Abstract
Recently, deep learning techniques have been used for low-dose CT (LDCT) reconstruction to reduce the radiation risk for patients. Despite the improvement in performance, the network models used for LDCT reconstruction are becoming increasingly complex and computationally expensive under the mantra of “deeper is better”. However, in clinical settings, lightweight models with a low computational cost and short reconstruction times are more popular. For this reason, this paper proposes a computationally efficient CNN model with a simple structure for sparse-view LDCT reconstruction. Inspired by super-resolution networks for natural images, the proposed model interpolates projection data directly in the sinogram domain with a fully convolutional neural network that consists of only four convolution layers. The proposed model can be used directly for sparse-view CT reconstruction by concatenating the classic filtered back-projection (FBP) module, or it can be incorporated into existing dual-domain reconstruction frameworks as a generic sinogram domain module. The proposed model is validated on both the 2016 NIH-AAPM-Mayo Clinic LDCT Grand Challenge dataset and The Lung Image Database Consortium dataset. It is shown that despite the computational simplicity of the proposed model, its reconstruction performance at lower sparsity levels (1/2 and 1/4 radiation dose) is comparable to that of the sophisticated baseline models and shows some advantages at higher sparsity levels (1/8 and 1/15 radiation dose). Compared to existing sinogram domain baseline models, the proposed model is computationally efficient and easy to train on small training datasets, and is thus well suited for clinical real-time reconstruction tasks.
1. Introduction
X-ray Computed Tomography (CT) has found extensive utilization in clinical practice. Compared with other radiological examinations, CT can provide higher resolution medical tomographic anatomical images. Although CT brings great convenience to patients in diagnosing diseases, the hazards of the radiation generated by CT scans of the human body have also gradually attracted attention [1]. In recent years, Low-Dose CT reconstruction (LDCT) has become a research hotspot [2].
Two main methods are used to reduce the CT radiation dose: reducing the tube current [3] and reducing the sampling views [4,5]. Both ways may introduce noise and artifacts that do not belong to tissue structures, resulting in a loss of image details and sharpness, which will affect the doctor’s diagnosis. To improve the reconstructed CT image quality, existing LDCT methods use core processing modules (e.g., denoising or interpolation) in the sinogram domain [6,7], image domain [8,9,10,11], or both domains simultaneously [12,13,14,15,16], or design new iterative reconstruction algorithms [17,18]. This work focuses on sinogram domain processing for sparse-view LDCT reconstruction, i.e., improving LDCT by enhancing the projection data in the sinogram domain.
In the last decade, with the revival of neural networks, LDCT reconstruction based on deep learning has gradually become a research hotspot. Chen et al. [19] applied a deep learning method to denoise LDCT images in the image domain, employing a simple network consisting of three convolution layers and an activation function. Since then, a number of networks originating from computer vision tasks have also been continuously used for LDCT reconstruction, including supervised [20,21,22,23] and unsupervised networks [15,16]. In [20], the well-known Residual Encoder–Decoder (RED) network was used for LDCT reconstruction. Generative Adversarial Networks (GANs) have also been introduced into LDCT. Yi et al. [21] proposed a conditional GAN for LDCT reconstruction, which was composed of generative, discriminant and sharp-detection networks.
In addition to image domain denoising, LDCT can be achieved by processing in the sinogram domain. In [22,23], convolutional neural networks were used directly to denoise the projected data and construct high-quality CT images through back projection. Lee et al. [6] introduced a deep U-Net approach for the purpose of interpolating the projected data. To solve the data consistency problem of sinograms, Lee et al. [8] proposed a sinogram synthesis method with prior knowledge constraints. Compared to image domain methods, sine domain methods are usually more efficient at coping with noise or insufficient projection data due to low doses. CT images reconstructed from low-quality sinograms usually contain more noise and artifacts [14], which are difficult to remove completely because it is hard to model their distributions in the image domain [5].
Recently, several dual-domain, end–end networks have been proposed, which act in both domains to cope with the low dose problem. Yuan et al. [24] proposed an end-to-end deep learning method which incorporates a fan beam projection layer to establish a connection between the two domains. In [12], a domain progressive 3D residual convolution network (DP-ResNet) was proposed for CT reconstruction in both the sinogram domain and the image domain. The dual-domain networks proposed in [12,24] have serial structures. Due to the sequential nature of the process, errors occurring in the sinogram domain have a tendency to accumulate and propagate in the image domain. Ge et al. [14] introduced a dual-domain network with a parallel structure in their work. This network was designed to combine two-domain information and avoid cumulative errors.
Guided by the saying “the deeper the better”, the structure of LDCT networks has a tendency to become increasingly complex, which means longer reconstruction times and higher computational costs. However, many clinical applications require fast (even real-time) reconstruction of CT images. From a clinical point of view, lightweight networks are more desirable. Inspired by the full convolutional network used for super resolution [25,26,27], we propose a simplified sinogram domain convolutional neural network model for sparse-view CT reconstruction, which contains only four convolutional layers of networks and is well suited for real-time CT reconstruction tasks. Our motivation stems from our recognition that the complexity of LDCT reconstruction networks needs to be reduced for the sake of clinical practice. The proposed network strikes a balance between image quality and computational efficiency, ensuring that LDCT reconstruction is performed quickly without compromising the quality of the reconstructed image.
The proposed network employs only four convolutional layers, where the first three layers are feature extraction modules and the final layer is a sub-view regression layer. By adjusting the parameters of the last layer, the network can be adapted to various levels of sparse-view LDCT reconstruction. In addition to being used directly for sinogram domain LDCT reconstruction, the proposed network can be incorporated into an existing dual domain framework as a lightweight sinogram domain module. The contributions of our work are as follows:
- An efficient fully convolutional network for LDCT reconstruction is proposed. This network can function as a stand-alone solution or as a lightweight component seamlessly integrated into existing dual-domain frameworks.
- We delve into an in-depth examination of the impacts of image domain loss and sinogram domain loss on the quality of reconstructed images.
The remainder of this work is summarized below. In Section 2, we delve into the theoretical foundations of the sparse-view LDCT problem, clarify the specific structure of the proposed model and introduce the concept of two different loss functions. Section 3 is dedicated to the presentation of experimental results. In Section 4, we discuss the results of the experiment. Finally, the conclusions are presented in Section 5.
2. Materials and Methods
2.1. The Sparse-View LDCT Problem
During a CT scan, the X-ray source rotates in a circle surrounding the body. The X-ray beam sent from the X-ray tube cuts across the body and is collected by the detector. The sinogram is obtained by capturing the attenuation values of X-rays as they pass through the scanned object during the CT scan. CT images can be recovered from full-view sinograms by CT reconstruction.
Assume that are the full-dose projection data (sinogram) acquired by a CT scan, where D is the the number of detectors and V is the number of projection views. is an underlying CT image of size . CT imaging can be represented as a linear mapping from the image domain to the sinogram domain:
where is the system-specific matrix of the imaging protocol used by the CT scanner, and and are column vectors reshaped by and , respectively.
CT reconstruction can be viewed as the inverse problem of Equation (6), i.e., estimating the image from projection data given the imaging system matrix . For full-dose CT, the system matrix is usually a tall matrix with column full rank (i.e., ), and the image can be theoretically solved by , where denotes the pseudo-inverse of the matrix . However, in practice, is difficult to compute analytically because the dimensionality of is too high. The classic FBP algorithm can be written as a linear mapping from the sinogram domain to the image domain,
where can be viewed as an approximation to . Iterative algorithms are also commonly used for full-dose CT, and are of non-closed form and gradually improve the intermediate image with deterministic and/or statistic knowledge [28].
Sparse-view LDCT is achieved by reducing the number of projected views in the sine diagram, i.e., increasing the angular separation between adjacent projection views. Let the full-dose sinogram consist of V projection views with a sampling step of 1, denoted as . The sparse-view sinogram at radiation dose can be expressed as , a subset of the columns of , where the sampling step and is divisible by V. Recovering a full-dose sinogram from a sparse-view sinogram is an ill-posed problem that requires some prior knowledge about sinograms. For this purpose, a full convolutional network is designed to learn this interpolated nonlinear mapping
and to predict the missing projection views , which are the complement of subset in , i.e., .
2.2. Structure of the Proposed Network
The proposed LDCT reconstruction scheme is presented in Figure 1. The LDCT reconstruction method consists of two modules: a sinogram domain network and an FBP module. The sinogram domain network learns the mapping in (3) to recover the full-view sinogram. The FBP module achieves the linear mapping in (2) with a convolutional layer and a fully-connected layer, both of which have fixed (pre-computed) weights. An image domain network can also be appended after the FBP module to form a dual-domain framework.
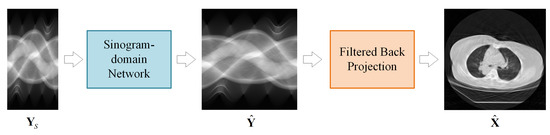
Figure 1.
Sinogram domain reconstruction scheme for sparse-view CT.
The detailed structure of the sinogram domain network is depicted in Figure 2, using the residual structure to predict the missing projection views . A linear interpolation module is designed to predict the linear component of , and a full convolution module consisting of four convolutional layers is employed to predict the nonlinear residuals between and its linear component. The first three convolutional layers of the full convolution module use a standard convolutional operation with , valid padding and tanh activation to extract sinogram features. The last convolutional layer is a projection-view regression layer with the number of channels which uses linear activation. To compensate for the feature map shrinkage due to valid padding, the sparse-view sinogram is first expanded before being input into the convolution module. Zeroes are interpolated in the detector dimension, and in the view dimension, since the projection views are sampled uniformly around a circle, the extended views (columns) can directly copy the original views (columns) of the same angles.
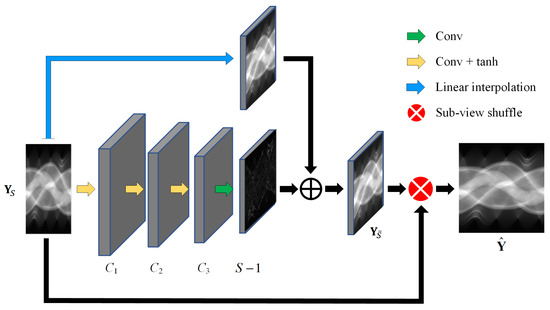
Figure 2.
Structure of the proposed sinogram domain network.
The method can flexibly handle CT reconstruction of different sparse-view levels by adjusting the parameters and view shuffling methods in the last layer. The use of a residual structure based on linear interpolation can effectively improve the efficiency of the model. The neural-network-based method implicitly learns a nonlinear mapping of sparse-view sinograms to full-view sinograms, making the reconstruction process more accurate and robust.
2.3. Loss Functions
For sparse-view LDCT reconstruction, designing the loss function in the sinogram domain is the most natural choice, which is calculated as:
where is the output from the network. is the expected output extracted from the full-dose sinogram.
Since the purpose of LDCT reconstruction is to produce precise CT images, it is logical to design the loss function in the image domain as well. Assuming that is the full-dose sinogram projected from the CT image with the imaging model in (6), the FBP reconstruction defined in (2) will inevitably introduce reconstruction error even if its input is the full-view sinogram , which means that a zero-deviation sinogram for the loss does not necessarily result in a CT reconstruction image with zero deviation from the original CT image, i.e., . To harmonize the sinogram network and the FBP module, we push back the loss calculation to the image domain,
where is the reconstructed CT image output by the FBP module and denotes the original full-dose CT image used to generate the sinogram domain training data. Since the FBP module is linear (see (2)), the errors in the image domain can be effectively propagated to the network layers in the sinogram domain using the back propagation algorithm.
Supervised training of the sinogram interpolation network with the image domain loss can compensate to some extent for the errors generated by FBP. In the subsequent experimental section, we quantitatively compare and evaluate the two loss functions defined in (3) and (4), testing their performances under different sparse levels. To the best of the authors’ knowledge, this study represents the first systematic investigation into the combined impact of sinogram interpolation and FBP reconstruction on LDCT reconstruction and into the ability of the sinogram domian network to reconcile FBP reconstruction errors at different sparse levels.
3. Results
3.1. Datasets and Experimental Setting
The proposed network was validated with two public datasets provided by The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) [29] and Mayo Clinic for the 2016 NIH-AAPM-Mayo Clinic Low Dose CT Grand Challenge [30].
Both datasets consist of 2000 images of the lungs from ten patients (200 images per patient). CT images are squares of size 512. The same data pre-processing method was applied to the two datasets. Full-view sinogram domain data were derived from full-view CT images by the forward projection procedure [31], which can be implemented using the Matlab 2020a function . The size of the full-view sinograms obtained by the fan-beam projection is , i.e., 605 detectors with 360 projection views (sampling interval = 1 degree). The sparse-view sinograms for sparse levels were obtained by downsampling the full-view sinograms in the view dimension with the corresponding sparse levels. For each dataset, 1800 samples were used to train the networks and the remaining 200 samples were used for testing. Figure 3a,b shows the samples from both datasets, which are from left to right, original (normal dose) CT images, full-view sinograms and sparse-view sinograms with various sparse levels ().
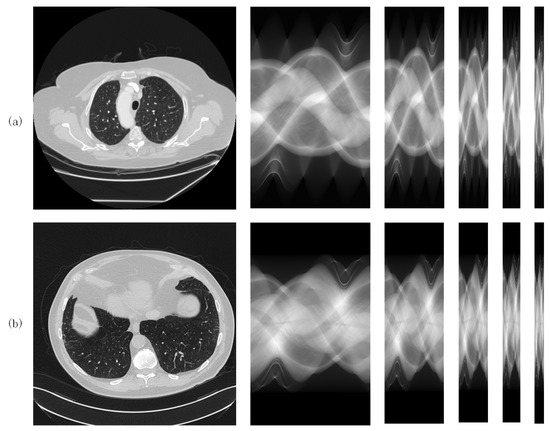
Figure 3.
CT samples from (a) the LIDC-IDR dataset and (b) the AAPM dataset.
The proposed network was compared with a linear interpolation method and two sinogram domain baseline networks: Res-Unet [24] and SDS-net [13]. Res-Unet has a classical network structure, and was used in a sinogram domain inpainting network in [24]. SDS-net is the sinogram domain subnetwork from the end-to-end network proposed in [13]. Both Res-Unet and SDS-net have a pre-linear-interpolation layer, and the interpolated full-view sinograms are then input to the sinogram domain network.
The hyperparameters for the baseline networks were set as described in their original papers. In the CT image domain, both the PSNR and the Structural Similarity (SSIM) metrics were calculated to quantitatively evaluate the quality of the reconstructed CT images, while in the sinogram domain, only the PSNR was calculated to quantitatively evaluate the difference between the estimated sinogram and the full-dose sinogram. This is because the estimated sinogram is an intermediate result in the CT reconstruction process, and is not visually inspected by humans and thus is not suitable for judging with a perception-based similarity metric such as SSIM.
In addition, the effects of different loss functions (sinogram domain loss and image domain loss ) on the performance of the sinogram domain interpolation model were compared.
All experiments were performed with a Tensorflow 2.3 @ Intel Xeon Gold 6154 CPU and NVIDIA GeForce RTX 3090 GPU. All networks were trained with the Adam algorithm.
3.2. Reconstruction Results
3.2.1. Training with the Sinogram Domain Loss
Figure 4 shows some test CT slices reconstructed by the four methods on the two datasets. The reconstructed images of the three machine-learning-based models are close and significantly better than those of linear interpolation. As the sparsity level s increases, the quality of all reconstructed CT images decreases significantly, and the linear interpolation method degrades the most significantly due to not using prior knowledge in the training set. Our network performs slightly better than the two baseline networks (SDS-net and Res-Unet) at higher sparse levels (). As shown in the local zooms corresponding to and 15, the proposed network is better in terms of edge preservation of CT images.
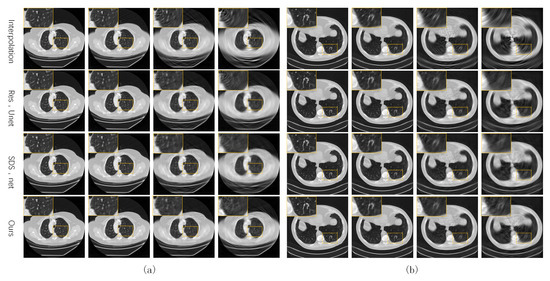
Figure 4.
Reconstructed CT images of the four methods from our own dataset (a) and the AAPM dataset (b).
The average PSNR values (dB) in the sinogram domain on the two datasets are given in Table 1, and reflect the similarity between the estimated sinogram () and the expected (full-view) sinogram (). Consistent with the results of human visual inspections, the three deep-learning-based methods significantly outperform linear interpolation in sinogram domain prediction.

Table 1.
Averaged PSNR (dB) in the sinogram domain trained with .
The proposed interpolation network performs comparably to the two baseline networks on both datasets, though its computational complexity is significantly lower than those of the two baselines (a comparison of computational efficiency will be given in Section 3.3). On the LIDC-IDRI dataset, when , the reconstruction results of our network are 60.31 dB and 52.24 dB, which are only 0.2 dB lower than the optimal results. On the AAPM dataset, when , the reconstruction results of our network are 56.84 dB and 50.12 dB. The highest difference compared to the optimal result is also only 0.6 dB.
Ours network demonstrates a slight advantage over the other deep-learning-based methods, particularly at higher levels of sparsity (). On the LIDC-IDRI dataset, the advantages of our model are more significant. For , it outperforms the optimal baseline model by 2.2 bB and 1.81 dB, respectively. Meanwhile, our model also maintains this advantage with the AAPM dataset, with the reconstruction results being higher than the optimal baseline model by 0.4 dB and 1.21 dB.
Table 2 and Table 3 show the average PSNR (dB) and SSIM (%) in the CT image domain, and this reflects the similarity between the reconstructed CT images () and the expected (full-dose) CT images (). In the CT image domain, combining both PSNR and SSIM metrics, the proposed network still performs well. The trends in the PSNR in the CT image domain and in the sinogram domain are consistent with both the AAPM and LIDC-IDRI datasets. When the sparsity level rises, the SSIM of the reconstructed image decreases rapidly. When , the optimal reconstruction results in SSIM are also only 78.84% and 66.53%, requiring subsequent denoising in the image domain.

Table 2.
Averaged PSNR (dB) in the CT image domain trained with .

Table 3.
Averaged SSIM (%) in the CT image domain trained with .
3.2.2. Training with the Image Domain Loss
The reconstruction results obtained by training the sinogram domain network using the image domain loss were also investigated and compared with those obtained using the sinogram domain loss .
The first two rows in Figure 5 show the reconstructed images obtained from our network trained with and their absolute difference from the full-dose CT results, respectively.
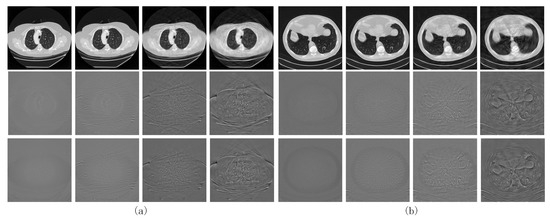
Figure 5.
CT images obtained by training the proposed network using the image domain loss () on (a) the LIDC-IDRI dataset and (b) the AAPM dataset. The first and second rows show the reconstructed image obtained by training with and its difference with the label, respectively. The third row shows the difference between the reconstructed image obtained by and the label.
The counterparts obtained via training with are shown in the last rows of Figure 4 and Figure 5, respectively. Comparing the quantized difference images shows that the image domain loss is better than the sinogram domain loss at lower sparse levels. As analyzed in Section 2.3, setting back the loss function to the image domain can to some extent coordinate the reconstruction error of the FBP module itself using the sinogram domain interpolation network, resulting in better CT reconstructed images. However, for higher sparsity levels, the results corresponding to image and sinogram domain losses tend to be consistent. To quantitatively evaluate the two loss functions, the PSNR- and SSIM-S (where S denotes the sparsity levels) curves for the CT images reconstructed by the proposed network trained with the two loss functions are compared in Figure 6a,b. The corresponding PSNR-S curves of the predicted sinogram are also compared in Figure 6c.
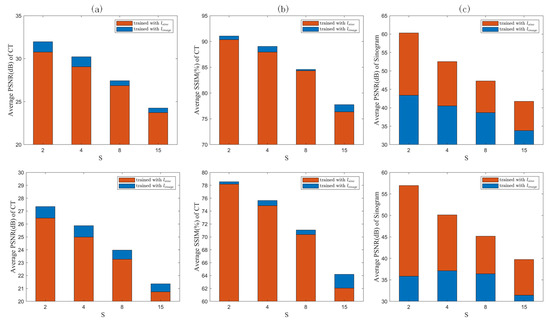
Figure 6.
Sinogram domain loss vs. image domain loss in terms of (a) PSNR and (b) SSIM for reconstructed CT images, and (c) PNSR for predicted sinograms.
It can be seen from Figure 6a,b that in terms of the reconstructed CT images, the image domain loss outperforms the sinogram domain loss at each sparsity level.
Since the feedback loop formed by the image domain loss contains the FBP module, the reconstruction error of the FBP itself can be effectively compensated for by optimizing the weights of the sinogram domain network. However, as shown in Figure 6c, in terms of the PSNR of the predicted sinogram, the sinogram domain loss is better than the image domain loss, indicating that the image domain loss sacrifices the prediction accuracy of the sinogram in order to coordinate the FBP module. However, in any case, the reconstructed CT images can benefit from the image domain loss. It should also be noted that the differences between image and sinogram domain losses decrease as the sparsity level S increases, both in terms of the PSNR of the reconstructed CT images and that of the predicted sinograms. Contrary to the gradual decrease in the PSNR, the SSIM increases significantly at .
3.3. Computational Efficiency
To evaluate the computational efficiency of the involved methods, the number of trainable parameters and the floating point operations (FLOPs) are shown in Table 4.

Table 4.
Computational efficiency of the three methods.
This represents the total number of floating point arithmetic operations performed by the model during inference or training, making it a key factor in measuring the computational resource requirements of a model. Higher FLOPs generally indicate more complex models that demand greater computational resources. The FLOPs of a convolutional layer can be calculated as:
where is the size of the convolution kernel. The size of the output feature map is . is the output channel size.
The results show that in comparison to the other two baseline networks, our network exhibits reduced parameter requirements and demands fewer floating point operations. Thus, our network is more computationally efficient.
4. Discussion
This paper introduces a computational efficient sinogram interpolation network for sparse-view CT reconstruction, which can work directly in series with an FBP module or be embedded into an existing dual-domain LDCT reconstruction framework. The proposed network has a very simple structure, consisting of only four convolutional layers and one residual connection, which makes it well suited for clinical real-time CT reconstruction tasks. The fully convolutional structure of the network allows it to adapt to sinograms of varying sizes and effectively handle sinograms with different levels of sparsity. We designed different loss functions for the network, the sinogram domain loss and image domain loss, and investigated the performance of the training of two different losses. The proposed network is validated using two publicly available datasets (LIDC-IDR dataset and AAPM dataset), and compared with the linear interpolation method and two baseline deep learning models. Despite the far simpler model structure than the baseline models, the proposed network performs comparably to the baselines on both datasets and even shows some advantages at higher sparsity levels. The experiments comparing the two loss functions show that the image domain loss can improve the final reconstructed CT images by sacrificing the prediction accuracy in the sinogram domain. As the sparsity level increases, the performance of both losses gradually increases. Although the performance gap between the two losses is large at low sparsity levels. As the sparsity level increases, the performance of both losses gradually approaches. Finally, the computational efficiency of the involved models is compared. The proposed network has significant advantages in terms of both the FLOPs and trainable parameters.
5. Conclusions
Low-Dose Computed Tomography (LDCT) has emerged as a valuable imaging modality in modern clinical practice. Historically, the prevailing notion has been guided by “the deeper, the better”, leading to the development of increasingly complex LDCT networks. However, clinical practice often requires rapid or real-time reconstruction of CT images. This paper proposes a simple structure and computationally efficient CNN model for sparse-view LDCT reconstruction. The proposed model offers versatility by seamlessly integrating the classic FBP module for direct use in sparse-view CT reconstruction. In addition, it can be incorporated into existing dual-domain reconstruction frameworks as a generic sinusoidal domain module, enhancing their computational efficiency. The experimental results show that the proposed network achieves a balance between computational burden and reconstruction quality at several sparse levels. Despite its computational simplicity, it exhibits a competitive reconstruction quality at lower sparse levels (1/2 and 1/4 radiation dose) and even outperforms sophisticated baseline models at higher sparse levels (1/8 and 1/15 radiation dose).
Author Contributions
Conceptualization, B.Y.; methodology, F.G.; software, F.G.; validation, F.G. and H.F.; formal analysis, F.G. and Z.Y.; investigation, F.G.; resources, C.L.; data curation, L.Y.; writing—original draft preparation, F.G.; writing—review and editing, B.Y.; visualization, C.L.; supervision, B.Y.; project administration, B.Y.; funding acquisition, W.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This document is the result of a research project funded by the Sichuan Science and Technology Support Program (2021YFQ0003, 2023YFSY0026, 2023YFH0004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://github.com/GFP-1011/efficientLDCT.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Hall, E.J.; Brenner., D.J. Cancer risks from diagnostic radiology. Br. J. Radiol. 2008, 81, 362–378. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, Q.; Liu, Y.; Cui, X.; Bai, Y.; Gui, Z. Low-dose CT statistical iterative reconstruction via modified MRF regularization. Comput. Methods Programs Biomed. 2016, 123, 129–141. [Google Scholar]
- Xie, Q.; Zeng, D.; Zhao, Q.; Meng, D.; Xu, Z.; Liang, Z.; Ma, J. Robust Low-Dose CT Sinogram Preprocessing via Exploiting Noise-Generating Mechanism. IEEE Trans. Med. Imaging 2017, 36, 2487–2498. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Yang, B.; Guo, F.; Zheng, W.; Poignet, P. Sparse-view CBCT reconstruction via weighted Schatten p-norm minimization. Opt. Express 2020, 28, 35469–35482. [Google Scholar] [CrossRef]
- Hu, D.; Liu, J.; Lv, T.; Zhao, Q.; Zhang, Y.; Quan, G.; Feng, J.; Chen, Y.; Luo, L. Hybrid-Domain Neural Network Processing for Sparse-View CT Reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 88–98. [Google Scholar] [CrossRef]
- Fu, J.; Dong, J.; Zhao, F. A Deep Learning Reconstruction Framework for Differential Phase-Contrast Computed Tomography with Incomplete Data. IEEE Trans. Image Process. 2020, 29, 2190–2202. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.; Kim, H.; Cho, B.; Cho, S. Deep-Neural-Network-Based Sinogram Synthesis for Sparse-View CT Image Reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 109–119. [Google Scholar] [CrossRef]
- Kan, E.; Min, J.; Ye, J.C. WaveNet: A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 2016, 44, e360–e375. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; Qiao, K.; Wang, L.; Yan, B.; Li, L.; Hu, G. Image Prediction for Limited-angle Tomography via Deep Learning with Convolutional Neural Network. arXiv 2016, arXiv:1607.08707. [Google Scholar]
- Liu, J.; Kang, Y.; Xia, Z.; Qiang, J.; Zhang, J.F.; Zhang, Y.; Chen, Y. MRCON-Net: Multiscale Reweighted Convolutional Coding Neural Network for Low-Dose CT Imaging. Comput. Methods Programs Biomed. 2022, 221, 106851. [Google Scholar] [CrossRef]
- Chan, Y.; Liu, X.; Wang, T.; Dai, J.; Xie, Y.; Liang, X. An attention-based deep convolutional neural network for ultra-sparse-view CT reconstruction. Comput. Biol. Med. 2023, 161, 106888. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Zhao, Q.; Liu, J.; Yang, W.; Yang, J.; Quan, G.; Chen, Y.; Shu, H.; Luo, L.; Coatrieux, J.L. Domain Progressive 3D Residual Convolution Network to Improve Low-Dose CT Imaging. IEEE Trans. Med. Imaging 2019, 38, 2903–2913. [Google Scholar] [CrossRef]
- Wang, W.; Xia, X.G.; He, C.; Ren, Z.; Lu, J.; Wang, T.; Lei, B. An End-to-End Deep Network for Reconstructing CT Images Directly From Sparse Sinograms. IEEE Trans. Comput. Imaging 2020, 6, 1548–1560. [Google Scholar] [CrossRef]
- Ge, R.; He, Y.; Xia, C.; Sun, H.; Zhang, Y.; Hu, D.; Chen, S.; Chen, Y.; Li, S.; Zhang, D. DDPNet: A Novel Dual-Domain Parallel Network for Low-Dose CT Reconstruction. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022: 25th International Conference, Singapore, 18–22 September 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 748–757. [Google Scholar]
- Zha, R.; Zhang, Y.; Li, H. NAF: Neural Attenuation Fields for Sparse-View CBCT Reconstruction. In Lecture Notes in Computer Science; Springer Nature: Cham, Switzerland, 2022; pp. 442–452. [Google Scholar] [CrossRef]
- Jung, C.; Lee, J.; You, S.; Ye, J.C. Patch-Wise Deep Metric Learning for Unsupervised Low-Dose CT Denoising. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 634–643. [Google Scholar]
- Hu, Z.; Gao, J.; Zhang, N.; Yang, Y.; Liu, X.; Zheng, H.; Liang, D. An improved statistical iterative algorithm for sparse-view and limited-angle CT image reconstruction. Sci. Rep. 2017, 7, 10747. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Chen, Y.; Zhang, J.; Zhang, W.; Sun, H.; Lv, Y.; Liao, P.; Zhou, J.; Wang, G. LEARN: Learned Experts’ Assessment-Based Reconstruction Network for Sparse-Data CT. IEEE Trans. Med. Imaging 2018, 37, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.; Zhang, W.; Liao, P.; Li, K.; Zhou, J.; Wang, G. Low-dose CT via convolutional neural network. Biomed. Opt. Express 2017, 8, 679–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.; Kalra, M.K.; Lin, F.; Chen, Y.; Liao, P.; Zhou, J.; Wang, G. Low-Dose CT with a Residual Encoder-Decoder Convolutional Neural Network. IEEE Trans. Med. Imaging 2017, 36, 2524–2535. [Google Scholar] [CrossRef]
- Yi, X.; Babyn, P. Sharpness-Aware Low-Dose CT Denoising Using Conditional Generative Adversarial Network. J. Digit. Imaging 2018, 31, 655–669. [Google Scholar] [CrossRef]
- Ghani, M.U.; Karl, W.C. Deep Learning-Based Sinogram Completion for Low-Dose CT. In Proceedings of the 2018 IEEE 13th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Aristi Village, Greece, 10–12 June 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Humphries, T.; Si, D.; Coulter, S.; Simms, M.; Xing, R. Comparison of deep learning approaches to low dose CT using low intensity and sparse view data. Med. Imaging 2019, 10948, 1048–1054. [Google Scholar]
- Yuan, H.; Jia, J.; Zhu, Z. SIPID: A deep learning framework for sinogram interpolation and image denoising in low-dose CT reconstruction. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1521–1524. [Google Scholar] [CrossRef]
- Tian, C.; Zhang, X.; Lin, J.C.W.; Zuo, W.; Zhang, Y.; Lin, C.W. Generative adversarial networks for image super-resolution: A survey. arXiv 2022, arXiv:2204.13620. [Google Scholar]
- Shi, W.; Caballero, J.; Huszár, F.; Totz, J.; Aitken, A.P.; Bishop, R.; Rueckert, D.; Wang, Z. Real-Time Single Image and Video Super-Resolution Using an Efficient Sub-Pixel Convolutional Neural Network. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1874–1883. [Google Scholar] [CrossRef]
- Tong, T.; Li, G.; Liu, X.; Gao, Q. Image Super-Resolution Using Dense Skip Connections. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 4809–4817. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, Y.; Ye, X.; Mou, X. Machine Learning for Tomographic Imaging; IOP Publishing: Bristol, UK, 2019. [Google Scholar] [CrossRef]
- Armato III, S.; Mclennan, G.; Bidaut, L.; McNitt-Gray, M.; Meyer, C.; Reeves, A.; Zhao, B.; Aberle, D.; Henschke, C.; Hoffman, E.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Moen, T.; Chen, B.; Holmes III, D.; Duan, X.; Yu, Z.; Yu, L.; Leng, S.; Fletcher, J.; McCollough, C. Low Dose CT Image and Projection Dataset. Med. Phys. 2020, 48, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Natterer, F. The Mathematics of Computerized Tomography; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2001. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).