Abstract
The glm R package is commonly used for generalized linear modeling. In this paper, we evaluate the ability of the glm package to predict binomial outcomes using logistic regression. We use single-cell RNA-sequencing datasets, after a series of normalization, to fit data into glm models repeatedly using 10-fold cross-validation over 100 iterations. Our evaluation criteria are glm’s Precision, Recall, F1-Score, Area Under the Curve (AUC), and Runtime. Scores for each evaluation category are collected, and their medians are calculated. Our findings show that glm has fluctuating Precision and F1-Scores. In terms of Recall, glm has shown more stable performance, while in the AUC category, glm shows remarkable performance. Also, the Runtime of glm is consistent. Our findings also show that there are no correlations between the size of fitted data and glm’s Precision, Recall, F1-Score, and AUC, except for Runtime.
1. Introduction
Supervised learning in the R programming language has been facilitated with built-in packages dedicated to processing data and building different types of models. Three types of logistic regression that are commonly used to build models in R include binary (binomial), multinomial, and ordinal regression. Binomial regression produces its prediction in a form of two types of outcomes, such as true or false, while multinomial regression has three or more types of outcome. Ordinal regression, on the other hand, deals with three or more ordered values [1]. GLM (generalized linear model) is a statistical model that allows the linear regression model to be extended to accommodate a wide range of responses and error distributions. The main idea of GLM is to use a link function as a way to relate the linear model to the response variable. GLM has many “families” (link functions) that can perform different types of regression, such as linear regression, logistic regression, and Poisson regression [2,3]. The evaluation of the glm R package remains inconclusive in terms of its performance, specifically, its Precision, Recall, F1-Score, Area Under the Curve (AUC), and Runtime [4,5,6,7]. Current studies evaluate generalized linear models in general but not in the R package itself [8,9]. Also, evaluating the performance of the glm R package using single-cell RNA-sequencing data helps to understand its applicability, strengths, and limitations in this very specific domain. In this paper, we choose to evaluate the glm package performance, since it is the most common R package for creating linear models, and we use “binomial” (nonlinear) regression because our data can only be classified into two types of outcomes: zeroes and ones.
Precision [10,11] is defined as the number of correctly predicted true values (TP) over the total number of falsely predicted true values (FP) and the number of correctly predicted true values. That is: .
Recall [11,12] is defined as the number of correctly predicted true values (TP) over the total number of values that are classified as false (FN) and the total number of correctly predicted true values. That is: .
The F1-Score [13,14,15], consequently, is derived as F1-Score = .
AUC [12,16] measures the ability of a specific model to distinguish between positive signals and negative signals. Models with higher AUC have better prediction. AUC can be calculated directly from the subject model’s ROC (receiver operating characteristic) curve, which includes the scores of both the true positive rate (represented, in the ROC plot, as the y-axis) and the false positive rate (represented as the x-axis) at different variations of thresholds.
When testing a dataset, similarities in input values often cause misclassification errors. Thus, we choose to run a cross-validation test where the datasets are randomly split into different folds (usually 10 folds). In each testing iteration, one fold is set as a testing set, and the rest of the remaining folds are set as a training set. The adjacent fold, then, in the next iteration, is set as the testing set, and the remaining folds are set as the training, set and so on.
A correlation between two variables means that a change in one variable will have an immediate effect on the other. The Pearson correlation coefficient, which is measured by dividing the covariance of two variables by the multiplication of the standard deviations of the first and second variables, is commonly used for this specific purpose. A linear correlation, either positive or negative, indicates a strong correlation.
Single-cell RNA-sequencing (scRNA-seq) is a technology used to express gene profiles at the level of the individual cell. It provides a better view of cellular heterogeneity within a complex biological system by measuring the RNA transcripts of individual cells. This is normally performed on a sample level; for example, a blood sample. In single-cell RNA-sequencing, it is performed on the individual cell level [17,18,19,20,21,22,23]. The level of gene expression provides information that can be used to study the causes of tumors and other diseases, such as fabric lung [24,25,26,27,28,29,30,31]. To deal with excessive data obtained from sequencing, supervised learning techniques are applied on cell annotations to give meaning to these data [32,33,34,35,36,37,38,39,40]. In our experiment, we use 22 scRNA-seq datasets from published studies [41] to evaluate the performance of the glm R package. These datasets are made available via the Conquer repository [42].
This paper is organized as follows. We will first describe the datasets used, then explain our procedures for evaluation and the analysis results of our measurement outcomes. Finally, we summarize our findings and derive our conclusion.
2. Materials and Design of Experiment
The evaluation of glm’s performance is not a straightforward process. Datasets must go through a normalization process to fit into the glm package. Memory overload, in some cases, can limit testing of additional datasets. Nevertheless, we have managed to utilize 22 datasets out of 40 from the targeted repository.
2.1. Datasets
The datasets used in this paper were extracted from the Conquer (consistent quantification of external RNA-sequencing data) repository that was developed by C. Soneson and M. Robinson at the University of Zurich, Switzerland [42]. Three organisms’ data were included in the repository—Homo sapiens, Mus musculus, and Danio rerio. Each dataset includes a different number of one organism’s cells. Protocols used to extract data from these cells were sequences—SMARTer C1, Smart-Seq2, SMART-Seq, Tang, Fluidigm C1Auto prep, and SMARTer. The data were split into two categories: gene-level and transcript-level. At the gene-level, there are four different types of measurements: TPM (transcripts per million abundance estimates for each gene), count (gene read counts), count_lstpm (length-scaled TPMs), and avetxlength (the average length of the transcripts expressed in each sample for each gene). In the transcript-level category, there are three different types of measurements: TPM (transcripts per million abundance estimates for each transcript), count (transcript read counts), and efflength (effective transcript lengths).
We have explored all datasets available in the Conquer repository. Unfortunately, not all datasets could be used, or even modified, to fit into the glm R package, for various reasons. For a dataset to fit our testing methodology, we have to verify that its samples can be separated into only two groups of common phenotypes; thus, each phenotype can be assigned either 1 or 0 in the same dataset. For example, the dataset GSE80032 was found to be unsuitable for our test because its samples include only one phenotype; using binomial regression, there would only be one class for all samples to fit in. The size of the dataset is also a significant factor in our test. A group of samples must not be so small that they generate misclassification or execution errors. Following these restrictions and based on our observation during dataset preparation, the ideal sample groups must have at least 30 samples to guarantee no misclassification or errors will be generated. In the end, 22 datasets fit perfectly into our test. Table 1 represents our selected datasets as they were represented in the Conquer repository with additional information.

Table 1.
List of selected datasets with their IDs, organisms from which cells were taken, a brief description, cell counts, and sequencing protocols (source: http://imlspenticton.uzh.ch:3838/conquer/ (accessed on 7 May 2023)), # means number of cells.
Accessing information in these datasets is not a straightforward process. We needed to have full control over these datasets in order to manipulate them. Thus, before we proceeded into our test, each dataset had to be normalized before it could fit into our method. In exploring the dataset categories, we found that some measurements (except for avetxlength) contained a large number of 0 values, which can jeopardize the integrity of the evaluation process. Thus, we chose avetxlength as our input data.
2.2. Evaluation Procedure
To access avetxlength’s data in each dataset, we closely followed the steps provided by the Conquer repository authors (using experiment()[“gene”] and assay()[avetxlength]). We downloaded the .rds file for each dataset and, by using the R programming language, we retrieved ExperimentList instances that contained RangedSummarizedExperiment for a gene-level object, which allowed us to access all available abundances; TPM (transcripts per million), gene count, length-scaled TPMs, and avetxlength. In our test, we chose genes’ avetxlength because it perfectly fits our model. The avetxlength data at this stage constitute a matrix, , where i represents samples and j represents genes.
To be able to use our binomial model, we individually explored each dataset to find two distinct phenotypic characteristics associated with the dataset’s sample groups that could be used to label our outputs. We denote the first characteristic as 1 and the other as 0. Thus, sample IDs are replaced by either 1 or 0. For example, in the EMTAB2805 dataset, different phenotypes are associated with each group of samples. We chose only two phenotypes based on stages of the cell cycle: G1 and G2M. As a result, IDs of the samples associated with the first stage, G1, were replaced by 1 (True), and IDs of the samples associated with the second stage, G2M, were replaced by 0 (False). We eliminated any extra samples associated with other cell cycle stages, if any. In some datasets, such as in GSE80032, there is only one phenotype; thus, classification is impossible. Table 2 shows the current status of the matrix.

Table 2.
matrix containing avetxlength data from the EMTAB2805 dataset.
We now rotate the dimensions of so that rows represent samples, and columns represent genes (Table 3). We then substitute each sample’s ID with either 1 or 0, depending on their original classification in their original dataset status (Table 4).

Table 3.
after rotation.

Table 4.
after substituting each sample’s ID with either 1 or 0.
In Table 5, we list all selected datasets after we identified the appropriate phenotypes (column 3) that were used in our test.

Table 5.
List of datasets used in our evaluation along with their chosen phenotypes that were eventually set at either 1 or 0, # refers to the number of 1s and 0s in the matrix.
To increase the integrity of our evaluation, we conducted a Wilcoxon test on our matrix to find the p-values associated with each gene.
At the time of testing, we ran into memory exhaustion errors caused by the large amount of genes that the matrix retained. Thus, we had to trim our matrix to include only 1000 genes that had the lowest p-values.
The final form of our input matrix is , where j ranges from 1 to n (the total number of phenotypes represented by either 1s or 0s) and i ranges from 1 to 1000 (genes with the lowest p-value).
At this point, our matrix has been completely normalized; thus, we can proceed into testing the glm R package over 100 iterations.
At the start of each testing iteration, we shuffle the rows (samples that were denoted by 1 and 0) of our matrix to prevent any bias that may occur from having identical sample IDs. Next, we conduct a 10-fold cross-validation test where we take 10% of the samples (rows) as our testing set and the remaining 90% as our training set. We measure the system’s time (which is our execution start time), and then we fit the training set into the glm package using its “binomial” function. Immediately after this step, we again measure the system’s time (which is our execution end time). By subtracting the end time from the start time, we obtain the glm runtime at that specific iteration.
The constructed model obtained from fitting glm is now used for prediction by employing the testing set. From this process, we can derive the confusion matrix that is the result of predicting the recently created model against the testing set. The confusion matrix is a matrix. In some cases where we have only a one-dimension matrix, we programmatically force the creation of a matrix to enable finding the actual and predicted values.
We now calculate the Precision by dividing the total number of true positive values by the total number of true positive values and the total number of false positive values. We also calculate the Recall from the same confusion matrix by dividing the total number of true positive values by the total number of true positive values and the total number of false negative values. At this point, we have both Precision and Recall; thus, we can calculate the F1-Score, which is Precision multiplied by Recall over the total value of Precision and Recall multiplied by 2.
Calculating the AUC requires additional steps. We must first compute the receiver operating characteristic (ROC) using the pROC R package and then, by using our prediction, we can extract the AUC measurement.
In the second iteration of our cross-validation testing, we take the fold adjacent to the first fold (that is the next 10% of the matrix) as our testing set and the remaining 90% as our training set. We repeat the same procedure applied to glm and obtain measurements of Runtime, Precision, Recall, F1-Score, and AUC. At the end of this 10-fold cross-validation testing, we have 10 measurements for each evaluation category. We collect these 10 measurements and calculate their means as our final score.
The previous testing procedure (starting from shuffling samples to collecting mean measurements of Runtime, Precision, Recall, F1-Score, and AUC) is repeated 100 times. Each time we collect the mean of all scores and plot our final results for visual analysis.
2.3. Code Repository
The evaluation codes were implemented in the R programming language. Scripts were deposited at https://github.com/Omar-Alaqeeli/glm (accessed on 7 May 2023). There are 22 R scripts. Each script retrieves and processes one dataset and fits it into the glm() package. Only the first script, titled EMTAB2805, contains the required package installation code lines. Datasets in their original status cannot be deposited into the same GitHub repository due to their excessive size, but they can be accessed at the Conquer repository link: http://imlspenticton.uzh.ch:3838/conquer/ (accessed on 7 May 2023). All codes were run on a personal computer using version 3.6.1. of R Studio. Table 6 shows the full specifications of the system used and R Studio.

Table 6.
System and R Studio specification details at the time of running the evaluation scripts.
3. Results and Analysis
In this section, we present the results obtained from running our Algorithm 1 on the chosen datasets using the glm package. Our analysis is based on the median and interquartile range of each dataset used. Outliers are excluded. For visualization, we chose boxplots because they includes all the aforementioned measurements.
| Algorithm 1 The complete algorithm for testing glm using Conquer datasets. |
|
3.1. Precision
Results in Figure 1 show that Precision varies across different datasets. The highest median value recorded was 0.94 when using GSE71585-GPL17021. Interestingly, this result also has the smallest interquartile range. On the other hand, the lowest median value recorded was 0.40 when using the GSE102299-smartseq2 dataset. The majority of Precision median values are in the range of 0.48 to 0.66, with 0.2 variation for 15 Precision values out of 22. Table 7 shows Precision’s medians, each associated with its corresponding dataset.
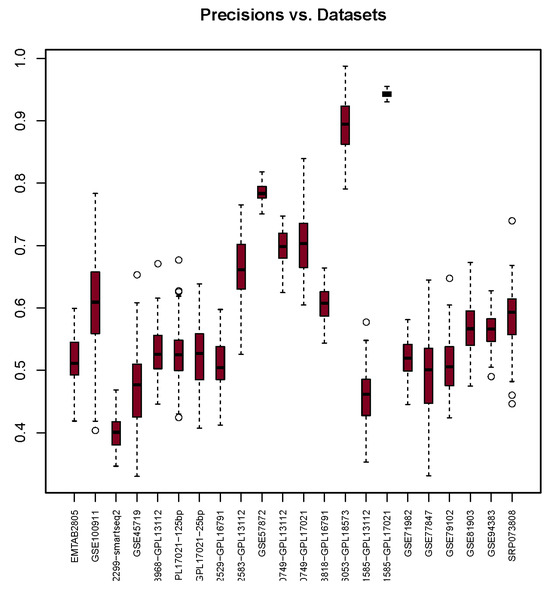
Figure 1.
Precision scores (y-axis) for each dataset (x-axis) used with the glm package. The bullets and dashed lines rrepresents extreme values distribution.

Table 7.
Median values in all evaluation categories (column numbers represent datasets).
3.2. Recall
The results of Recall depicted in Figure 2 highlight that the highest median value recorded is 0.75 when using the GSE71585-GPL17021 dataset. Notably, this result exhibits the smallest interquartile range among all datasets. On the other hand, the lowest medians, 0.50, are recorded when using GSE102299-smartseq2, GSE48968-GPL17021-125bp, and GSE77847. Nearly all median values presented in Table 7 range from 0.5 to 0.59. Only two datasets, GSE66053-GPL18573 and GSE71585-GPL17021, demonstrate medians that exceed 0.59, which may be due to the disparity between the number of 0s and 1s in these datasets.
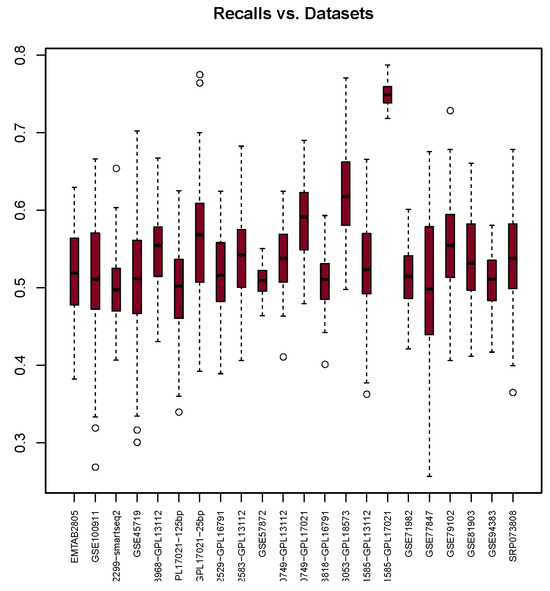
Figure 2.
Recall scores (y-axis) for each dataset (x-axis) used with the glm package. The bullets and dashed lines rrepresents extreme values distribution.
The result suggests that glm is reliable when it comes to identifying type II error rates. The lower the median, the better the model.
3.3. F1-Score
The F1-Score results depicted in Figure 3 indicate that the dataset GSE71585-GPL17021 achieved the highest median, with a value of 0.83. On the other hand, the GSE102299-smartseq2 dataset exhibits the smallest interquartile range and the lowest median value of 0.44. Among all F1-Scores in Table 7, 13 F1-Score values fall within the range of 0.50 to 0.55. The remaining F1-Scores are close to this range, with the exception of GSE66053-GPL18573, which has a median of 0.72.
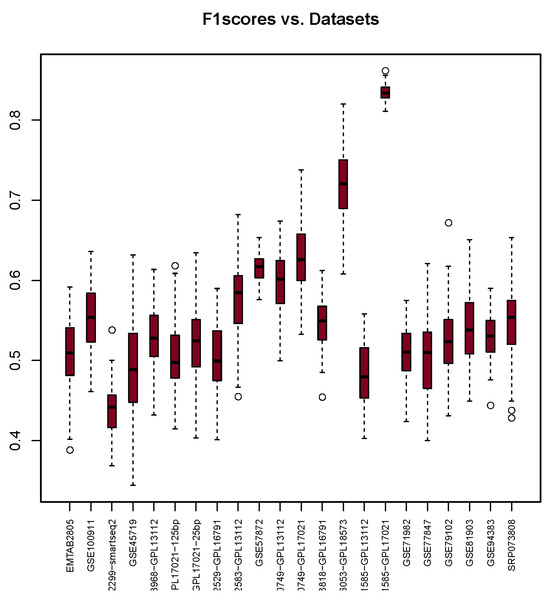
Figure 3.
F1-Scores (y-axis) for each dataset (x-axis) used with the package. The bullets and dashed lines rrepresents extreme values distribution.
3.4. Area under the Curve
The AUC median values shown in Figure 4 provide insights into glm’s prediction performance. It shows that the highest median is 0.76 in the SRP073808 dataset, and the lowest median is 0.62 in the GSE60749-GPL13112 dataset. Out of 22, 19 of the median scores fall within the range of 0.72 to 0.76, which indicates consistent performance. These AUC scores also demonstrate a similar interquartile range as well as, roughly, the same size of interquartile range. Only one AUC score deviates notably from this range: that of the GSE60749-GPL13112 dataset.
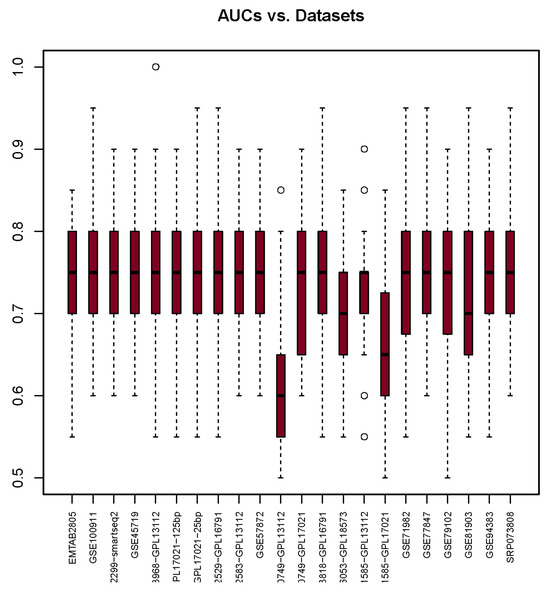
Figure 4.
AUC scores (y-axis) for each dataset (x-axis) used with the glm package. The bullets and dashed lines represents extreme values distribution.
3.5. Runtime
The Runtime analysis (in seconds) depicted in Figure 5 shows that the highest median value is 29.58 and the lowest is 0.53. The majority of the median scores (18 datasets) fall within the range of 0.53 to 5.35. However, there are four datasets (GSE57872, GSE66053-GPL18573, GSE71585-GPL17021, and GSE48968-GPL17021-125bp) that demonstrate significantly higher runtime values of 11.72, 16.13, 19.87, and 29.58, respectively. It is unclear why these datasets took a longer time, as there are no correlations found between the size of these datasets and Runtime.
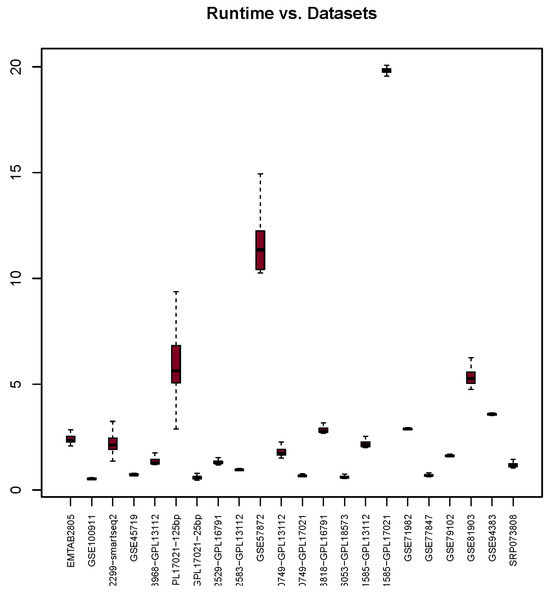
Figure 5.
Runtime scores (y-axis) for each dataset (x-axis) used with the glm package. Dashed lines represents extreme values distribution.
4. Discussion and Conclusions
Based on our analysis of the results presented in the previous section, we can deduce the following regarding each evaluation category. Regarding glm’s Precision, there is a noticeable fluctuation in the results. That can be seen with the median value of fitting the GSE71585-GPL17021 dataset. Nevertheless, it has the smallest interquartile range, which indicates that it has the most accurate testing because its scores mostly do not fluctuate. Other interquartile ranges show some similarities in size, but their medians vary. Thus, when it comes to Precision, glm shows minimal performance.
When considering Recall results, glm shows more stable performance. The majority of median scores are concentrated within a narrow range of 0.09, which highlights the excellent performance of glm. The high score of GSE71585-GPL17021’s Recall is expected, as it aligns with its high Precision score mentioned earlier. While the large size of GSE71585-GPL17021 could potentially contribute to this high score, it is important to note that GSE57872, which is larger than GSE71585-GPL17021, does not exhibit the same pattern. Therefore, the high Recall score of GSE71585-GPL17021 cannot be solely attributed to its size.
F1-Scores, on the other hand, show a fluctuating performance. This is largely connected to the glm’s Precision because F1-Scores are derived from it. This is especially obvious when looking at both the Precision and F1-Score figures. Additionally, median values of Precision and F1-Scores, in Table 7, are nearly similar. Therefore, glm also has minimal performance when it comes to F1-Score.
In the AUC category, glm shows remarkable performance. The AUC medians across the vast majority of tests are almost the same, and their interquartile ranges have the same length. This indicates that glm has a stable performance regardless of the size of the input data.
The analysis of Runtime (in seconds) results indicates that the glm package demonstrates a consistent performance. The fluctuation of runtime occurs within a narrow time range of approximately 0.25 s. However, four datasets deviate significantly from this time range. This could be due to the size of GSE57872 and GSE71585-GPL17021, which may contribute to their divergent runtimes, but this assumption is proven false when considering the size of the other two datasets, GSE71585-GPL17021 and GSE81903. Hence, further investigation is required to understand the reasons behind these outliers.
It is intuitive to suspect that the size of the fitted data may influence the performance of glm. In our datasets, although the number of columns is consistent, 1000, the number of rows varies significantly. To visually investigate the influence of the size of the data in each evaluation category, we plot the relation between size and collected median scores. Figure 6 shows that there are no correlations between the size of the fitted data and glm’s Precision, Recall, F1-Score, and AUC, except for a minimal effect on Runtime.
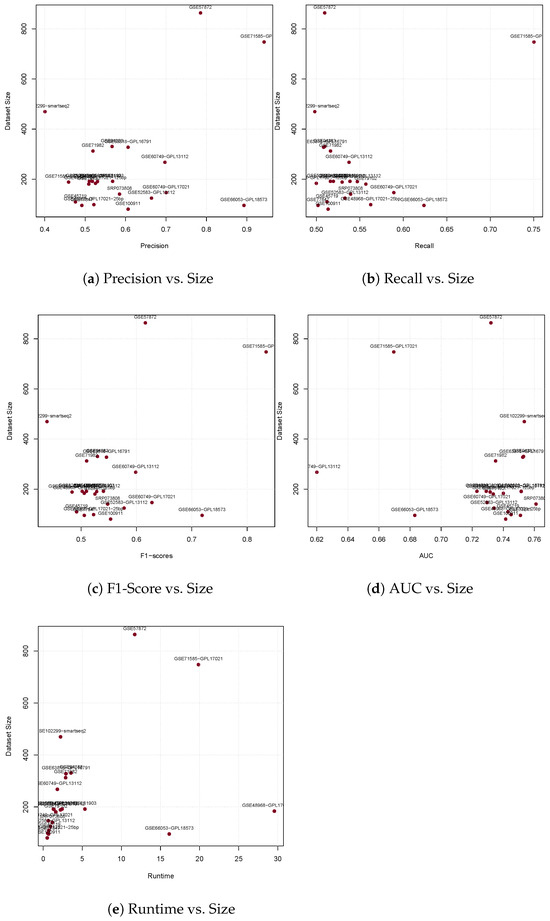
Figure 6.
Evaluation scores for all measurements versus dataset sizes.
In further research, we plan to investigate the performance of glm in various categories using different “families” (link functions) for different types of regression, including Gaussian, gamma, Poisson, and quasi functions. This investigation will involve utilizing different datasets that require regression models with more than two labels or classes. By expanding our analysis to these different regression types, we will gain a more comprehensive understanding of glm’s performance across diverse scenarios.
Author Contributions
Conceptualization, O.A. and R.A.; methodology, O.A.; software, O.A.; validation, R.A.; formal analysis, O.A.; writing—original draft preparation, O.A.; writing—review and editing, R.A.; funding acquisition, R.A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research, Imam Mohammad Ibn Saud Islamic University (IMSIU), Saudi Arabia, for funding this research work through Grant No. (221409004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Evaluation codes are available at https://github.com/Omar-Alaqeeli/glm (accessed on 11 May 2023). The Conquer repository can be found at http://imlspenticton.uzh.ch:3838/conquer/ (accessed on 11 May 2023).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AUC | Area under the Curve |
| scRNA-seq | Single-cell Ribonucleic Acid Sequencing |
| ROC | Receiver Operating Characteristic |
References
- Cucchiara, A. Applied Logistic Regression. Technometrics 1992, 34, 358–359. [Google Scholar] [CrossRef]
- Dunn, P.K.; Smyth, G.K. Generalized Linear Models with Examples in R; Springer: Berlin/Heidelberg, Germany, 2018; Volume 53. [Google Scholar]
- Rutherford, A. ANOVA and ANCOVA: A GLM Approach; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Guisan, A.; Weiss, S.B.; Weiss, A.D. GLM versus CCA spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Stefánsson, G. Analysis of groundfish survey abundance data: Combining the GLM and delta approaches. ICES J. Mar. Sci. 1996, 53, 577–588. [Google Scholar] [CrossRef]
- Pepe, M.S. An interpretation for the ROC curve and inference using GLM procedures. Biometrics 2000, 56, 352–359. [Google Scholar] [CrossRef]
- Tran, M.N.; Nguyen, N.; Nott, D.; Kohn, R. Bayesian deep net GLM and GLMM. J. Comput. Graph. Stat. 2020, 29, 97–113. [Google Scholar] [CrossRef]
- Potts, S.E.; Rose, K.A. Evaluation of GLM and GAM for estimating population indices from fishery independent surveys. Fish. Res. 2018, 208, 167–178. [Google Scholar] [CrossRef]
- Calcagno, V.; de Mazancourt, C. glmulti: An R Package for Easy Automated Model Selection with (Generalized) Linear Models. J. Stat. Softw. 2010, 34, 1–29. [Google Scholar] [CrossRef]
- Bi, J.; Kuesten, C. Type I error, testing power, and predicting precision based on the GLM and LM models for CATA data–Further discussion with M. Meyners and A. Hasted. Food Qual. Prefer. 2023, 106, 104806. [Google Scholar] [CrossRef]
- Xiong, Y. Building text hierarchical structure by using confusion matrix. In Proceedings of the 2012 5th International Conference on BioMedical Engineering and Informatics, Chongqing, China, 16–18 October 2012; pp. 1250–1254. [Google Scholar]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning, Honolulu, HI, USA, 23–29 July 2006; pp. 233–240. [Google Scholar]
- Caelen, O. A Bayesian interpretation of the confusion matrix. Ann. Math. Artif. Intell. 2017, 81, 429–450. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, J.; Zhao, X. Estimating the uncertainty of average F1 scores. In Proceedings of the 2015 International Conference on the Theory of Information Retrieval, Northampton, MA, USA, 27–30 September 2015; pp. 317–320. [Google Scholar]
- Zhang, D.; Wang, J.; Zhao, X.; Wang, X. A Bayesian hierarchical model for comparing average F1 scores. In Proceedings of the 2015 IEEE International Conference on Data Mining, Atlantic City, NJ, USA, 14–17 November 2015; pp. 589–598. [Google Scholar]
- Myerson, J.; Green, L.; Warusawitharana, M. Area under the curve as a measure of discounting. J. Exp. Anal. Behav. 2001, 76, 235–243. [Google Scholar] [CrossRef]
- Habermann, A.C.; Gutierrez, A.J.; Bui, L.T.; Yahn, S.L.; Winters, N.I.; Calvi, C.L.; Peter, L.; Chung, M.I.; Taylor, C.J.; Jetter, C.; et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1972. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Nolte, L.; Reyes, M. Segmentation of brain tumor images based on atlas-registration combined with a Markov-Random-Field lesion growth model. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 2018–2021. [Google Scholar] [CrossRef]
- Pliner, H.A.; Shendure, J.; Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 2019, 16, 983–986. [Google Scholar] [CrossRef] [PubMed]
- Seyednasrollah, F.; Laiho, A.; Elo, L.L. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief. Bioinform. 2013, 16, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; The Wadsworth & Brooks/Cole Statistics/Probability Series; Wadsworth & Brooks/Cole Advanced Books & Software: Monterey, CA, USA, 1984. [Google Scholar]
- Grubinger, T.; Zeileis, A.; Pfeiffer, K.P. evtree: Evolutionary Learning of Globally Optimal Classification and Regression Trees in R. J. Stat. Softw. Artic. 2014, 61, 1–29. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, D.; Yang, Q.; Lv, X.; Huang, W.; Zhou, Z.; Wang, Y.; Zhang, Z.; Yuan, T.; Ding, X.; et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020, 11, 6322. [Google Scholar] [CrossRef]
- Adams, T.S.; Schupp, J.C.; Poli, S.; Ayaub, E.A.; Neumark, N.; Ahangari, F.; Chu, S.G.; Raby, B.A.; DeIuliis, G.; Januszyk, M.; et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1983. [Google Scholar]
- Nawy, T. Single-cell sequencing. Nat. Methods 2014, 11, 18. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Jaakkola, M.K.; Seyednasrollah, F.; Mehmood, A.; Elo, L.L. Comparison of methods to detect differentially expressed genes between single-cell populations. Brief. Bioinform. 2016, 18, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, B.; Nelson, C.E.; Nabavi, S. Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinform. 2019, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Krzak, M.; Raykov, Y.; Boukouvalas, A.; Cutillo, L.; Angelini, C. Benchmark and Parameter Sensitivity Analysis of Single-Cell RNA Sequencing Clustering Methods. Front. Genet. 2019, 10, 1253. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Zhang, Y.; Enge, M.; Caneda, C.; Shuer, L.M.; Hayden Gephart, M.G.; Barres, B.A.; Quake, S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 2015, 112, 7285–7290. [Google Scholar] [CrossRef]
- Seyednasrollah, F.; Rantanen, K.; Jaakkola, P.; Elo, L.L. ROTS: Reproducible RNA-seq biomarker detector—Prognostic markers for clear cell renal cell cancer. Nucleic Acids Res. 2015, 44, e1. [Google Scholar] [CrossRef]
- Elo, L.L.; Filen, S.; Lahesmaa, R.; Aittokallio, T. Reproducibility-Optimized Test Statistic for Ranking Genes in Microarray Studies. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008, 5, 423–431. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015, 25, 1860–1872. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Alaqeeli, O.; Xing, L.; Zhang, X. Software Benchmark—Classification Tree Algorithms for Cell Atlases Annotation Using Single-Cell RNA-Sequencing Data. Microbiol. Res. 2021, 12, 20022. [Google Scholar] [CrossRef]
- Soneson, C.; Robinson, M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 2018, 15, 255–261. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).