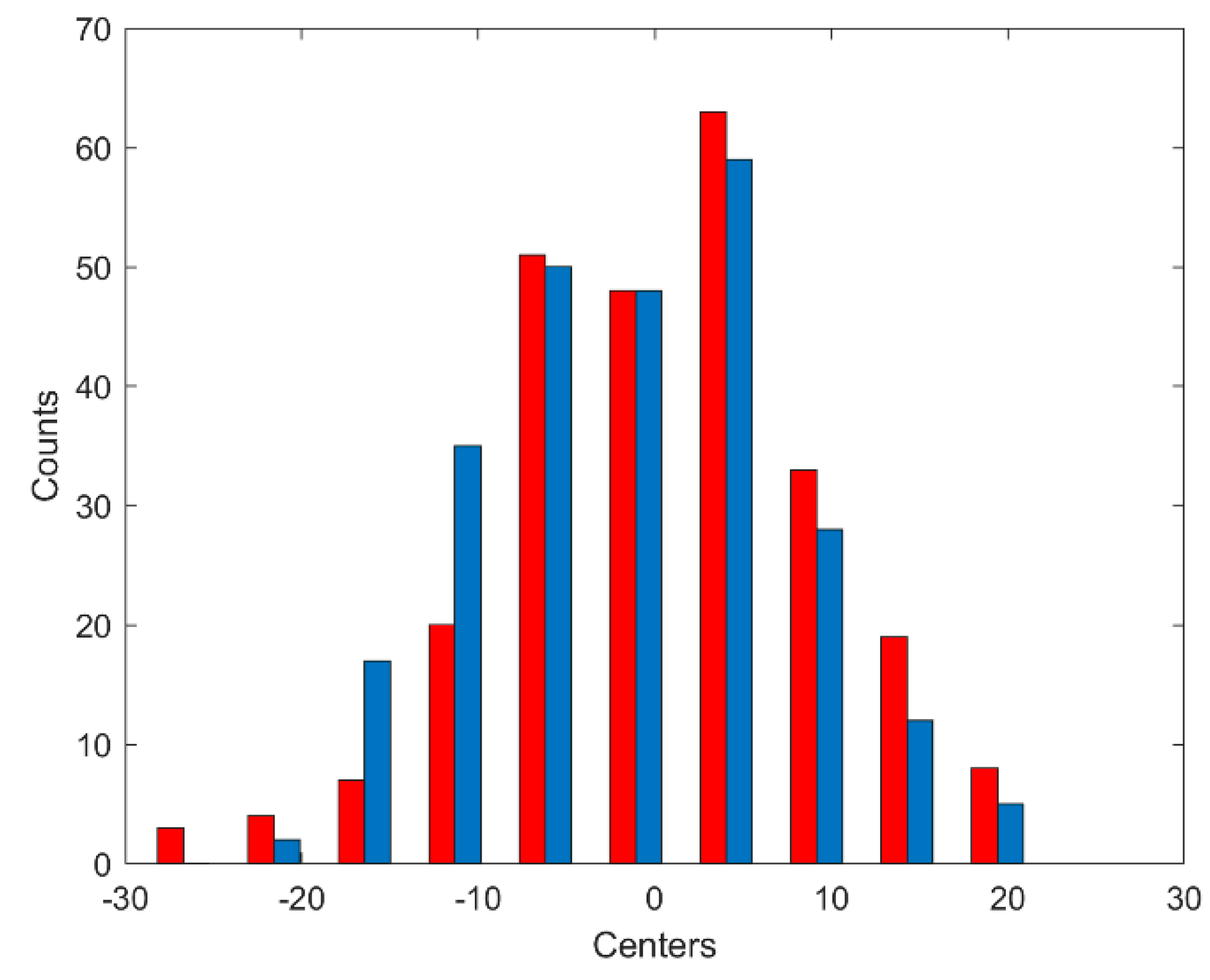
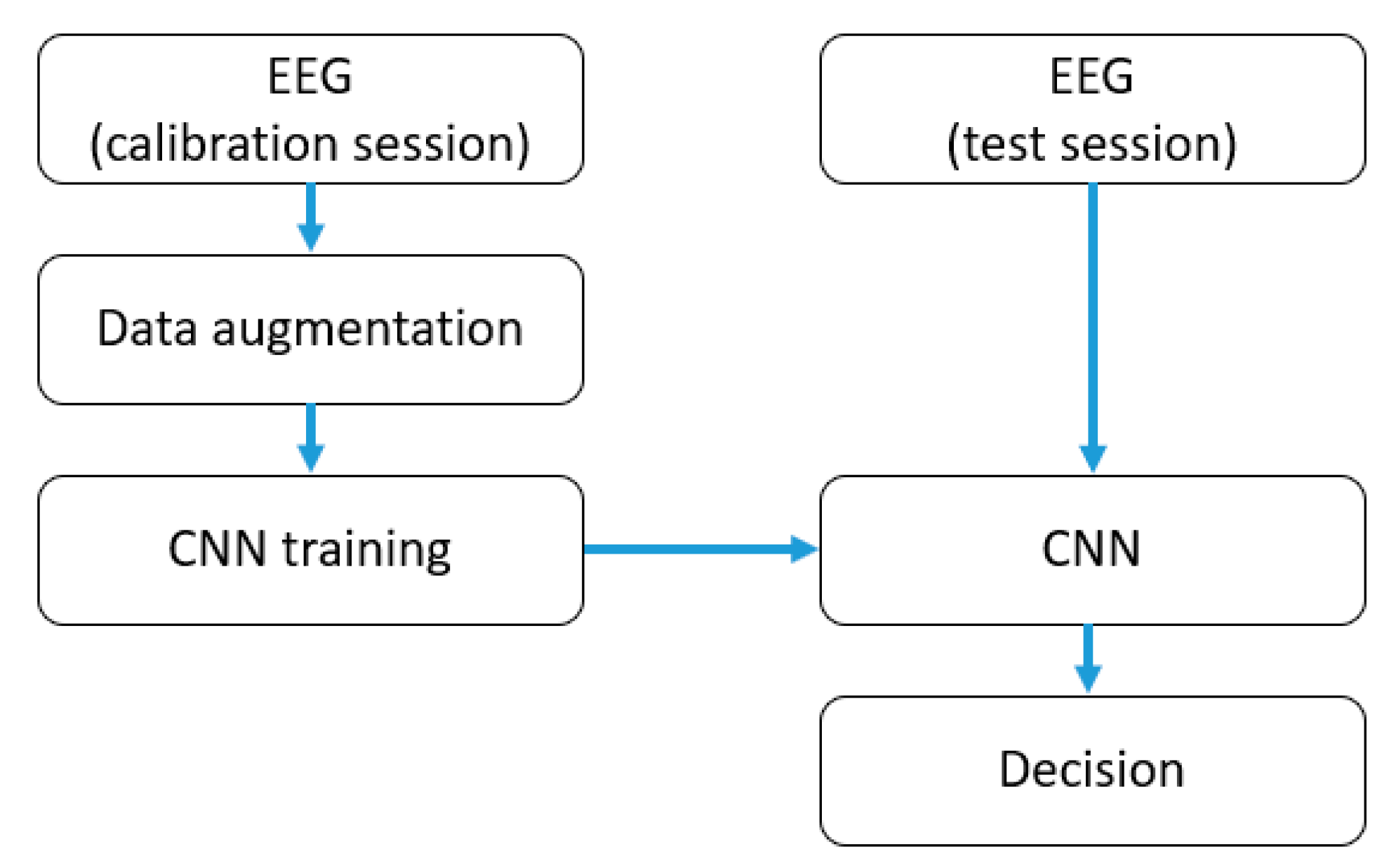
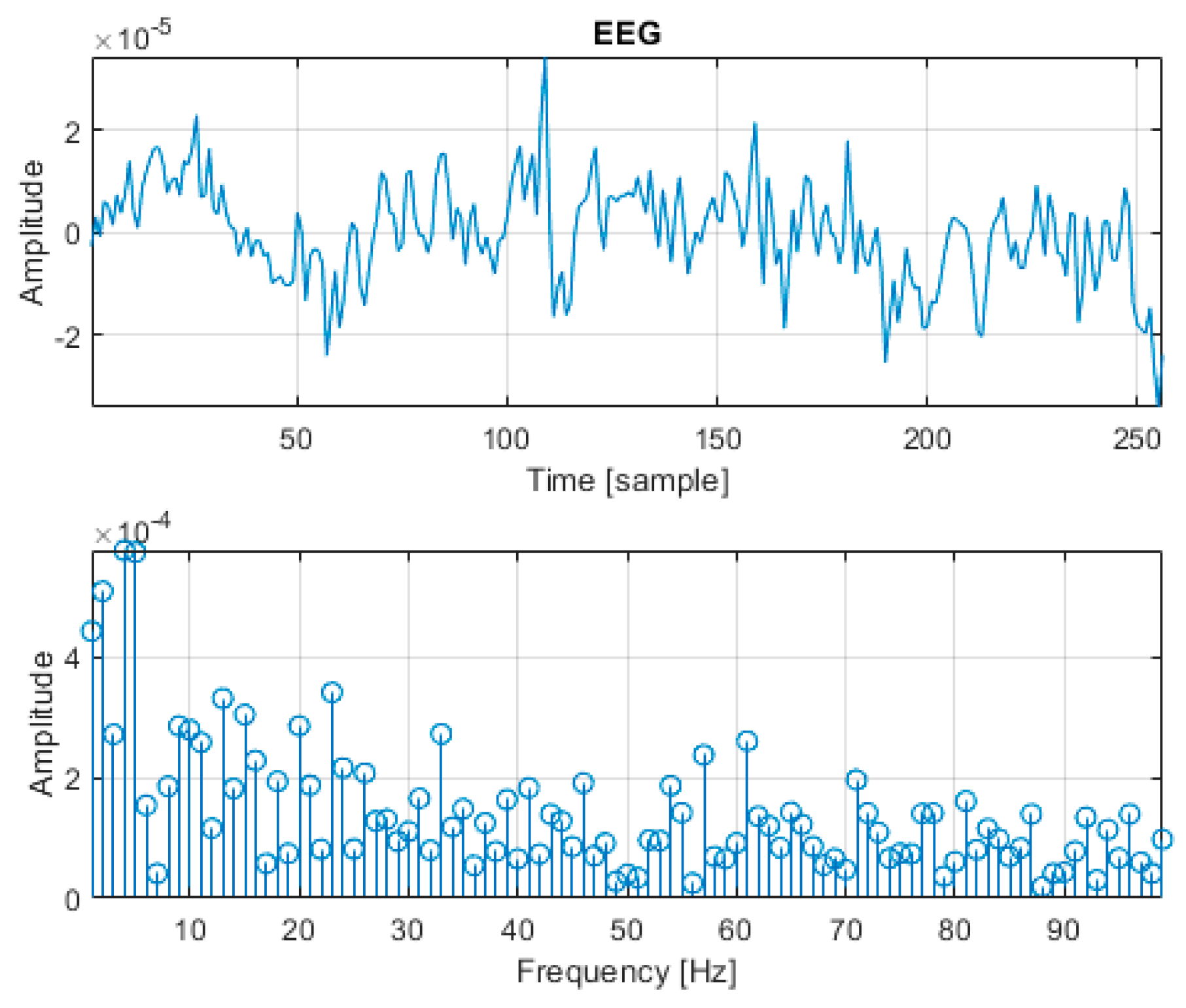
3.1. Data Augmentation
- 1.
- Create a new zero-time vector Sa of length L. This vector corresponds to the newly generated EEG signal for time samples from 0 to L × T − T (with step T).
- 2.
- In a loop, for each value of frequency k = 0 to fs/2, perform the following:
- a.
- Choose an Ar value randomly from the range 〈−0.82; 0.82〉,
- b.
- Choose a φr value randomly from the range 〈−2〉
- c.
- Choose a Pkr element randomly from the Pk set
- d.
- Update vector Sa according to the formula:
- 3.
- Add a vector R of length L to the vector Sa containing values chosen randomly from the range –ε to ε, where = 〈−1.84 × 10−8; 1.84 × 10−8〉
3.2. Convolutional Neural Network
3.3. Classical SSVEP Detection Methods
- 1.
- All frequencies between 1 and 40 Hz were extracted.
- 2.
- Only the frequencies of possible stimulations and their second and third harmonics were extracted. For the frequencies of 5, 6, 7, 8 Hz, these were, respectively: 5, 6, 7, 8, 10, 12, 14, 16, 15, 18, 21, and 24 Hz.
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAM | Adaptive Moment Estimation |
| AR-BCI | Augmented Reality based Brain–Computer Interface |
| BCI | Brain–Computer Interface |
| CCA | Canonical Correlation Analysis |
| CNN | Convolutional Neural Network |
| DFT | Discrete Fourier Transform |
| DNN | Deep Neural Network |
| EEG | Electroencephalography |
| EMG | Electromyography |
| ERD/ERS | Event-Related Desynchronization/Event-Related Synchronization |
| FBCCA | Filter Bank Canonical Correlation Analysis |
| FBCNN | Filter Bank Convolutional Neural Network |
| FB-EEGNet | Filter Bank EEGNet |
| GAN | Generative Adversarial Network |
| ITR | Information Transfer Rate |
| K-NN | K-Nearest Neighbors |
| LDA | Linear Discriminant Analysis |
| LDA-SFS | Linear Discriminant Analysis with Sequential Feature Selection |
| LDA-T | Linear Discriminant Analysis with t-test |
| MPL | Multi-layer Perceptron |
| P300 | P300 Wave |
| QDA | Quadratic Discriminant Analysis |
| QDA-SFS | Quadratic Discriminant Analysis with Sequential Feature Selection |
| QDA-T | Quadratic Discriminant Analysis with t-test |
| SDG | Stochastic Gradient Descent |
| SSVEP | Steady State Visually Evoked Potentials |
| SVM | Support Vector Machine |
| MLP | Multilayer perceptron |
| sMP | simplified Matching Pursuit |
References
- Gu, X.; Cao, Z.; Jolfaei, A.; Xu, P.; Wu, D.; Jung, T.-P.; Lin, C.-T. EEG-Based Brain-Computer Interfaces (BCIs): A Survey of Recent Studies on Signal Sensing Technologies and Computational Intelligence Approaches and Their Applications. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1645–1666. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, D.; Li, C.; Qi, S. Brain–Computer Interface Speller Based on Steady-State Visual Evoked Potential: A Review Focusing on the Stimulus Paradigm and Performance. Brain Sci. 2021, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Norizadeh Cherloo, M.; Mijani, A.M.; Zhan, L.; Daliri, M.R. A Novel Multiclass-Based Framework for P300 Detection in BCI Matrix Speller: Temporal EEG Patterns of Non-Target Trials Vary Based on Their Position to Previous Target Stimuli. Eng. Appl. Artif. Intell. 2023, 123, 106381. [Google Scholar] [CrossRef]
- Ramkumar, S.; Amutharaj, J.; Gayathri, N.; Mathupriya, S. A Review on Brain Computer Interface for Locked in State Patients. Mater. Today Proc. 2021. SSN 2214-7853. [Google Scholar] [CrossRef]
- Choi, W.-S.; Yeom, H.-G. Studies to Overcome Brain–Computer Interface Challenges. Appl. Sci. 2022, 12, 2598. [Google Scholar] [CrossRef]
- Abibullaev, B.; Kunanbayev, K.; Zollanvari, A. Subject-Independent Classification of P300 Event-Related Potentials Using a Small Number of Training Subjects. IEEE Trans. Hum.-Mach. Syst. 2022, 52, 843–854. [Google Scholar] [CrossRef]
- Edlinger, G.; Allison, B.Z.; Guger, C. How Many People Can Use a BCI System? In Clinical Systems Neuroscience; Kansaku, K., Cohen, L.G., Birbaumer, N., Eds.; Springer: Tokyo, Japan, 2015; pp. 33–66. ISBN 978-4-431-55037-2. [Google Scholar]
- Mu, J.; Grayden, D.B.; Tan, Y.; Oetomo, D. Comparison of Steady-State Visual Evoked Potential (SSVEP) with LCD vs. LED Stimulation. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 2946–2949. [Google Scholar]
- Wang, J.; Bi, L.; Fei, W. Multitask-Oriented Brain-Controlled Intelligent Vehicle Based on Human–Machine Intelligence Integration. IEEE Trans. Syst. Man. Cybern. Syst. 2022, 53, 2510–2521. [Google Scholar] [CrossRef]
- Waytowich, N.R.; Krusienski, D.J. Multiclass Steady-State Visual Evoked Potential Frequency Evaluation Using Chirp-Modulated Stimuli. IEEE Trans. Hum.-Mach. Syst. 2016, 46, 593–600. [Google Scholar] [CrossRef]
- Lin, B.-S.; Wang, H.-A.; Huang, Y.-K.; Wang, Y.-L.; Lin, B.-S. Design of SSVEP Enhancement-Based Brain Computer Interface. IEEE Sens. J. 2021, 21, 14330–14338. [Google Scholar] [CrossRef]
- Brennan, C.; McCullagh, P.; Lightbody, G.; Galway, L.; McClean, S.; Stawicki, P.; Gembler, F.; Volosyak, I.; Armstrong, E.; Thompson, E. Performance of a Steady-State Visual Evoked Potential and Eye Gaze Hybrid Brain-Computer Interface on Participants With and Without a Brain Injury. IEEE Trans. Hum.-Mach. Syst. 2020, 50, 277–286. [Google Scholar] [CrossRef]
- Castillo, J.; Müller, S.; Caicedo, E.; Bastos, T. Feature Extraction Techniques Based on Power Spectrum for a SSVEP-BCI. In Proceedings of the 2014 IEEE 23rd International Symposium on Industrial Electronics (ISIE), Istanbul, Turkey, 1–4 June 2014; pp. 1051–1055. [Google Scholar]
- Shao, X.; Lin, M. Filter Bank Temporally Local Canonical Correlation Analysis for Short Time Window SSVEPs Classification. Cogn. Neurodyn. 2020, 14, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Kołodziej, M.; Majkowski, A.; Rak, R.J. Simplified Matching Pursuit as a New Method for SSVEP Recognition. In Proceedings of the 2016 39th International Conference on Telecommunications and Signal Processing (TSP), Vienna, Austria, 27–29 June 2016; pp. 346–349. [Google Scholar]
- Waytowich, N.R.; Faller, J.; Garcia, J.O.; Vettel, J.M.; Sajda, P. Unsupervised Adaptive Transfer Learning for Steady-State Visual Evoked Potential Brain-Computer Interfaces. In Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Budapest, Hungary, 9–12 October 2016; pp. 004135–004140. [Google Scholar]
- Müller-Putz, G.R.; Scherer, R.; Brauneis, C.; Pfurtscheller, G. Steady-State Visual Evoked Potential (SSVEP)-Based Communication: Impact of Harmonic Frequency Components. J. Neural Eng. 2005, 2, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Jiao, Y.; Miao, Y.; Zuo, C.; Wang, X.; Cichocki, A.; Jin, J. Efficient Representations of EEG Signals for SSVEP Frequency Recognition Based on Deep Multiset CCA. Neurocomputing 2020, 378, 36–44. [Google Scholar] [CrossRef]
- Lahane, P.; Jagtap, J.; Inamdar, A.; Karne, N.; Dev, R. A Review of Recent Trends in EEG Based Brain-Computer Interface. In Proceedings of the 2019 International Conference on Computational Intelligence in Data Science (ICCIDS), Chennai, India, 21–23 February 2019; pp. 1–6. [Google Scholar]
- Osowski, S.; Cichocki, A.; Lempitsky, V.; Poggio, T. Deep Learning: Theory and Practice. Bull. Pol. Acad. Sci. Tech. Sci. 2018, 66, 757–759. [Google Scholar]
- Shen, C.; Nguyen, D.; Zhou, Z.; Jiang, S.B.; Dong, B.; Jia, X. An Introduction to Deep Learning in Medical Physics: Advantages, Potential, and Challenges. Phys. Med. Biol. 2020, 65, 05TR01. [Google Scholar] [CrossRef]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, e7068349. [Google Scholar] [CrossRef]
- Israsena, P.; Pan-Ngum, S. A CNN-Based Deep Learning Approach for SSVEP Detection Targeting Binaural Ear-EEG. Front. Comput. Neurosci. 2022, 16, 868642. [Google Scholar] [CrossRef]
- Kwak, N.-S.; Müller, K.-R.; Lee, S.-W. A Convolutional Neural Network for Steady State Visual Evoked Potential Classification under Ambulatory Environment. PLoS ONE 2017, 12, e0172578. [Google Scholar] [CrossRef]
- Ma, P.; Dong, C.; Lin, R.; Ma, S.; Jia, T.; Chen, X.; Xiao, Z.; Qi, Y. A Classification Algorithm of an SSVEP Brain-Computer Interface Based on CCA Fusion Wavelet Coefficients. J. Neurosci. Methods 2022, 371, 109502. [Google Scholar] [CrossRef]
- Guney, O.B.; Oblokulov, M.; Ozkan, H. A Deep Neural Network for SSVEP-Based Brain-Computer Interfaces. IEEE Trans. Biomed. Eng. 2022, 69, 932–944. [Google Scholar] [CrossRef]
- Ravi, A.; Manuel, J.; Heydari, N.; Jiang, N. A Convolutional Neural Network for Enhancing the Detection of SSVEP in the Presence of Competing Stimuli. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6323–6326. [Google Scholar]
- Waytowich, N.; Lawhern, V.J.; Garcia, J.O.; Cummings, J.; Faller, J.; Sajda, P.; Vettel, J.M. Compact Convolutional Neural Networks for Classification of Asynchronous Steady-State Visual Evoked Potentials. J. Neural Eng. 2018, 15, 066031. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, J.; Kesavadas, T. Convolutional Correlation Analysis for Enhancing the Performance of SSVEP-Based Brain-Computer Interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, A.; Washizawa, Y. Steady-State Visual Evoked Potential Classification Using Complex Valued Convolutional Neural Networks. Sensors 2021, 21, 5309. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Qiu, S.; Ma, X.; Wu, C.; Li, J.; Wang, S.; He, H. A CNN-Based Comparing Network for the Detection of Steady-State Visual Evoked Potential Responses. Neurocomputing 2020, 403, 452–461. [Google Scholar] [CrossRef]
- Xing, J.; Qiu, S.; Wu, C.; Ma, X.; Li, J.; He, H. A Comparing Network for the Classification of Steady-State Visual Evoked Potential Responses Based on Convolutional Neural Network. In Proceedings of the 2019 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA), Tianjin, China, 14–16 June 2019; pp. 1–6. [Google Scholar]
- Maharana, K.; Mondal, S.; Nemade, B. A Review: Data Pre-Processing and Data Augmentation Techniques. Glob. Transit. Proc. 2022, 3, 91–99. [Google Scholar] [CrossRef]
- He, C.; Liu, J.; Zhu, Y.; Du, W. Data Augmentation for Deep Neural Networks Model in EEG Classification Task: A Review. Front. Hum. Neurosci. 2021, 15, 765525. [Google Scholar] [CrossRef] [PubMed]
- Kalaganis, F.P.; Laskaris, N.A.; Chatzilari, E.; Nikolopoulos, S.; Kompatsiaris, I. A Data Augmentation Scheme for Geometric Deep Learning in Personalized Brain–Computer Interfaces. IEEE Access 2020, 8, 162218–162229. [Google Scholar] [CrossRef]
- Wang, F.; Zhong, S.; Peng, J.; Jiang, J.; Liu, Y. Data Augmentation for EEG-Based Emotion Recognition with Deep Convolutional Neural Networks. In Proceedings of the MultiMedia Modeling; Schoeffmann, K., Chalidabhongse, T.H., Ngo, C.W., Aramvith, S., O’Connor, N.E., Ho, Y.-S., Gabbouj, M., Elgammal, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 82–93. [Google Scholar]
- Chiang, K.-J.; Wei, C.-S.; Nakanishi, M.; Jung, T.-P. Boosting Template-Based SSVEP Decoding by Cross-Domain Transfer Learning. J. Neural Eng. 2021, 18, 016002. [Google Scholar] [CrossRef]
- Yao, H.; Liu, K.; Deng, X.; Tang, X.; Yu, H. FB-EEGNet: A Fusion Neural Network across Multi-Stimulus for SSVEP Target Detection. J. Neurosci. Methods 2022, 379, 109674. [Google Scholar] [CrossRef]
- Duart, X.; Quiles, E.; Suay, F.; Chio, N.; García, E.; Morant, F. Evaluating the Effect of Stimuli Color and Frequency on SSVEP. Sens. 2020, 21, 117. [Google Scholar] [CrossRef]
- Hui, S.; Żak, S.H. Discrete Fourier Transform and Permutations. Bull. Pol. Acad. Sciences. Tech. Sci. 2019, 675, 130874. [Google Scholar] [CrossRef]
- Bin, G.; Gao, X.; Yan, Z.; Hong, B.; Gao, S. An Online Multi-Channel SSVEP-Based Brain-Computer Interface Using a Canonical Correlation Analysis Method. J. Neural Eng. 2009, 6, 046002. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Zhang, C.; Higashi, H. SSVEP Frequency Detection Methods Considering Background EEG. In Proceedings of the The 6th International Conference on Soft Computing and Intelligent Systems, and The 13th International Symposium on Advanced Intelligence Systems, Kobe, Japan, 20–24 November 2012; pp. 1138–1143. [Google Scholar]
- Jović, A.; Brkić, K.; Bogunović, N. A Review of Feature Selection Methods with Applications. In Proceedings of the 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 1200–1205. [Google Scholar]
- Wang, M.; Liu, G. A Simple Two-Sample Bayesian t-Test for Hypothesis Testing. Am. Stat. 2016, 70, 195–201. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J. Feature Selection for Image Classification Based on a New Ranking Criterion. J. Comput. Commun. 2015, 3, 74–79. [Google Scholar] [CrossRef]
- Tahir, M.A.; Bouridane, A.; Kurugollu, F. Simultaneous Feature Selection and Feature Weighting Using Hybrid Tabu Search/K-Nearest Neighbor Classifier. Pattern Recognit. Lett. 2007, 28, 438–446. [Google Scholar] [CrossRef]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A Review of Classification Algorithms for EEG-Based Brain–Computer Interfaces: A 10 Year Update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.W.; Zou, G.; Morrow, S.A.; Fridman, S.; Racosta, J.M. Wilcoxon-Mann-Whitney Odds Ratio: A Statistical Measure for Ordinal Outcomes Such as EDSS. Mult. Scler. Relat. Disord. 2022, 59, 103516. [Google Scholar] [CrossRef]
- Nakanishi, M.; Wang, Y.; Chen, X.; Wang, Y.-T.; Gao, X.; Jung, T.-P. Enhancing Detection of SSVEPs for a High-Speed Brain Speller Using Task-Related Component Analysis. IEEE Trans. Biomed. Eng. 2018, 65, 104–112. [Google Scholar] [CrossRef]
- Maÿe, A.; Mutz, M.; Engel, A.K. Training the Spatially-Coded SSVEP BCI on the Fly. J. Neurosci. Methods 2022, 378, 109652. [Google Scholar] [CrossRef]
- Kołodziej, M.; Majkowski, A.; Tarnowski, P.; Rak, R.J.; Rysz, A. A New Method of Cardiac Sympathetic Index Estimation Using a 1D-Convolutional Neural Network. Bull. Pol. Acad. Sciences. Tech. Sci. 2021, 69, 136921. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, D.; Zeng, X. HeartID: A Multiresolution Convolutional Neural Network for ECG-Based Biometric Human Identification in Smart Health Applications. IEEE Access 2017, 5, 11805–11816. [Google Scholar] [CrossRef]
- Furui, A.; Hayashi, H.; Nakamura, G.; Chin, T.; Tsuji, T. An Artificial EMG Generation Model Based on Signal-Dependent Noise and Related Application to Motion Classification. PLoS ONE 2017, 12, e0180112. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Du, Y.; Zhang, R. A CNN-Based Multi-Target Fast Classification Method for AR-SSVEP. Comput. Biol. Med. 2022, 141, 105042. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Qiu, S.; Xing, J.; He, H. A CNN-Based Compare Network for Classification of SSVEPs in Human Walking. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 2986–2990. [Google Scholar]
- Nguyen, T.-H.; Chung, W.-Y. A Single-Channel SSVEP-Based BCI Speller Using Deep Learning. IEEE Access 2019, 7, 1752–1763. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, T.; Tian, Y.; Jiang, X. Filter Bank Convolutional Neural Network for SSVEP Classification. IEEE Access 2021, 9, 147129–147141. [Google Scholar] [CrossRef]



| No. | Name of Layer | Parameters |
|---|---|---|
| 1 | Input Layer | 256 × 3 × 1 signals with zero-center normalization |
| 2 | Convolution_1 | 32 filters of size 8 × 3 with stride [1 1] and padding ‘same’ |
| 3 | Batch Normalization_1 | Batch normalization with 32 channels |
| 4 | ReLU_1 | ReLU |
| 5 | Convolution_2 | 64 filters of size 16 × 3 with stride [1 1] and padding |
| 6 | Batch Normalization_2 | Batch normalization with 64 channels |
| 7 | ReLU_2 | ReLU |
| 8 | Convolution_3 | 128 filters of size 32 × 1 with stride [1 1] and padding |
| 9 | Batch Normalization_3 | Batch normalization with 128 channels |
| 10 | ReLU_3 | ReLU |
| 11 | Convolution_4 | 128 filters of size 64 × 1 with stride [1 1] and padding |
| 12 | Batch Normalization_4 | Batch normalization with 128 channels |
| 13 | ReLU_4 | ReLU |
| 14 | Fully Connected | 4 fully connected layer |
| 15 | Softmax | Softmax |
| 16 | Classification Output | Crossentropyex |
| Algorithm | Execution Time |
|---|---|
| Data set augmentation | 12.115 s |
| CCA algorithm for classification (does not require training) | 0.112 s |
| sMP algorithm for classification (does not require training) | 0.311 s |
| Training the CNN for 50 epochs | 145 min 8 s |
| Training the MLP | 11.1 s |
| Training the LDA | 14.3 s |
| Training the QDA | 17.2 s |
| Training LDA with SFS/t-test feature selection | 31.4 s/19.2 s |
| Training QDA with SFS/t-test feature selection | 41.8 s/22.5 s |
| Method | CNN | CCA | sMP | MLP | QDA | LDA | QDA | LDA | QDA-SFS | LDA-SFS | QDA-T | LDA-T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input | EEG raw | DFT 1–40 Hz | DFT 1–40 Hz | DFT 1–40 Hz | DFT 5 Hz, 6 Hz, 7 Hz, 8 Hz, 10 Hz, 12 Hz, 14 Hz, 16 Hz, 15 Hz, 18 Hz, 21 Hz, 24 Hz | |||||||
| Training the classifier on the generated data | Y | N | N | Y | Y | Y | N | N | N | N | N | N |
| Feature selection | - | - | - | - | - | - | - | - | SFS 25 features | SFS 25 features | t-test 14 features | t-test 14 features |
| Accuracy | ||||||||||||
| User S01 | 0.81 | 0.75 | 0.58 | 0.62 | 0.62 | 0.75 | 0.65 | 0.66 | 0.62 | 0.63 | 0.68 | 0.76 |
| User S02 | 0.88 | 0.54 | 0.51 | 0.61 | 0.61 | 0.59 | 0.56 | 0.40 | 0.48 | 0.47 | 0.61 | 0.50 |
| User S03 | 0.42 | 0.40 | 0.29 | 0.30 | 0.27 | 0.31 | 0.23 | 0.33 | 0.26 | 0.33 | 0.18 | 0.22 |
| User S04 | 0.75 | 0.54 | 0.54 | 0.70 | 0.65 | 0.68 | 0.58 | 0.65 | 0.56 | 0.65 | 0.59 | 0.56 |
| User S05 | 0.75 | 0.63 | 0.61 | 0.63 | 0.63 | 0.65 | 0.62 | 0.58 | 0.61 | 0.59 | 0.66 | 0.62 |
| Mean value | 0.72 | 0.57 | 0.51 | 0.57 | 0.55 | 0.59 | 0.53 | 0.52 | 0.51 | 0.53 | 0.54 | 0.53 |
| F1-score | ||||||||||||
| User S01 | 0.79 | 0.59 | 0.46 | 0.51 | 0.48 | 0.59 | 0.56 | 0.53 | 0.51 | 0.51 | 0.55 | 0.60 |
| User S02 | 0.87 | 0.41 | 0.33 | 0.49 | 0.50 | 0.48 | 0.45 | 0.28 | 0.31 | 0.33 | 0.48 | 0.33 |
| User S03 | 0.29 | 0.30 | 0.19 | 0.18 | 0.18 | 0.23 | 0.15 | 0.24 | 0.12 | 0.25 | 0.11 | 0.14 |
| User S04 | 0.60 | 0.40 | 0.42 | 0.56 | 0.54 | 0.55 | 0.46 | 0.55 | 0.43 | 0.54 | 0.47 | 0.45 |
| User S05 | 0.60 | 0.51 | 0.50 | 0.50 | 0.52 | 0.51 | 0.50 | 0.46 | 0.50 | 0.48 | 0.53 | 0.49 |
| Mean value | 0.63 | 0.44 | 0.38 | 0.44 | 0.44 | 0.47 | 0.42 | 0.41 | 0.37 | 0.42 | 0.42 | 0.40 |
| Subject | CNN | CCA |
|---|---|---|
| S01 | 59.8 | 47.5 |
| S02 | 76.8 | 16.5 |
| S03 | 5.95 | 4.6 |
| S04 | 47.5 | 16.5 |
| S05 | 47.5 | 27.7 |
| Mean | 42.0 | 19.9 |
| EEG Signal (Std) | Noise (Std) | CNN | CCA | |
|---|---|---|---|---|
| I | EEG (0.87 × 10−5) | - | 0.81 | 0.75 |
| II | EEG (0.87 × 10−5) | 5.99 × 10−6 | 0.69 | 0.54 |
| III | EEG (0.87 × 10−5) | 1.60 × 10−5 | 0.59 | 0.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kołodziej, M.; Majkowski, A.; Rak, R.J.; Wiszniewski, P. Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data. Appl. Sci. 2023, 13, 13350. https://doi.org/10.3390/app132413350
Kołodziej M, Majkowski A, Rak RJ, Wiszniewski P. Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data. Applied Sciences. 2023; 13(24):13350. https://doi.org/10.3390/app132413350
Chicago/Turabian StyleKołodziej, Marcin, Andrzej Majkowski, Remigiusz J. Rak, and Przemysław Wiszniewski. 2023. "Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data" Applied Sciences 13, no. 24: 13350. https://doi.org/10.3390/app132413350
APA StyleKołodziej, M., Majkowski, A., Rak, R. J., & Wiszniewski, P. (2023). Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data. Applied Sciences, 13(24), 13350. https://doi.org/10.3390/app132413350






