Abstract
MicroRNAs (miRNAs) are small RNA molecules consisting of approximately 22 nucleotides; they regulate gene expression and are employed in the development of therapeutics for intractable diseases. Predicting the association between miRNAs and genes is crucial for understanding their roles in molecular processes. miRNA–gene associations have been studied using deep learning methods, but these methods present various constraints. Through addressing the limitations of previous methods, this study aimed to achieve better performance than the state-of-the-art (SOTA) methods for studying miRNA–gene associations. We constructed the most extensive embedded dataset to date, comprising 717,728 miRNA–gene pairs, specifically designed for our deep learning model. Further, we applied an embedding method used for protein embedding for transforming our gene sequence data. Moreover, we constructed a sophisticated negative dataset based on three distance criteria, unlike most studies that randomly designate negative data. Leveraging the data and insights from these approaches, we built a deep learning model with the best performance among SOTA miRNA–gene studies (area under the receiver operating characteristic curve = 0.9834). In addition, we conducted a case study using the learned model to predict potential positive data. We also aimed to identify miRNAs closely associated with a gene linked to various cancers.
1. Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules comprising approximately 22 nucleotides; they can regulate gene expression and are employed in the development of therapeutics for intractable diseases [1]. Unlike conventional RNAs, miRNAs function to control protein expression by binding to messenger RNAs (mRNAs) and regulating gene expression through RNA interference. The human miRNA sequence consists of the four nucleotide bases: adenine (A), uracil (U), cytosine (C), and guanine (G), which are crucial for the regulation of gene expression. Currently, there are approximately 2000 known species of human miRNAs that play a key role in survival. miRNAs play pivotal roles in diverse biological processes, including development, signal transduction, immunity, nervous system formation, cell proliferation, differentiation, and death. miRNAs have been used to develop innovative therapeutics for various intractable diseases, including cancer. They have been detected in body fluids, such as blood, urine, and saliva, and are used as diagnostic biomarkers for various diseases. Owing to improved comprehension of their role through basic research and also their clinical importance, miRNAs have increasingly been the subject of studies in the past few years [2].
miRNAs have been recognized as playing important roles in the pathogenesis of several diseases. The identification of disease-specific miRNAs, especially for diseases with ineffective therapies, is valuable. Consequently, predicting miRNA–disease associations has gained attention in research. From 2017 to 2023, 29 studies were conducted to predict the association of miRNAs with different diseases [3]. Acquiring information on molecules related to microRNAs and diseases is essential for miRNA–disease association studies [4]. Over 2000 types of miRNAs have been recognized in humans; however, the features of each one are not completely understood because of complex associations with related molecules and, thus, must be studied further. Previously, determining the association between miRNAs and genes using biological experiments was complicated and challenging in traditional wet labs; these evaluations were time-consuming and expensive. Recently, the prediction of various genomic associations has been undertaken using computational methods that can consider and process a large quantity of biological information [5]. Various embedding methods and learning algorithms applied to open datasets can be utilized to construct mathematical in silico models for predicting the potential relationships between miRNAs and their interacting molecules [4].
Deep learning models are well-suited for handling complex biological information, identifying features, and predicting relationships. Prediction of the association between miRNAs and genes using a deep learning model involves the following steps. First, a dataset is constructed to train the deep learning model. Three datasets are essential to convert biological datasets into input data for deep learning model association prediction: (a) a dataset evidencing a proven association between miRNAs and genes, (b) a dataset possessing biological information on miRNAs, and (c) a dataset possessing biological information on genes. At this stage of the conversion, a researcher should choose the biological information to utilize, such as sequence, structure information, etc. Once these datasets have been constructed, a labeling step for supervised learning is essential. Constructing a training dataset involves both positive and negative labeling. Positive labeling information exists formally; however, negative labeling information does not exist in nature and so must be carefully curated. The next step is data embedding, in which each feature is extracted and vectorized to use the dataset to train the deep learning model. Once embedding is complete according to the labeled data, the data are used for training to create a deep learning model that predicts associations. Recently, various deep learning studies have been conducted using this approach.
First, there are several studies dedicated to predicting the association between miRNAs and genes. The SG-LSTM-FRAME model [4] was developed to predict the association between miRNAs and genes; it utilizes both sequential and mechanistic features of miRNAs and genes to predict their relationship. miRNA–gene relationship information was obtained from miRTarBase [6], and sequence information was obtained from miRBase [7] and biomaRt [8]. Negative data were generated from positive data by setting a threshold using Euclidean distance [9] and cosine similarity [10] to create a negative sample. Each piece of sequence information was embedded using Doc2Vec [11], and the miRNA–gene relationship information was embedded using Role2Vec [12]. The embedded information was used to learn and predict the association between miRNAs and genes using long short-term memory (LSTM) [13]. miTAR [14] uses both spatial and sequential features between miRNAs and genes through a CNN [15] and a bidirectional RNN (Bi-RNN) [16]. DeepMirTar [17] and miRAW [18] were used for miRNA–gene relationship information and miRBase was used for miRNA sequence information. The DeepMirTar dataset was used for negative data. The sequence information was converted into a five-dimensional vector of {A, U, G, C, N} characters in the embedding layer. A CNN was used to learn the spatial features of the miRNA:Target, and a Bi-RNN was used to learn the sequential features of the miRNA:Target. SRG-Vote [19] was implemented using the existing SG-LSTM-WHOLE [4] dataset. After embedding the sequence information using Doc2Vec, the miRNA–gene relationship information was embedded using various methods, such as Role2Vec, graph convolutional network (GCN) [20], and node2vec [21]. Accordingly, the highest area under the receiver operating characteristic curve (AUC) was achieved using a GCN, and GCN embedding was applied using the embedding generated by Role2Vec as a feature of the node. After that, the experiment was conducted by comparing Bi-LSTM [22] and LSTM.
Second, there are studies aimed at predicting the association between miRNAs and target mRNAs. Inferring miRNA targets based on restricted Boltzmann machines (IMTRBM) [23], using machine learning, employed RBM to predict miRNA targets. They obtained the miRNA–gene relationship data from miRTarBase and BioGRID [24] and the corresponding sequence information from miRBase and NCBI [25]. A weighted miRNA–mRNA interaction network was manually constructed to utilize the prediction results of multiple methods, weighting miRNA–mRNA pairs according to their frequency. A weighted network was used to train the model and make predictions using restricted Boltzmann machines. DeepTarget [26] is a recurrent neural network (RNN) [27] framework for miRNA–mRNA relationship prediction. It was constructed using a miRNA target-related dataset from miRecords [28] and miRNA sequence information from miRBase. Negative data for the experiment were randomly generated using the miRanda algorithm after random sequence generation using the Fisher–Yates shuffle algorithm. Sequence information was encoded as a four-dimensional dense vector that was randomly initialized and trained using the gradient descent method, and the encoded information was trained using an RNN-based autoencoder to predict the interaction between miRNA and mRNA. DeepMirTar predicts miRNA–mRNA relationships. Information on the miRNA–mRNA relationship was obtained from mirMark and CLASH data, and sequence information was obtained from miRBase and the UCSC Genome Browser. The negative data were randomly sampled using the miRanda algorithm. Each piece of sequence information was embedded using one-hot encoding, and an autoencoder was used for prediction.
Lastly, there is a study predicting the association between miRNAs and long non-coding RNA (lncRNA). LncMirNet [29] was developed to predict the interaction between lncRNA and miRNA; a dataset of the lncRNA–miRNA relationship was obtained from lncRNASNP2, and sequence information was obtained from miRBase and GENCODE [30]. Negative data were shuffled from positive pairs using the Knuth–Durstenfeld shuffle algorithm and then randomly selected and used as negative samples if there was no positive interaction. Each piece of sequence information was embedded using K-mer, CTD, and Doc2Vec, and the lncRNA–miRNA relationship was embedded using Role2Vec. A convolutional neural network (CNN) was trained with the embedded data to learn and predict potential interactions between lncRNAs and miRNAs. The related studies in terms of the correlation data used, respective sequence data, number of positive and negative datasets, method of generating the negative dataset, computational model used, and prediction performance are summarized in Table 1.

Table 1.
Summary of related studies.
Previous studies have several limitations. The first constraint is the number of datasets used to generate the model. The largest dataset size among the related studies was 31,080. Larger datasets are increasingly required to generate high-performance predictive models. The second constraint is the embedding method for the feature extraction of each data item. More sophisticated embedding methods are required to extract important features from data. The third constraint is the negative data generation method. Most previous studies have randomly generated negative data [14,17,25,27]. However, ideally, negative data used in constructing a deep learning model must be methodically generated using sophisticated criteria. Finally, an efficient deep learning model for association prediction is needed. As biological data comprise sequences, it would be advantageous to leverage deep learning models that are specialized for time-series data.
In this study, we propose a method for predicting the association between miRNAs and genes using a deep learning model. Predicting the association between miRNAs and genes is crucial for an extensive understanding of the underlying molecular processes. To generate the best-performing miRNA–gene association prediction model, we (1) generated the largest training dataset of existing miRNA–gene datasets, (2) applied optimal embedding methods to extract features of miRNA and gene sequences, (3) generated negative datasets according to three distance criteria, and (4) implemented the optimal association prediction deep learning model (AUC = 0.9832). In addition, we evaluated the applicability of the proposed method by conducting several case studies using the proposed model.
2. Materials and Methods
2.1. Dataset Construction
Training deep learning models requires a large quantity of data. This is particularly important for biological research. miRNAs and genes are difficult to observe directly, and there are limitations to understanding their characteristics and properties through wet lab experiments. Therefore, computational methods have been adopted, and most of them have used open datasets to construct the datasets. However, open datasets cannot be used in their current state, and researchers must combine and extract data to suit their specific research purposes. In this study, using a deep learning model that predicts the association between human miRNAs and genes, we propose that the best way to improve a model’s performance is to train it with a large amount of association data; therefore, we built a large number of datasets through the following process.
Three datasets were used to construct the data for this study. (a) First, datasets with a proven positive relationship between human miRNAs and genes were obtained from the miRTarBase [6]. The miRTarBase is a curated database of miRNA–gene interactions containing data on 502,652 miRNA–gene pairs. We filtered out duplicate pairs, after which 380,634 pairs remained. However, we cannot utilize all these relationship datasets unless we have comprehensive biological information for every miRNA and gene element within the dataset. Thus, to make use of a large number of datasets, we decided to use nucleotide sequence information, which is the most widely available among any other biological data. Details of the datasets are discussed in the subsections that follow. (b) Second, miRBase [7] was chosen as the miRNA sequence database, and (c) biomaRt [8] was chosen as the gene sequence database. miRBase contains the sequence and annotation information of miRNAs. It also provides extensive information on miRNAs, including genomic coordinates and deep sequencing expression data. biomaRt is an open-gene dataset developed by the European Bioinformatics Institute. The main advantage of biomaRt is that users can easily extract gene sequence information without any programming knowledge or complex database structure and can segment the data according to user-specified criteria. The gene sequence information was acquired using the biomaRt R package (Version 2.58.0). Pairs containing miRNAs and genes for which no sequence information exists in miRBase and biomaRt were excluded from the 380,634 pairs. This process resulted in 2656 miRNA and 14,319 gene sequences, and 358,864 positive relationship datasets (Table 2). Therefore, we built the largest number of positive datasets among miRNA–gene association deep learning model studies.

Table 2.
Data produced by leveraging open datasets.
2.2. Embedding Method
The miRNA sequence, composed of nucleotide bases A, U, C, and G, plays a crucial role in gene expression regulation. Likewise, gene sequences, which dictate protein synthesis, are formed from the bases A, T, C, and G within DNA. In a deep learning model, the proper representation of miRNAs and genes is crucial as it has a significant impact on the predictive performance of the model. In many previous studies, one-hot encoding was used to represent the sequence data [17]. One-hot encoding is specialized for representing categorical data, in which each category of data is represented by a unique index. However, one-hot encoding does not express the relationships among data and has the disadvantage that the dimension of the vector is equal to the number of data types, which increases the dimensionality. With the advancement in natural language processing, various embedding techniques have been developed. Among them are Word2Vec [31] and Doc2Vec [11]. Word2Vec is a technique recently employed for embedding sequence data encompassing gene sequences. Word2Vec is an algorithm that generates the embedding of words by using the surrounding words to predict the center word (CBOW) or by using the center word to predict the surrounding words (skip-gram). Word2Vec has limitations in that it is not sensitive to context and word order; therefore, it was deemed unsuitable for this study, as it predicts associations using miRNA and gene sequence information alone. Doc2Vec can be seen as an extension of Word2Vec, designed to embed an entire document within a continuous vector space. It employs two primary architectures: distributed memory (DM) and distributed bag of words (DBOW). DM predicts words in light of the document’s sequential order, while DBOW estimates the document as a whole, sidelining individual words and relying on the overall context. Consequently, when it comes to capturing the holistic meaning and structure of a sequence, Doc2Vec often surpasses Word2Vec.
In this study, we utilized a Doc2Vec-based embedding method, protein2Vec [32], which more effectively captures the order and flow of sequence data. protein2Vec is an embedding method for the better prediction of specific properties of proteins and has excellent embedding qualities. We assumed that protein2Vec, which is based on Doc2Vec, would be effective for embedding not only protein sequences but also nucleotide sequences; hence, we employed it to embed the data in this study. We embedded 2656 miRNAs and 14,319 genes in 64 dimensions (Figure 1).
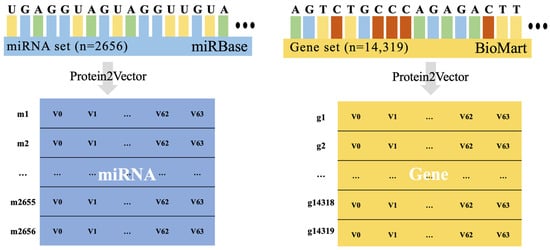
Figure 1.
The structural representation of the original sequence dataset for miRNA and genes. This figure also explains the embedding method employed in this study, showcasing how the data is transformed into vector values (64 dimensions). Additionally, this figure depicts the number of miRNA and gene sequences utilized in the research.
2.3. Negative Dataset
As mainstream biological research has moved from wet labs to groundbreaking bioinformatic-based models, one of the major challenges has been negative data. Most association–prediction studies that employ deep learning are binary classification problems. Although there is considerable experimentally validated and open positive data, there are very limited validated negative data (no interaction) [33]. However, to optimize the performance of a binary classification deep learning model, having a balanced dataset is often highly beneficial. Most related studies use random pairs that do not exist in positive pairs and consider them as negative data. However, we introduced a distance-based filtering method to generate a sophisticated negative dataset. In a recent study [4], negative data were generated using the Euclidean distance and cosine similarity as the distance criteria. Our study introduces the Mahalanobis distance [34] as an additional criterion building upon this concept. The data in this study comprised miRNA–gene pairs, all of which existed in a vector space through embedding. The miRNAs and genes that spread in the vector space are separated by a distance, which is an important metric. In natural language processing, the distance in this vector space is considered a measure of similarity. The following three distances were used to construct sophisticated negative data: (1) The Euclidean distance, which represents the straight-line distance between two points and is the shortest distance between two points. It is frequently used to compare the similarity between two data points, with a lower distance indicating a higher similarity. (2) The cosine similarity that measures the cosine angle between two vectors. It is frequently used in text analytics to measure document similarities. (3) The Mahalanobis distance measures the distance between two points, considering the covariance structure of the data. Euclidean distance treats all dimensions equally, while Mahalanobis distance takes into account the variance and correlation of the data to calculate the distance. This is useful for detecting outliers or clustering in multivariate data. We utilized these three distance criteria to calculate the distances between miRNA and gene pairs in the positive set. The average of these calculated distances was then used as a threshold for constructing the negative dataset:
Based on these three distance criteria, the negative dataset was constructed as follows: First, 2656 miRNAs and 14,319 genes in the positive dataset were matched for all the cases. A total of 38,031,264 datasets (2656 × 14,319) were generated, and 358,864 positive datasets were removed. Then, we removed pairs that were closer than the three average distances calculated earlier (smaller values for the Euclidean distance and Mahalanobis distance and higher values for the cosine similarity) because we assumed that they did not show the characteristics of negative data (Figure 2). After filtering, we obtained a set of 4,932,554 negative candidates. We have illustrated the counts of instances filtered as negative candidates through each of the three distance criteria using a Venn diagram (Figure 3). From this set of negative candidates, we randomly selected 358,864 values as negative data to balance the quantity of positive data (Figure 4 and Table 3). These negative data exhibited negative features that are more distant than the average distance of the positive dataset on the three distance measures of similarity.
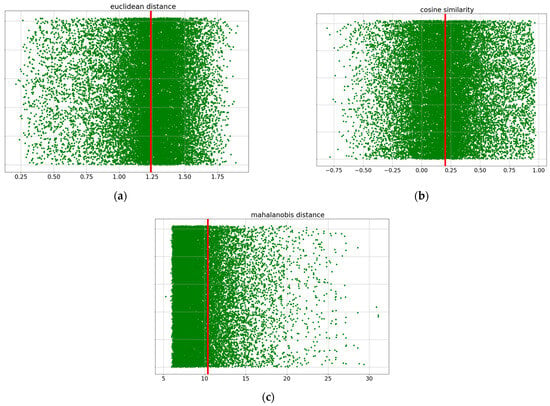
Figure 2.
Distribution of data around the threshold for each of the three distance criteria before filtering. (a) The calculated average Euclidean distance of the positive data is 1.245299, denoted by the red line. The data to the left of the red line were deleted to obtain a set of negative candidates. (b) The calculated average cosine similarity of the positive data is 0.20212, shown by the red line. The data to the right of the red line were deleted to obtain a set of negative candidates. (c) The calculated average Mahalanobis distance of the positive data is 10.403354, shown by the red line. The data to the left of the red line were deleted to obtain a set of negative candidates.
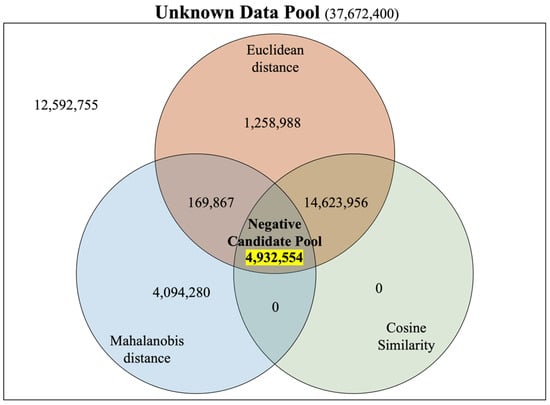
Figure 3.
To construct a sophisticated negative set, we utilized three distinct distance measures for filtering within the Unknown Data Pool. The Venn diagram represents the quantities of instances that were filtered as candidates for the negative set according to each distance criterion. As a result, a Negative Candidate Pool consisting of 4,932,554 instances was generated. This signifies that the Euclidean distance, cosine similarity, and Mahalanobis distance are essential for the construction of our sophisticated negative set.
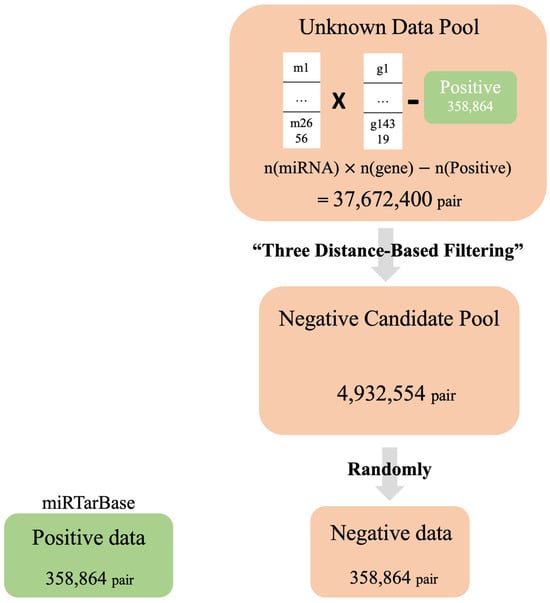
Figure 4.
For the positive data set, we employed a subset of experimentally validated miRNA–gene interaction pairs derived from the miRTarBase. To construct the Unknown Data Pool, we generated a comprehensive set of possible interactions by combining all miRNAs and genes used in the positive data set, excluding any pairs that overlap with the positive data set to avoid redundancy. Subsequently, we initiated Three Distance-Based Filtering to establish a Negative Candidate Pool. From this Negative Candidate Pool, we randomly selected a number of pairs equal to that of the positive set to form our final negative data set, thereby ensuring a balanced representation of both classes for the training process.

Table 3.
Process of generating negative data generation and the names of the corresponding datasets.
2.4. Deep Learning Model
In this study, we worked with sequence data generated from miRNA–gene pairs. To accurately capture the complex sequence features of miRNAs and genes for the prediction of their mutual associations, we employed specialized deep learning models that are tailored for the analysis of sequential data. RNNs (recurrent neural networks) [27] are a type of deep learning architecture tailored for sequential data. And LSTM (long short-term memory) [13], an advanced type of RNN, is designed to overcome the vanishing gradient issue inherent in basic RNNs. Therefore, we employed LSTM and Bi-LSTM [22] models, leveraging their ability to effectively remember and utilize long-range sequential information, which is crucial for understanding the complex interactions between miRNA and gene sequences. A pivotal component of the LSTM model is the cell state. The cell state undergoes an update process, which can be summarized as follows:
- Begin by multiplying the previous cell state value (Ct−1) with the output of the forget gate (ft), determining what information is to be discarded.
- Update the current cell state value by multiplying it with the output of the input gate. This operation gauges how much new information should be stored.
- Add the updated current cell state value to the output of the input gate. This step identifies the information to be retained.
A visual representation of this process can be observed in Figure 5.
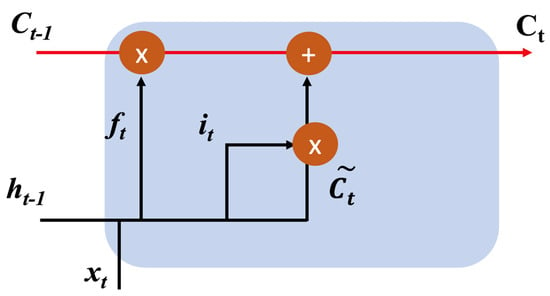
Figure 5.
Sequence of cell state updates, which is the heart of the LSTM cell. This part solves the long-term dependency problem of traditional RNNs. This cell state decides what information to keep and what information to discard. Cell state allows LSTMs to remember their relationship to previous data points in the sequence and use this context to better understand and predict current and future data points.
Bi-LSTM is a deep learning architecture that extends LSTM to model bidirectional information within sequence data. Traditional LSTM processes input data from front to back. In contrast, Bi-LSTM processes input data in both forward and backward directions, ensuring that the model has access to future contexts when making predictions at a given point in the sequence. Consequently, combining bidirectional information improves the model’s generalization performance and reduces overfitting. However, due to the doubled parameter count inherent to its bidirectional nature, Bi-LSTM is computationally intensive and learns slower than traditional LSTM.
3. Results
3.1. Dataset
Data organization is the most important aspect of deep learning model research. The quantity of data in a large dataset of good quality (data labeling is accurate, and the embedding of the data is representative of the features) has a significant effect on the performance and generalization ability of a deep learning model.
Using the methods described in the previous section, we generated the largest quantity of miRNA–gene association data (717,728) to train and evaluate our deep learning models. Our dataset comprises positive and negative miRNA–gene sequence data in a 1:1 ratio, and the sequence data consists of 2656 miRNAs and 14,319 genes. The overall dataset used in this study is listed in Table 4.

Table 4.
Our data configuration.
3.2. LSTM and Bi-LSTM Experiment
We conducted experiments to evaluate the performance of our deep learning models using our dataset to determine the model best suited for our specific dataset. The number of LSTM layers was set to three. For the loss function, cross-entropy was used to calculate the degree of loss by converting the predicted value to a value between 0 and 1. Cross-entropy is used primarily in classification problems. The Adam optimization function, which is characterized by a lack of gradient scaling effect on the step size during training, was used for optimization. The epoch size was 200, and the batch size was set to 128. The number of Bi-LSTM layers was set to two. For the loss function, binary cross-entropy was used to calculate the degree of loss by converting the predicted value to a value between 0 and 1. In addition, cross-entropy and Adam optimization were used for both models. Furthermore, the Robust Scaler was used to improve the model convergence and performance. The Robust Scaler uses the median and interquartile range (IQR) of the data to perform scaling, which provides more robust scaling for data with outliers. The epoch size was 200, and the batch size was set to 128 [35].
A total of 717,728 data elements were divided into training and test data in an 8:2 ratio. Each data element represents 128-dimensional miRNA–gene data. The LSTM model has an AUC of 0.98, and the Bi-LSTM model has an AUC of 0.936 (Table 5). We found the LSTM model to be more suitable for this study. The training time of the Bi-LSTM model, which is considerably more computationally intensive owing to the large quantity of data, is also longer. Bi-LSTMs are designed to capture both past and future context in sequence data, potentially providing a richer representation. However, given our balanced dataset and our prediction performance results, we determined that a simpler LSTM model was more appropriate.

Table 5.
AUC scores achieved by the LSTM and Bi-LSTM models.
3.3. K-Fold Cross-Validation and Model Performance
K-fold cross-validation is a statistical method for evaluating the performance of machine learning models. It works by dividing a dataset into k-folds and then training and evaluating the model multiple times, using each fold as the test data and the remaining folds as the training data. This method yields statistically reliable results for evaluating the performance of the model. In bioinformatic deep learning studies, where data are relatively scarce, k-fold cross-validation is often used to demonstrate the generalization performance of a model.
We conducted a k-fold cross-validation to determine the generalization performance and statistical confidence of our model. We used the same 717,728 datasets as those in the aforementioned experiment with a fold of 5. We obtained test AUC values of 0.9819, 0.9807, 0.9834, 0.9732, and 0.9826 with an average AUC of 0.9804. The AUC and convergence metrics of the best-performing model are shown in Figure 6.
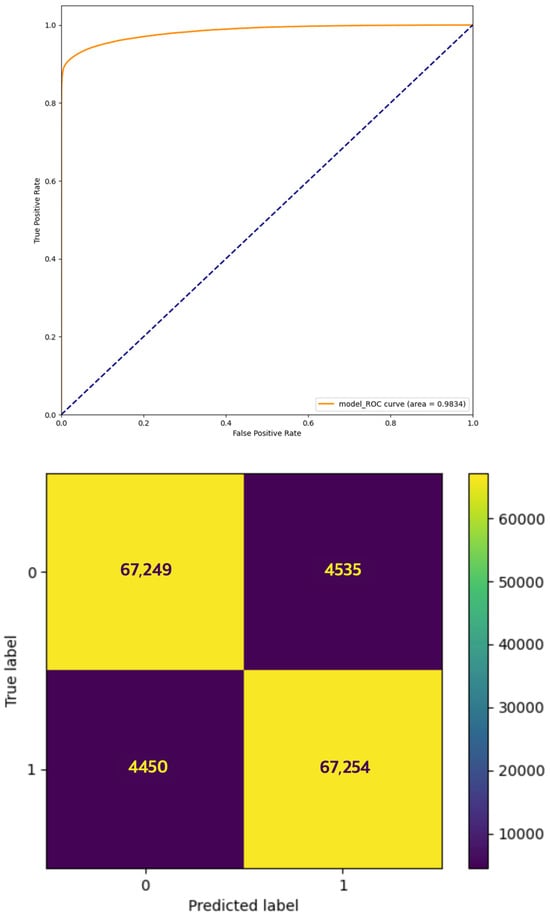
Figure 6.
Our model’s highest achieved AUC (0.9834) and corresponding confusion matrix. AUC is the most widely used evaluation metric in this field and represents the overall performance of the model, illustrating the relationship between the true positive rate (TPR, sensitivity) for the positive class and the false positive rate (FPR, 1—specificity) across different classification thresholds. As the AUC approaches 1, it is interpreted that the model is better at distinguishing the positive class from the negative class.
We used the AUC as a performance metric for the prediction model because it is the most widely used metric in the field and represents the overall performance of the model. The area under the receiver operating characteristic curve (AUC) is a measure that illustrates the relationship between the true positive rate (TPR, or sensitivity) for positive classes and the false positive rate (FPR, or 1—specificity) for negative classes at different classification thresholds. An AUC value closer to 1 indicates a superior prediction model. According to a review paper published in 2022 [3], which investigated 29 recent studies on miRNA–disease association prediction, among all the performance metrics, AUC is the only one presented as a performance measure in all 29 studies. We also present metrics such as precision, recall, accuracy, and F1 score. The performance values are summarized in Table 6. The highest AUC achieved is 0.9834 (Figure 6), which represents the best performance among state-of-the-art miRNA–gene association prediction deep learning studies; furthermore, it underscores the appropriateness of our data generation method and its capability to accurately represent the association between miRNAs and genes.

Table 6.
The table of five-fold cross-validation prediction performance results.
3.4. Validation of the Model
A case study was conducted to demonstrate the utility and performance of the proposed model using the “Unknown Data Pool” (Table 3) generated in this study. The aim was to identify unknown positive candidates in addition to known positive data from miRTarBase. From the “Unknown Data Pool”, positive and negative data used for model training and evaluation were excluded. We predicted the association between all remaining miRNA–gene data pairs and identified the top 20 pairs based on their association scores from our model. The results are summarized in Table 7. We identified several pairs that were not designated as positive data in this research but are considered positive pairs in other datasets, specifically TargetScan [36] and miRWalk [37]. We also predicted candidate pairs that might have a positive relationship that was not identified in open datasets or other sources. This finding underscores the genuine value of our study, highlighting the potential to uncover novel pairs of associations using our data and models.

Table 7.
Top 20 miRNA candidates associated with genes from the Unknown Data Pool.
We also identified miRNAs that are potentially associated with genes known to cause cancer. BRCA2 is a human gene strongly associated with the risk of breast and ovarian cancers. BRCA2 codes for a protein that plays an important role in repairing DNA damage and helps regulate the cell cycle, preventing errors during DNA replication. Cells with unrepaired DNA damage can become malignant and develop cancer [38]. Women and men with BRCA2 mutations have an increased risk of breast cancer, ovarian cancer, and prostate cancer. Therefore, we predicted miRNAs associated with the BRCA2 using our model. Based on the association prediction score of our model, we present the top 10 miRNAs associated with BRCA2 (Table 8). We identified one miRNA associated with BRCA2 that is also supported by TargetScan. While not explicitly stated to have a direct association with BRCA2, we observed that miRNAs related to diseases closely associated with BRCA2, such as ovarian cancer, breast cancer, and prostate cancer, were included in the top 10 rankings. According to GeneCaRNA [39], in this experiment, hsa-miR-503-5p was ranked highest in predicted association with BRCA2, which is related to prostate cancer. hsa-miR-125a-3p is a key miRNA causing ovarian and breast cancers, hsa-miR-665 is associated with Breast Large Cell Neuroendocrine Carcinoma, and hsa-let-7e-5p is an miRNA related to cervical cancer. The BRCA2-associated miRNAs predicted in this study can serve as a foundation for future research.

Table 8.
Top 10 candidate miRNAs associated with BRCA2 from the Unknown Data Pool.
4. Discussion
4.1. Dataset
In bioinformatics, biological information is often used to create networks and sequences. While sequence information on several miRNAs and genes is plentiful, there is a lack of network-related information. Our research goal was to improve the prediction performance of deep learning models, so instead of reducing the dataset, we tried to generate as much relevant data as possible. Therefore, we used only sequence information in this study, not network information. We hypothesized that a model trained on extensive miRNA–gene relationship data would have superior generalization performance and would be able to capture the characteristics of each miRNA and gene.
We also conducted additional experiments to determine the impact of a negative dataset on the performance of deep learning models. Keeping the 358,864 positive data points constant, we substituted the negative data points with random pairs that were not present in the positive set in equal numbers. The AUC for the model trained with random negatives was 0.91, which is 0.07 lower than that achieved when using our established criteria for negative data (see Table 9). This suggests that our method of labeling negatives based on the three distance criteria improves model performance.

Table 9.
Performance changes due to negative dataset differences.
Additionally, we present the distribution of the positive and negative data used in this study in a PCA (principal component analysis) three-dimensional space, as shown in Figure 7.
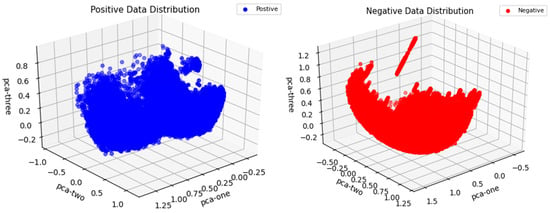
Figure 7.
Distribution of positive and negative data in the PCA three-dimensional space of the study.
4.2. Comparison with Other Related Studies
Based on the criteria outlined in Table 1, we compared our study with the related studies in Table 10. (1) The primary distinction is the volume of data. While the SRG-Vote [19] model used data from 1947 miRNAs and 6823 genes, we used data from 2656 miRNAs and 14,319 genes. However, we focused on using only sequence information, while the SRG-Vote method learned from both sequence and network features. (2) Another difference lies in negative data labeling. Most previous studies have used randomly generated negative data; however, based on the distance criterion filtering proposed by SG-LSTM [4], we applied the Mahalanobis distance, which considers the covariance structure in addition to the Euclidean distance and cosine similarity. (3) Another distinction was the embedding method applied using protein2Vec [32]. Further, we applied a distinct embedding method based on protein2Vec, which, although based on Doc2Vec [11], produces better embedding results because protein sequences are more similar to nucleotide sequences than typical documents. Finally, our LSTM model that demonstrated the best performance (AUC = 0.9834) was based on meticulously curated data. It outperformed the Bi-LSTM model and also predicted the degree of association of unlabeled data.

Table 10.
Related studies on predicting miRNA–gene associations and miGAP.
4.3. Future Work
In the future, we aim to integrate additional biological information for training our deep learning model. We also intend to apply a wider variety of deep learning models and conduct more comprehensive case studies. Furthermore, we will conduct research to predict entities related to various diseases for the treatment of incurable and intractable diseases. To conduct research on the treatment of incurable diseases, we plan to carry out studies sequentially predicting the association between genes and diseases, as well as the association between mRNA and diseases. Furthermore, we ultimately aim to undertake deep learning research on drug repurposing for the treatment of incurable diseases based on miRNA information.
5. Conclusions
In this study, we propose a unique deep learning approach to predict associations between human miRNAs and genes. The performance of our method was measured using five-fold cross-validation, achieving the highest AUC of 0.9834 and an average AUC of 0.9804. This method outperformed the SOTA miRNA–gene association prediction methods. The outstanding performance of the proposed model can be attributed to the following:
- Extensive training data: our model benefited from training on the largest sequence dataset ever constructed for miRNA–gene associations.
- Optimal data embedding: we employed a sophisticated vectorization technique, transforming complex miRNA and gene sequence features. This was based on a model specifically designed for protein sequence embedding.
- Logical negative data construction: by considering Euclidean distance, cosine similarity, and Mahalanobis distance, we defined a set of criteria that allowed for the logical construction of negative data.
- Optimized model architecture: drawing from the above data, we designed an effective miRNA–gene LSTM deep learning model.
We also conducted two case studies using our method. Initially, to identify potentially associated pairs, we performed experiments to predict pairs with unknown associations. Furthermore, we conducted experiments to predict miRNAs closely associated with BRCA2, a gene linked to various cancers. Through these experiments, we demonstrate that our research exhibits outstanding generalization performance and has a wide range of practical applications.
There are limitations in this study as well. Specifically, when selecting the negative set randomly from the Negative Candidate Pool to match the number of positives, the data that is randomly chosen as the negative data can have a minor impact on performance; however, this is considered an inevitable part of deep learning research that requires the prediction of associations, as data with a clear negative relationship are almost non-existent in the field of bioinformatics. The method of setting negatives in this way is expected to significantly influence the advancement of future bioinformatics deep learning research.
Author Contributions
Conceptualization, S.Y. and K.L.; methodology, S.Y. and I.H.; software, S.Y. and I.H.; validation, S.Y., I.H., J.C. and H.Y.; formal analysis, I.H. and H.Y.; investigation, H.Y. and J.C.; resources, S.Y., I.H., J.C. and H.Y.; data curation, I.H. and J.C.; writing—original draft preparation, S.Y.; writing—review and editing, S.Y. and K.L.; visualization, J.C. and H.Y.; supervision, K.L.; project administration, K.L.; funding acquisition, K.L. and S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (No. 2021R1F1A106151313).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data produced or examined during this research can be found in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Peng, Q. A deep ensemble model to predict miRNA-disease association. Sci. Rep. 2017, 7, 14482. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, L.; Chen, X. Updated review of advances in micrornas and complex diseases: Towards systematic evaluation of computational models. Brief. Bioinform. 2022, 23, bbac407. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Luo, J.; Pan, C.; Liu, Y. SG-LSTM-FRAME: A computational frame using sequence and geometrical information via LSTM to predict miRNA–gene associations. Brief. Bioinform. 2021, 22, 2032–2042. [Google Scholar] [CrossRef] [PubMed]
- Deepthi, K.; Jereesh, A.; Liu, Y. A deep learning ensemble approach to prioritize antiviral drugs against novel coronavirus SARS-CoV-2 for COVID-19 drug repurposing. Appl. Soft Comput. 2021, 113, 107945. [Google Scholar]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Danielsson, P.-E. Euclidean distance mapping. Comput. Graph. Image Process. 1980, 14, 227–248. [Google Scholar] [CrossRef]
- Rahutomo, F.; Kitasuka, T.; Aritsugi, M. Semantic cosine similarity. In Proceedings of the 7th International Student Conference on Advanced Science and Technology ICAST, Seoul, Republic of Korea, 29–30 October 2012; Volume 4, p. 1. [Google Scholar]
- Le, Q.; Mikolov, T. Distributed representations of sentences and documents. In Proceedings of the International Conference on Machine Learning, Beijing, China, 21–26 June 2014; pp. 1188–1196. [Google Scholar]
- Ahmed, N.K.; Rossi, R.; Lee, J.B.; Willke, T.L.; Zhou, R.; Kong, X.; Eldardiry, H. Learning role-based graph embeddings. arXiv 2018, arXiv:1802.02896. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Zhao, X.; Barbazuk, W.B.; Lee, J.-H. miTAR: A hybrid deep learning-based approach for predicting miRNA targets. BMC Bioinform. 2021, 22, 96. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Wen, M.; Cong, P.; Zhang, Z.; Lu, H.; Li, T. DeepMirTar: A deep-learning approach for predicting human miRNA targets. Bioinformatics 2018, 34, 3781–3787. [Google Scholar] [CrossRef]
- Pla, A.; Zhong, X.; Rayner, S. miRAW: A deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PLoS Comput. Biol. 2018, 14, e1006185. [Google Scholar] [CrossRef]
- Xie, W.; Zheng, Z.; Zhang, W.; Huang, L.; Lin, Q.; Wong, K.-C. SRG-vote: Predicting miRNA-gene relationships via embedding and LSTM ensemble. IEEE J. Biomed. Health Inform. 2022, 26, 4335–4344. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Graves, A.; Jaitly, N.; Mohamed, A.-r. Hybrid speech recognition with deep bidirectional LSTM. In Proceedings of the 2013 IEEE Workshop on Automatic Speech Recognition and Understanding, Olomouc, Czech Republic, 8–12 December 2013; pp. 273–278. [Google Scholar]
- Liu, Y.; Luo, J.; Ding, P. Inferring microRNA targets based on restricted Boltzmann machines. IEEE J. Biomed. Health Inform. 2018, 23, 427–436. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI biosystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Baek, J.; Park, S.; Yoon, S. deepTarget: End-to-end learning framework for microRNA target prediction using deep recurrent neural networks. In Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Seattle, WA, USA, 2–5 October 2016; pp. 434–442. [Google Scholar]
- Medsker, L.R.; Jain, L.C. Recurrent Neural Networks: Desing and Application; CRC Press: Boca Raton, FL, USA; London, UK; New York, NY, USA; Washington, DC, USA, 2001. [Google Scholar]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA–target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, Y.; Lin, Y.; Shao, D.; He, K.; Huang, L. LncMirNet: Predicting LncRNA–miRNA interaction based on deep learning of ribonucleic acid sequences. Molecules 2020, 25, 4372. [Google Scholar] [CrossRef] [PubMed]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Yang, K.K.; Wu, Z.; Bedbrook, C.N.; Arnold, F.H. Learned protein embeddings for machine learning. Bioinformatics 2018, 34, 2642–2648. [Google Scholar] [CrossRef]
- Fang, Y.; Pan, X.; Shen, H.-B. Recent Deep Learning Methodology Development for RNA–RNA Interaction Prediction. Symmetry 2022, 14, 1302. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D. The Mahalanobis Distance, Chemometrics and Intelligent Laboratory Systems; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Ahn, H. Performance Evaluation of a Feature-Importance-based Feature Selection Method for Time Series Prediction. J. Inf. Commun. Converg. Eng. 2023, 21, 82–89. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Mavaddat, N.; Peock, S.; Frost, D.; Ellis, S.; Platte, R.; Fineberg, E.; Evans, D.G.; Izatt, L.; Eeles, R.A.; Adlard, J. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. JNCI J. Natl. Cancer Inst. 2013, 105, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Barshir, R.; Fishilevich, S.; Iny-Stein, T.; Zelig, O.; Mazor, Y.; Guan-Golan, Y.; Safran, M.; Lancet, D. GeneCaRNA: A comprehensive gene-centric database of human non-coding RNAs in the GeneCards suite. J. Mol. Biol. 2021, 433, 166913. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).