The Unmanned Aerial Vehicle (UAV)-Based Hyperspectral Classification of Desert Grassland Plants in Inner Mongolia, China
Abstract
:1. Introduction
- (1)
- Based on an improved depth-separable convolution to improve the nonlinear fitting ability of the model, this study proposed a streamlined 2D-CNN (SL-CNN) model for desert grassland plant taxa classification. This model effectively explored lightweight convolution in desert grassland species classification research and could achieve the efficient and high-precision monitoring of grassland species.
- (2)
- The model used improved convolutional block attention (CBAM-F) to effectively focus on important channel features and key spatial information and improved the model’s feature refinement capability by adaptively learning feature map channels and spatial relationships. It was combined with residual block convolution (RBC-F) to fuse the feature data and improve the model classification performance.
- (3)
- Using the variance and Frobenius norm2 feature band selection methods, we could efficiently reduce the dimensionality of the data, enhance the computational efficiency of the model, retain important information for classification tasks, and effectively alleviate data redundancy in hyperspectral images.
2. Materials and Methods
2.1. UAV Hyperspectral Remote Sensing System
2.2. Study Area
2.3. Data Acquisition
2.4. Data Preprocessing
2.4.1. Feature Classification
2.4.2. Data Labeling
3. CNN Model Construction
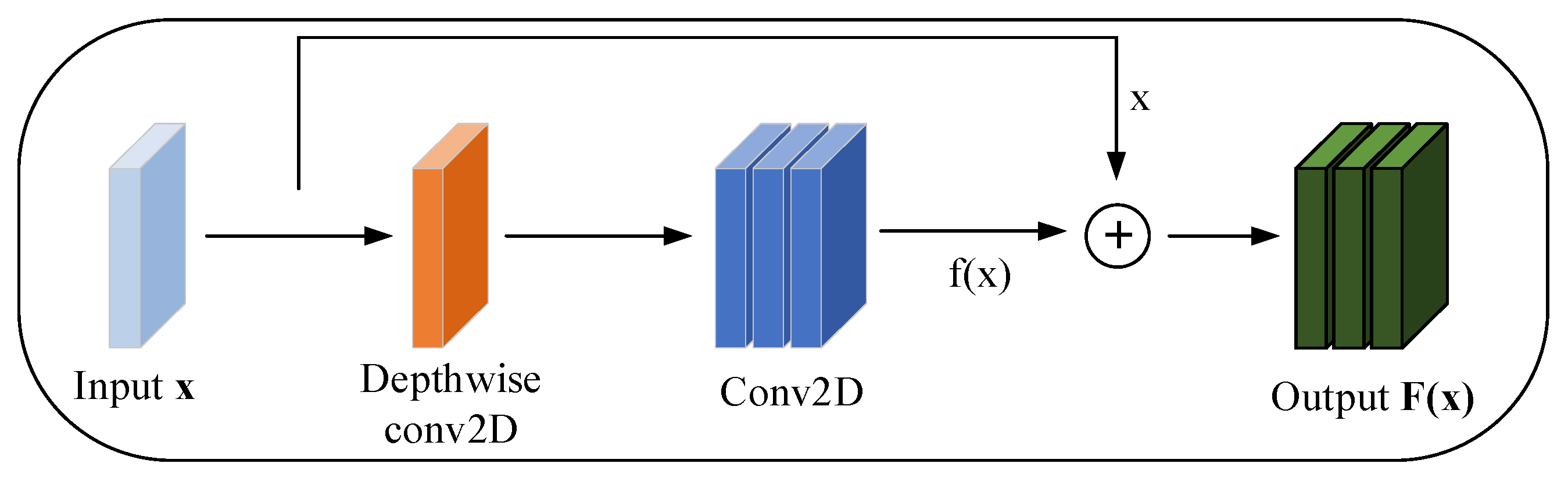
3.1. Improved Depth-Separable Convolution
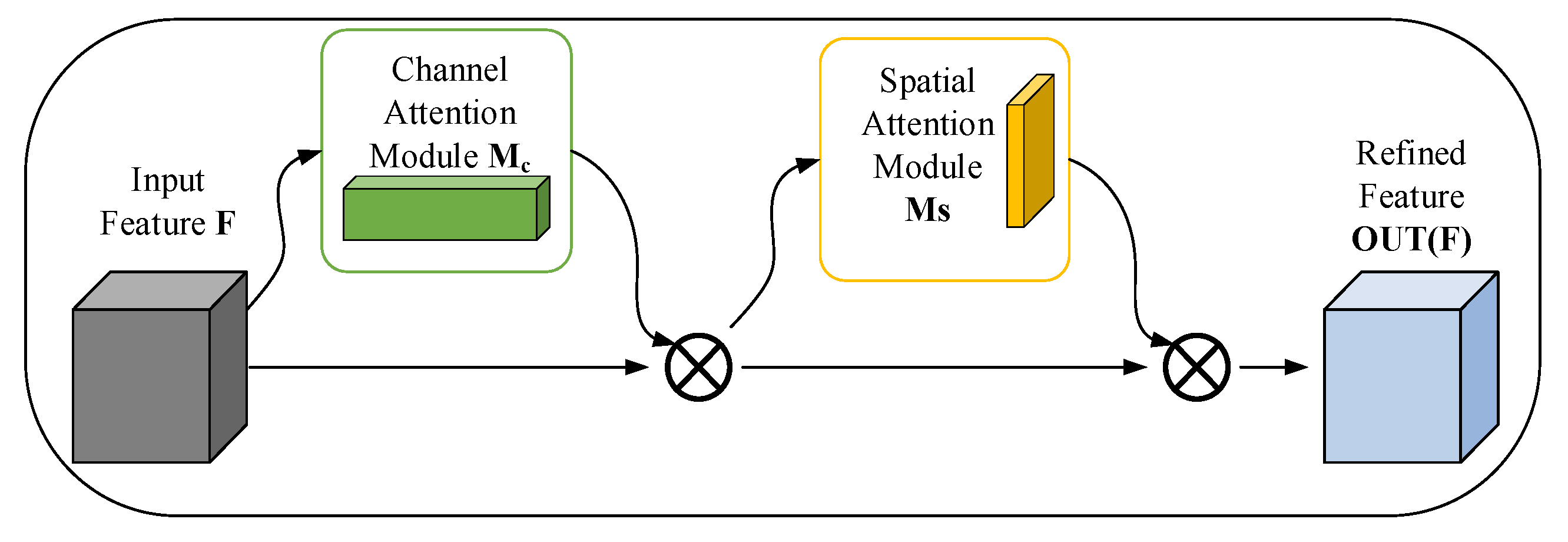
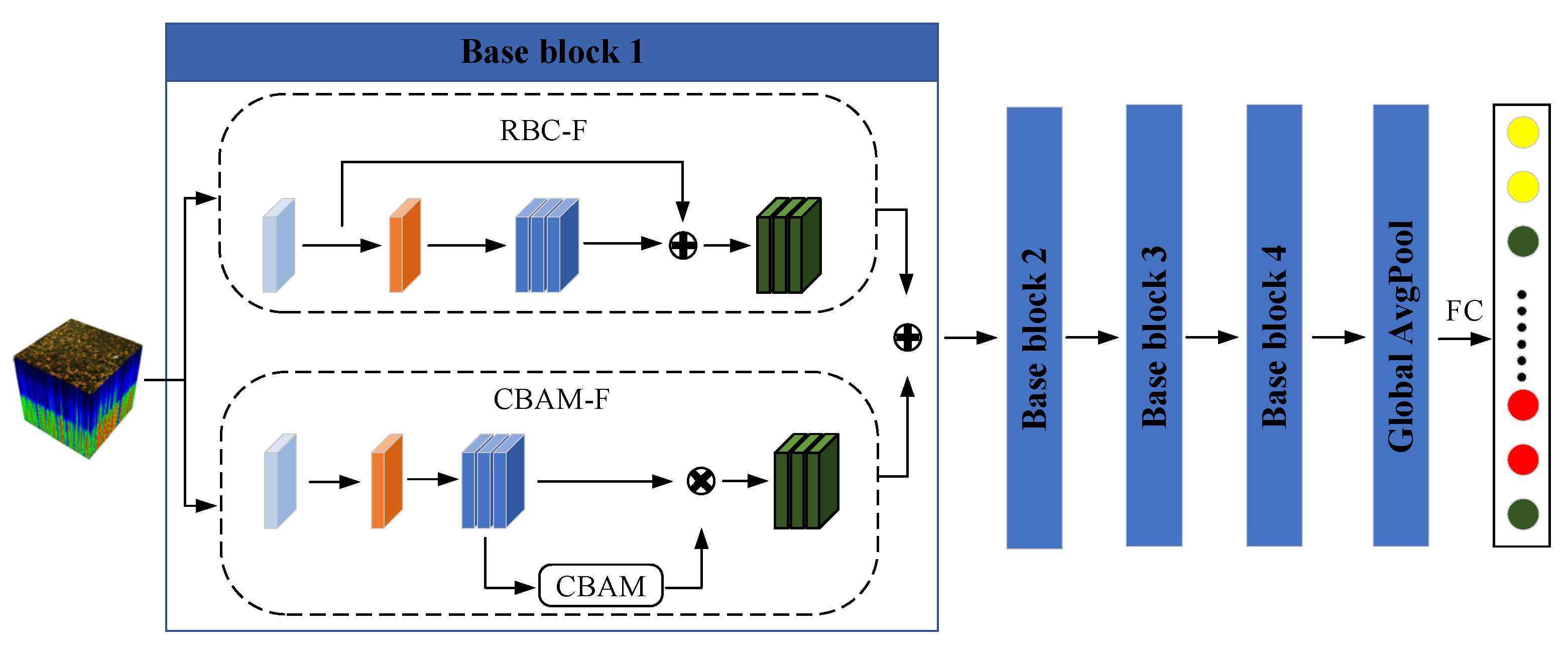
3.2. Convolutional Block Attention Feature Refinement Module (CBAM-F)
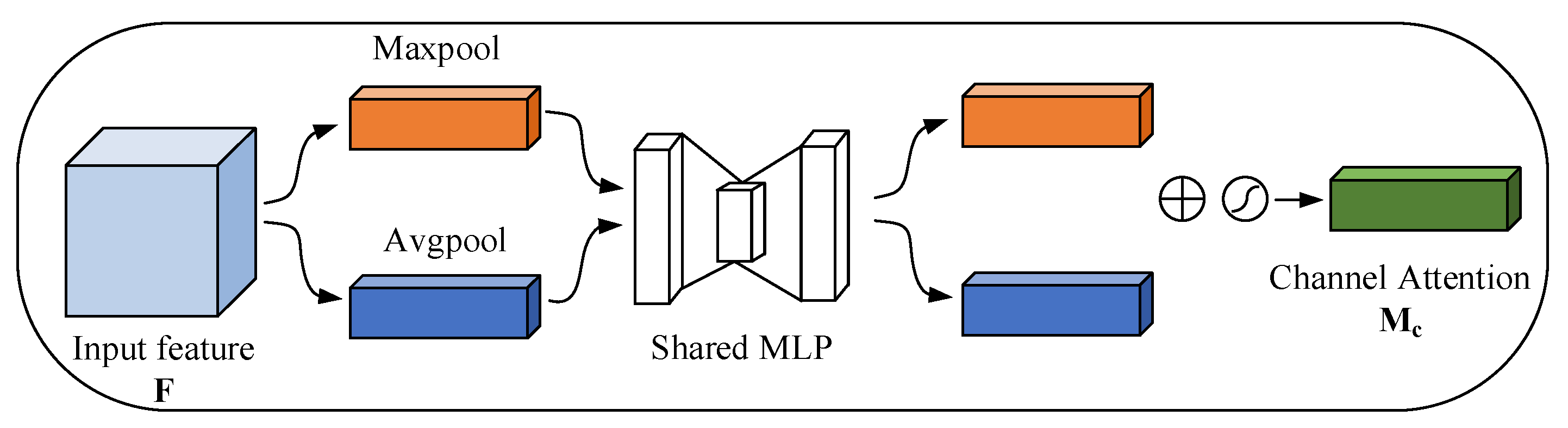
3.2.1. Channel Attention
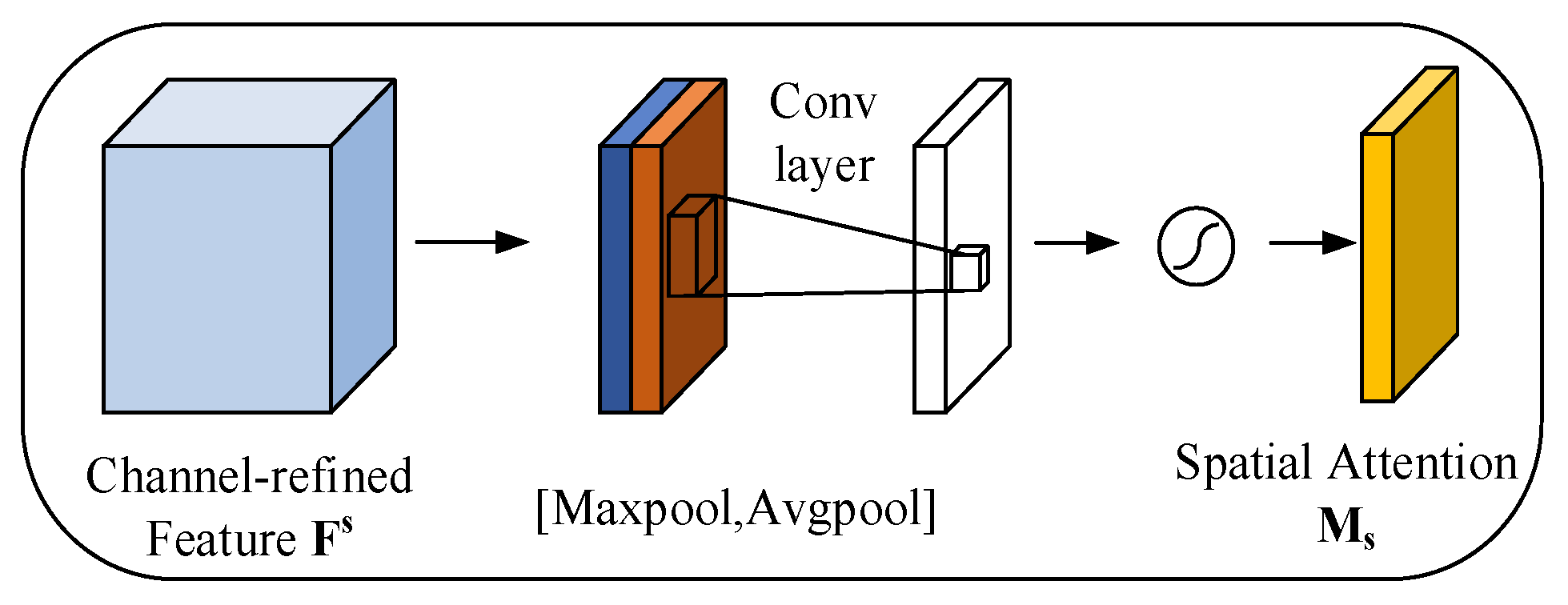
3.2.2. Spatial Attention
3.3. Residual Block Convolution Feature Enhancement Module (RBC-F)
3.4. Streamlined 2D-CNN Model (SL-CNN)
4. Results and Discussion
4.1. Waveband Processing
4.2. Parameter Optimization
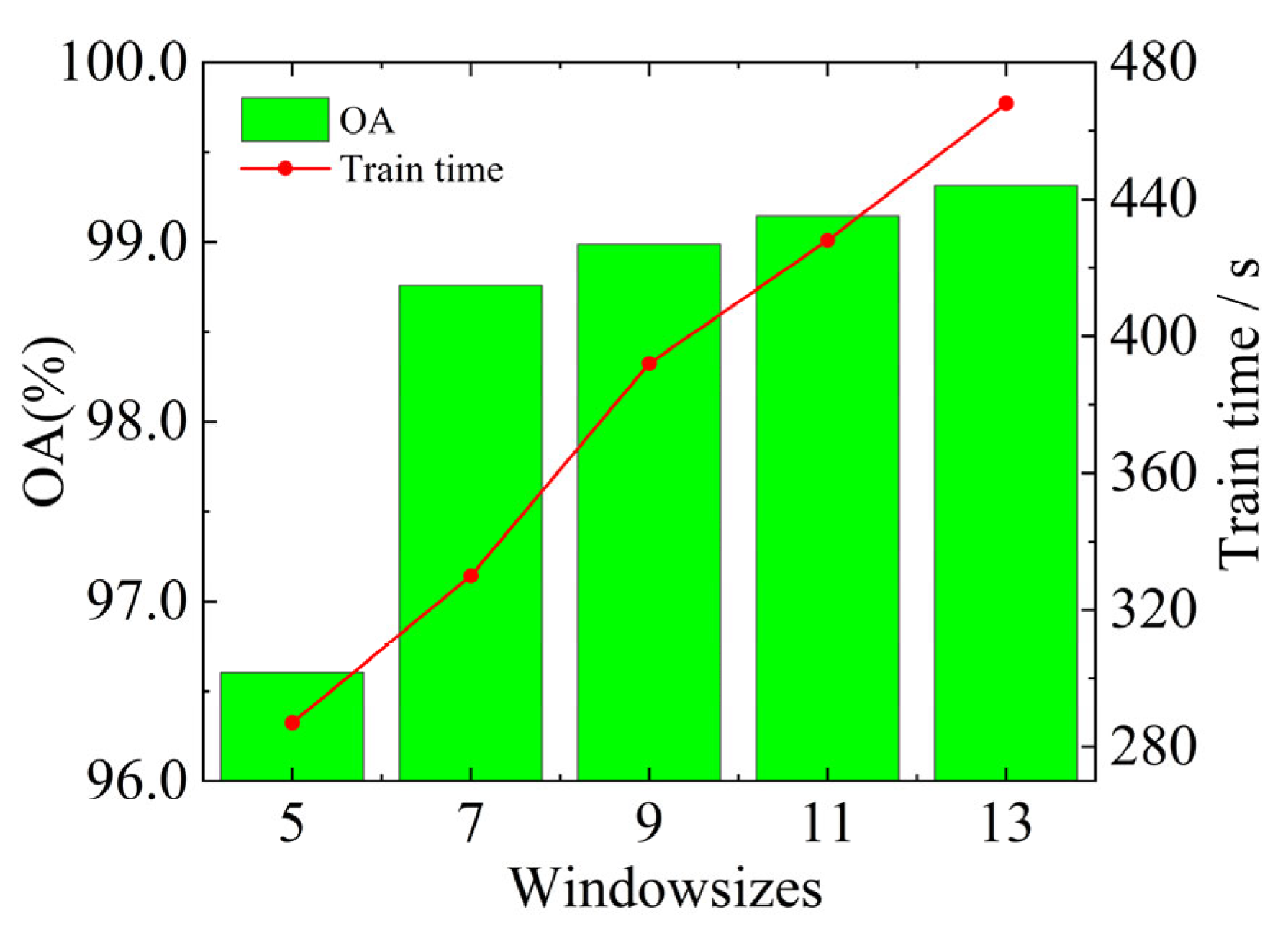
4.2.1. Window Size Selection
4.2.2. Learning Rate Selection
4.2.3. Batch Size Optimization
4.2.4. Optimization of the Number of Base Blocks
4.3. Comparison of Ablation Experiments
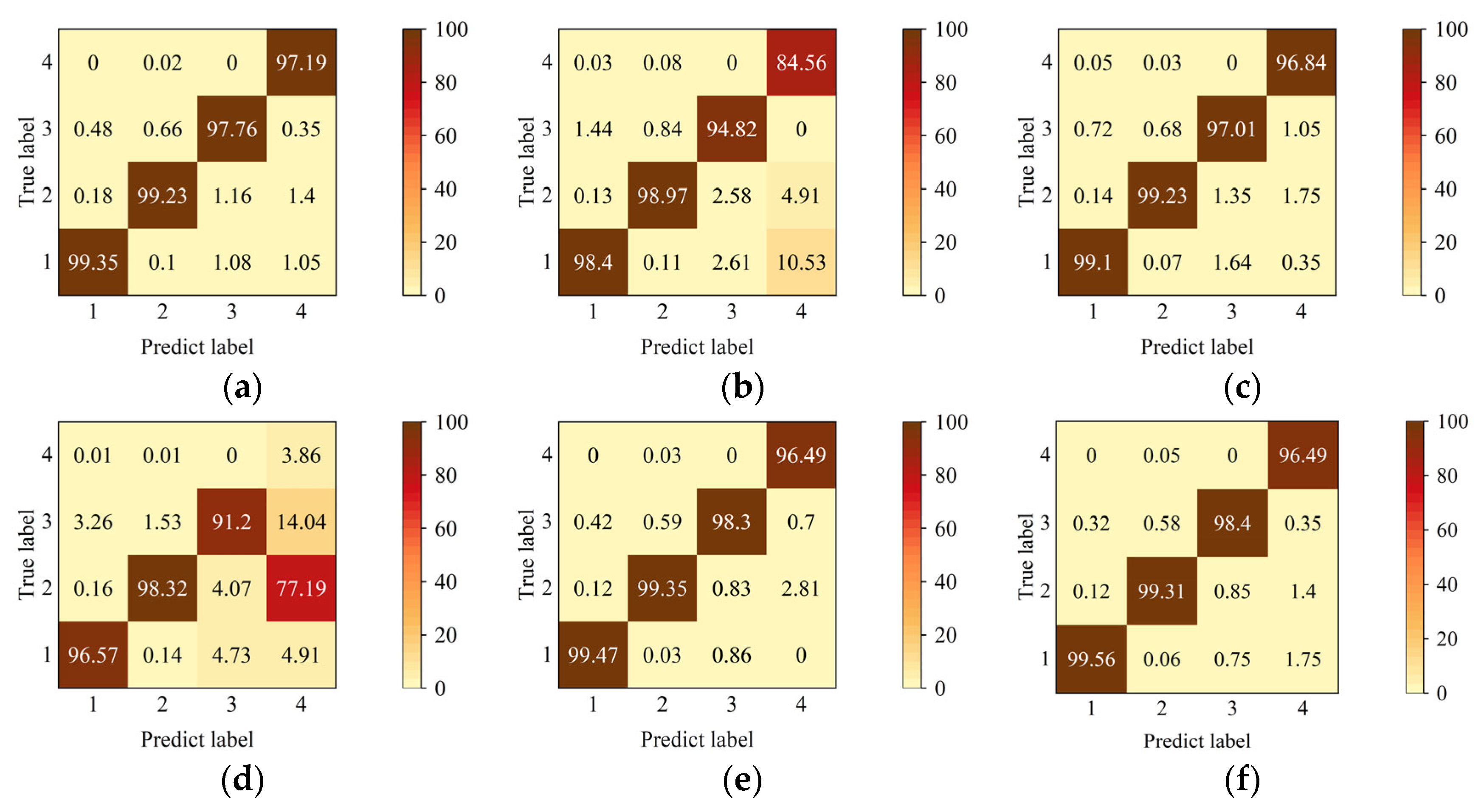
4.4. Experimental Results
4.5. Discussion
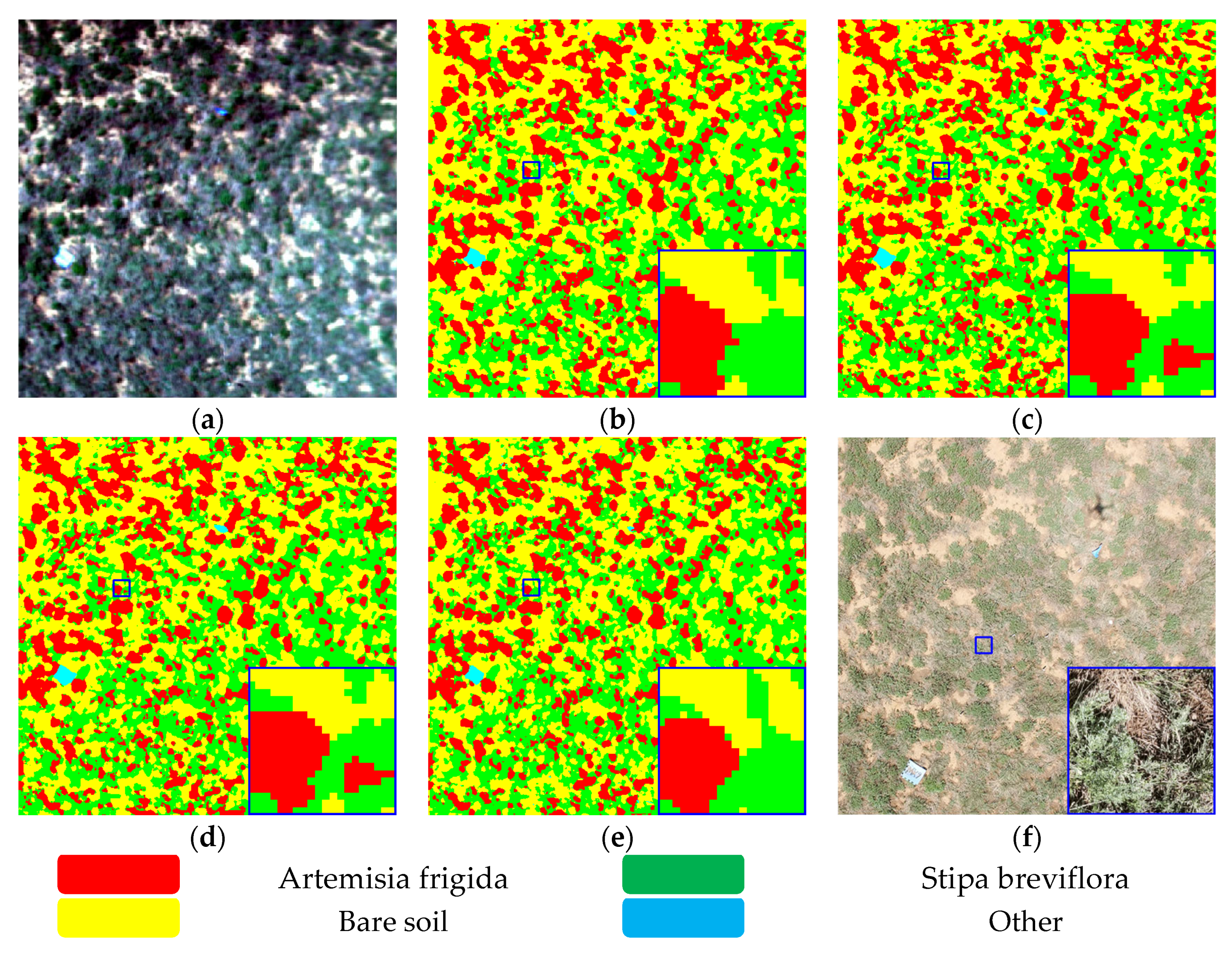
4.6. Data Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, J.; Ma, B.; Lu, X. Grazing enhances soil nutrient effects: Trade-offs between aboveground and belowground biomass in alpine grasslands of the Tibetan Plateau. Land Degrad. Dev. 2018, 29, 337–348. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Yang, X.; Sun, Y.; Song, N. Sustainable application of GF-6 WFV satellite data in desert steppe: A village-scale grazing study in China. Front. Environ. Sci. 2023, 11, 57. [Google Scholar] [CrossRef]
- Tsafack, N.; Fattorini, S.; Benavides Frias, C.; Xie, Y.; Wang, X.; Rebaudo, F. Competing vegetation structure indices for estimating spatial constrains in carabid abundance patterns in chinese grasslands reveal complex scale and habitat patterns. Insects 2020, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Xu, X.; Gao, C.; Yu, Z.; Wang, X.; Wang, C. Effects of grazing on total nitrogen and stable nitrogen isotopes of plants and soil in different types of grasslands in Inner Mongolia. Acta Prataculturae Sin. 2021, 30, 208–214. [Google Scholar]
- Wang, M.; Zhang, C. Climate Change in Inner Mongolia Grassland and the Effects on Pastural Animal Husbandry. Grassl. Prataculture 2013, 25, 5–12. [Google Scholar]
- Men, X.; Lv, S.; Hou, D.; Wang, Z.; Li, Z.; Han, G.; Sun, H.; Wang, B.; Wang, Z. Effects of grazing intensity on the density and spatial distribution of Cleistogenes songorica in desert steppe. Acta Agrestia Sin. 2022, 30, 3106–3112. [Google Scholar]
- Dong, S.; Shang, Z.; Gao, J.; Boone, R.B. Enhancing sustainability of grassland ecosystems through ecological restoration and grazing management in an era of climate change on Qinghai-Tibetan Plateau. Agric. Ecosyst. Environ. 2020, 287, 106684. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.-Y.; Li, L.; Wei, J.-Q.; Shi, F.-Z.; Yao, H.-Y.; Liu, L. Plant Harvesting Impacts on Soil Water Patterns and Phenology for Shrub-encroached Grassland. Water 2018, 10, 736. [Google Scholar] [CrossRef]
- Guo, Q.; Hu, T.; Ma, Q.; Xu, K.; Yang, Q.; Sun, Q.; Li, Y.; Su, Y. Advances for the new remote sensing technology in ecosystem ecology research. Chin. J. Plant Ecol. 2020, 44, 418–435. [Google Scholar] [CrossRef]
- Li, G.; Chen, C.; Li, J.; Peng, J. Advances in applying low-altitude unmanned aerial vehicle remote sensing in grassland ecological monitoring. Acta Ecol. Sin. 2023, 43, 6889–6901. [Google Scholar]
- Li, C.; Han, W.; Peng, M. Improving the spatial and temporal estimating of daytime variation in maize net primary production using unmanned aerial vehicle-based remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102467. [Google Scholar] [CrossRef]
- Lyu, X.; Li, X.; Dang, D.; Dou, H.; Wang, K.; Lou, A. Unmanned Aerial Vehicle (UAV) Remote Sensing in Grassland Ecosystem Monitoring: A Systematic Review. Remote Sens. 2022, 14, 1096. [Google Scholar] [CrossRef]
- Daud, S.M.S.M.; Yusof, M.Y.P.M.; Heo, C.C.; Khoo, L.S.; Singh, M.K.C.; Mahmood, M.S.; Nawawi, H. Applications of drone in disaster management: A scoping review. Sci. Justice 2022, 62, 30–42. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent advances of hyperspectral imaging technology and applications in agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Rueda-Ayala, V.P.; Peña, J.M.; Höglind, M.; Bengochea-Guevara, J.M.; Andújar, D. Comparing UAV-based technologies and RGB-D reconstruction methods for plant height and biomass monitoring on grass ley. Sensors 2019, 19, 535. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Zhao, H.; Zhang, S.; He, J.; Yang, X.; Xiao, X. Identification of plant species in an alpine steppe of Northern Tibet using close-range hyperspectral imagery. Ecol. Inform. 2021, 61, 101213. [Google Scholar] [CrossRef]
- Wan, L.; Zhu, J.; Du, X.; Zhang, J.; Han, X.; Zhou, W.; Li, X.; Liu, J.; Liang, F.; He, Y. A model for phenotyping crop fractional vegetation cover using imagery from unmanned aerial vehicles. J. Exp. Bot. 2021, 72, 4691–4707. [Google Scholar] [CrossRef]
- Sa, I.; Popović, M.; Khanna, R.; Chen, Z.; Lottes, P.; Liebisch, F.; Nieto, J.; Stachniss, C.; Walter, A.; Siegwart, R. WeedMap: A large-scale semantic weed mapping framework using aerial multispectral imaging and deep neural network for precision farming. Remote Sens. 2018, 10, 1423. [Google Scholar] [CrossRef]
- Zhang, Y.; Ta, N.; Guo, S.; Chen, Q.; Zhao, L.; Li, F.; Chang, Q. Combining spectral and textural information from UAV RGB images for leaf area index monitoring in Kiwifruit Orchard. Remote Sens. 2022, 14, 1063. [Google Scholar] [CrossRef]
- Yu, R.; Ren, L.; Luo, Y. Early detection of pine wilt disease in Pinus tabuliformis in North China using a field portable spectrometer and UAV-based hyperspectral imagery. For. Ecosyst. 2021, 8, 44. [Google Scholar] [CrossRef]
- Hu, G.; Yao, P.; Wan, M.; Bao, W.; Zeng, W. Detection and classification of diseased pine trees with different levels of severity from UAV remote sensing images. Ecol. Inform. 2022, 72, 101844. [Google Scholar] [CrossRef]
- Imran, H.A.; Gianelle, D.; Rocchini, D.; Dalponte, M.; Martín, M.P.; Sakowska, K.; Wohlfahrt, G.; Vescovo, L. VIS-NIR, Red-Edge and NIR-Shoulder Based Normalized Vegetation Indices Response to Co-Varying Leaf and Canopy Structural Traits in Heterogeneous Grasslands. Remote Sens. 2020, 12, 2254. [Google Scholar] [CrossRef]
- Sun, G.; Jiao, Z.; Zhang, A.; Li, F.; Fu, H.; Li, Z. Hyperspectral image-based vegetation index (HSVI): A new vegetation index for urban ecological research. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102529. [Google Scholar] [CrossRef]
- Baloloy, A.B.; Blanco, A.C.; Ana, R.R.C.S.; Nadaoka, K. Development and application of a new mangrove vegetation index (MVI) for rapid and accurate mangrove mapping. ISPRS J. Photogramm. Remote Sens. 2020, 166, 95–117. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Campos-Taberner, M.; Moreno-Martínez, Á.; Walther, S.; Duveiller, G.; Cescatti, A.; Mahecha, M.D.; Muñoz-Marí, J.; García-Haro, F.J.; Guanter, L.; et al. A unified vegetation index for quantifying the terrestrial biosphere. Sci. Adv. 2021, 7, eabc7447. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yang, W.; Feng, M.; Sun, H.; Wang, C. Development of winter wheat yield estimation models based on hyperspectral vegetation indices. Chin. J. Ecol. 2022, 41, 1433–1440. [Google Scholar]
- Jae-Hyun, R.; Jeong, H.; Cho, J. Performances of Vegetation Indices on Paddy Rice at Elevated Air Temperature, Heat Stress, and Herbicide Damage. Remote Sens. 2020, 12, 2654. [Google Scholar]
- Wang, X.; Dong, J.; Baoyin, T.; Bao, Y. Estimation and Climate Factor Contribution of Aboveground Biomass in Inner Mongolia’s Typical/Desert Steppes. Sustainability 2019, 11, 6559. [Google Scholar] [CrossRef]
- He, W.; Yu, L.; Yao, Y. Estimation of plant leaf chlorophyll content based on spectral index in karst areas. Guihaia 2022, 42, 914–926. [Google Scholar]
- Pamungkas, S. Analysis Of Vegetation Index For Ndvi, Evi-2, And Savi For Mangrove Forest Density Using Google Earth Engine In Lembar Bay, Lombok Island. IOP Conference Series. Earth Environ. Sci. 2023, 1127, 012034. [Google Scholar] [CrossRef]
- Umut, H.; NIJAT, K.; Chen, C.; Mamat, S. Estimation of Winter Wheat LAI Based on Multi-dimensional Hyperspectral Vegetation Indices. Trans. Chin. Soc. Agric. Mach. 2022, 53, 181–190. [Google Scholar]
- Zhu, X.; Bi, Y.; Liu, H.; Pi, W.; Zhang, X.; Shao, Y. Study on the Identification Method of Rat Holes in Desert Grasslands Based on Hyperspectral Images. Chin. J. Soil Sci. 2020, 51, 263–268. [Google Scholar]
- Yang, H.; Du, J.; Ruan, P.; Zhu, X.; LIiu, H.; Wang, Y. Vegetation Classification of Desert Steppe Based on Unmanned Aerial Vehicle Remote Sensing and Random Forest. Trans. Chin. Soc. Agric. Mach. 2021, 52, 186–194. [Google Scholar]
- Zhang, S.; Gong, Y.; Wang, J. The Development of Deep Convolution Neural Network and Its Applications on Computer Vision. Chin. J. Comput. 2019, 42, 453–482. [Google Scholar]
- Zhang, T.; Du, J.; Zhu, X.; Gao, X. Research on Grassland Rodent Infestation Monitoring Methods Based on Dense Residual Networks and Unmanned Aerial Vehicle Remote Sensing. J. Appl. Spectrosc. 2023, 89, 1220–1231. [Google Scholar] [CrossRef]
- Pi, W.; Du, J.; Liu, H.; Zhu, X. Desertification glassland classification and three-dimensional convolution neural network model for identifying desert grassland landforms with unmanned aerial vehicle hyperspectral remote sensing images. J. Appl. Spectrosc. 2020, 87, 309–318. [Google Scholar] [CrossRef]
- Wei, D.; Liu, K.; Xiao, C.; Sun, W.; Liu, W.; Liu, L.; Huang, X.; Feng, C. A Systematic Classification Method for Grassland Community Division Using China’s ZY1-02D Hyperspectral Observations. Remote Sens. 2022, 14, 3751. [Google Scholar] [CrossRef]
- Zhu, X.; Bi, Y.; Du, J.; Gao, X.; Zhang, T.; Pi, W.; Zhang, Y.; Wang, Y.; Zhang, H. Research on deep learning method recognition and a classification model of grassland grass species based on unmanned aerial vehicle hyperspectral remote sensing. Grassl. Sci. 2023, 69, 3–11. [Google Scholar] [CrossRef]
- Song, G.; Wang, Q. Species classification from hyperspectral leaf information using machine learning approaches. Ecol. Inform. 2023, 76, 102141. [Google Scholar] [CrossRef]
- Zhang, T.; Bi, Y.; Du, J.; Zhu, X.; Gao, X. Classification of desert grassland species based on a local-global feature enhancement network and UAV hyperspectral remote sensing. Ecol. Inform. 2022, 72, 101852. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Comput. Res. Repos. 2015, abs/1502.03167. [Google Scholar]
- Fang, B.; Li, Y.; Zhang, H.; Chan, J.C.-W. Hyperspectral images classification based on dense convolutional networks with spectral-wise attention mechanism. Remote Sens. 2019, 11, 159. [Google Scholar] [CrossRef]
- Han, G.; He, M.; Gao, M.; Yu, J.; Liu, K.; Liang, Q. Insulator Breakage Detection Based on Improved YOLOv5. Sustainability 2022, 14, 6066. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Q.; Du, B.; Huang, X.; Tang, Y.Y.; Tao, D. Simultaneous spectral-spatial feature selection and extraction for hyperspectral images. IEEE Trans. Cybern. 2016, 48, 16–28. [Google Scholar] [CrossRef]
- Yuan, Y.; Zheng, X.; Lu, X. Discovering diverse subset for unsupervised hyperspectral band selection. IEEE Trans. Image Process. 2016, 26, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Song, W.; Wang, G.; Zeng, G.; Tian, F. Image recovery and recognition: A combining method of matrix norm regularisation. IET Image Process. 2019, 13, 1246–1253. [Google Scholar] [CrossRef]
- Fisher, R.A. XV.—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Trans. R. Soc. Edinb. 2012, 52, 399–433. [Google Scholar] [CrossRef]
- Pi, W.; Du, J.; Bi, Y.; Gao, X.; Zhu, X. 3D-CNN based UAV hyperspectral imagery for grassland degradation indicator ground object classification research. Ecol. Inform. 2021, 62, 101278. [Google Scholar] [CrossRef]


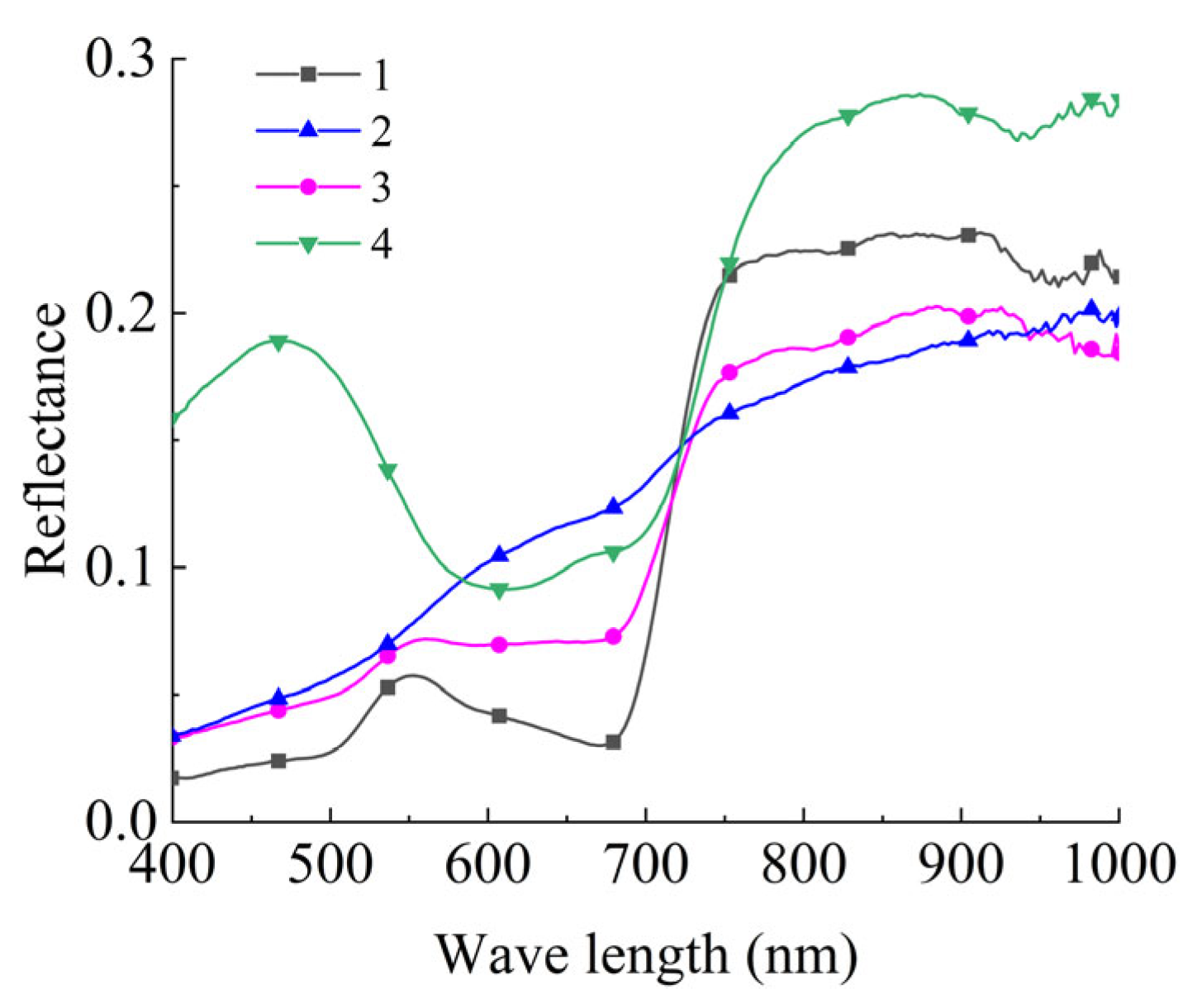




| Category | Number (N) | |||
|---|---|---|---|---|
| NO. | Name | Training | Validation | Testing |
| 1 | Artemisia frigida | 9075 | 3870 | 13,003 |
| 2 | Bare soil | 13,785 | 5901 | 19,683 |
| 3 | Stipa breviflora | 4782 | 2079 | 6830 |
| 4 | Other | 218 | 91 | 285 |
| 5 | Total | 27,860 | 11,941 | 39,801 |
| Selection of Different Wave Numbers | ||||||
|---|---|---|---|---|---|---|
| Class | 1 | 2 | 3 | 4 | PCA | Full-band |
| Bands | 126–136 | 116–146 | 106–156 | 96–166 | 11 | 256 |
| Test loss (%) | 0.022 | 0.023 | 0.024 | 0.020 | 0.026 | 0.051 |
| Kappa × 100 | 98.735 | 98.637 | 98.617 | 98.788 | 98.524 | 96.858 |
| OA (%) | 99.216 | 99.156 | 99.143 | 99.249 | 99.085 | 98.053 |
| Train time (s) | 367 | 382 | 383 | 471 | 427 | 585 |
| Base Block | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| OA (%) | 98.188 | 98.912 | 99.216 | 99.342 |
| Train time (s) | 215 | 277 | 367 | 791 |
| Memory (MB) | 1.98 | 5.22 | 16.30 | 59.10 |
| Model params (MB) | 0.28 | 1.18 | 4.73 | 18.82 |
| RBC-F | CBAM-F | Kappa × 100 | OA (%) | AA (%) | Test loss (%) | F1 |
|---|---|---|---|---|---|---|
| ✓ | 98.386 | 99.000 | 98.382 | 0.028 | 0.983 | |
| ✓ | 98.154 | 98.857 | 98.387 | 0.031 | 0.980 | |
| ✓ | ✓ | 98.735 | 99.216 | 98.442 | 0.022 | 0.985 |
| Class | ResNet34 | GoogLeNet | DenseNet121 | MLP | 2D-CNN | SL-CNN |
|---|---|---|---|---|---|---|
| Kappa × 100 | 98.073 | 96.716 | 98.032 | 93.271 | 98.690 | 98.735 |
| OA (%) | 98.807 | 97.967 | 98.791 | 95.849 | 99.188 | 99.216 |
| AA (%) | 96.809 | 94.187 | 98.047 | 72.487 | 98.403 | 98.442 |
| Train time (s) | 2015 | 600 | 1491 | 25 | 557 | 367 |
| Memory (MB) | 247.0 | 93.4 | 92.2 | 21.7 | 55.8 | 16.3 |
| Model params(MB) | 81.38 | 28.76 | 27.02 | 7.21 | 17.91 | 4.73 |
| GDIF-3D-CNN | DIS-O | LGFEN | SL-CNN | |
|---|---|---|---|---|
| Kappa × 100 | 95.933 | 95.033 | 98.293 | 98.735 |
| OA (%) | 97.480 | 96.922 | 98.942 | 99.216 |
| AA (%) | 96.972 | 93.304 | 98.158 | 98.442 |
| Train time (s) | 49 | 63 | 233 | 367 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Bi, Y.; Du, J.; Zhang, T.; Gao, X.; Jin, E. The Unmanned Aerial Vehicle (UAV)-Based Hyperspectral Classification of Desert Grassland Plants in Inner Mongolia, China. Appl. Sci. 2023, 13, 12245. https://doi.org/10.3390/app132212245
Wang S, Bi Y, Du J, Zhang T, Gao X, Jin E. The Unmanned Aerial Vehicle (UAV)-Based Hyperspectral Classification of Desert Grassland Plants in Inner Mongolia, China. Applied Sciences. 2023; 13(22):12245. https://doi.org/10.3390/app132212245
Chicago/Turabian StyleWang, Shengli, Yuge Bi, Jianmin Du, Tao Zhang, Xinchao Gao, and Erdmt Jin. 2023. "The Unmanned Aerial Vehicle (UAV)-Based Hyperspectral Classification of Desert Grassland Plants in Inner Mongolia, China" Applied Sciences 13, no. 22: 12245. https://doi.org/10.3390/app132212245
APA StyleWang, S., Bi, Y., Du, J., Zhang, T., Gao, X., & Jin, E. (2023). The Unmanned Aerial Vehicle (UAV)-Based Hyperspectral Classification of Desert Grassland Plants in Inner Mongolia, China. Applied Sciences, 13(22), 12245. https://doi.org/10.3390/app132212245






