A Biomedical Relation Extraction Method Based on Graph Convolutional Network with Dependency Information Fusion
Abstract
:1. Introduction
- (1)
- Obtain the dependency syntax tree of biomedical data labeled with entities and propose to use a local-global pruning strategy to filter out interfering information and improve accuracy;
- (2)
- Construct dependency type matrix for dependency information fusion on the pruned dependency tree so as to incorporate dependency information into the graph convolutional model. Calculating attention weights allows the model to distinguish the important dependency types in the dependency tree for predicting relations, thus improving the relation extraction performance.
2. Related Work
3. Method Design
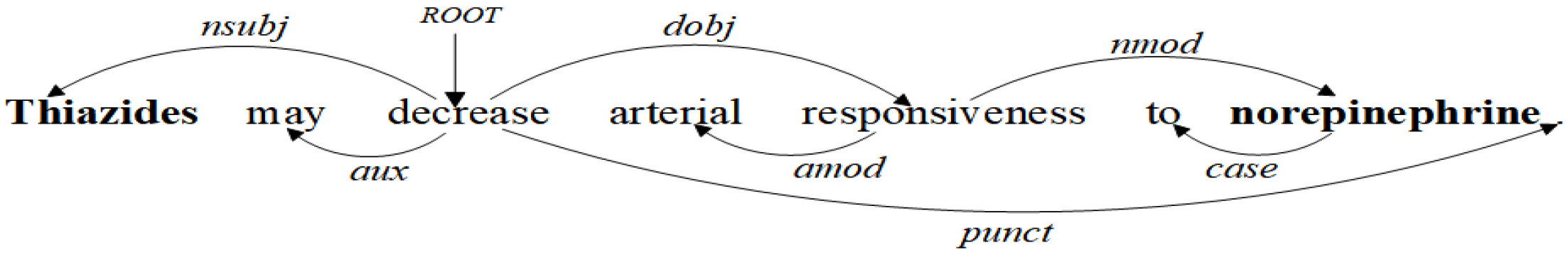
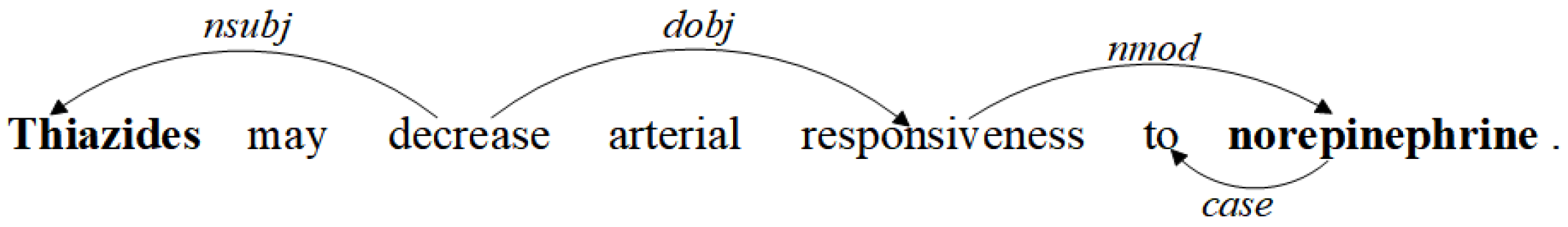
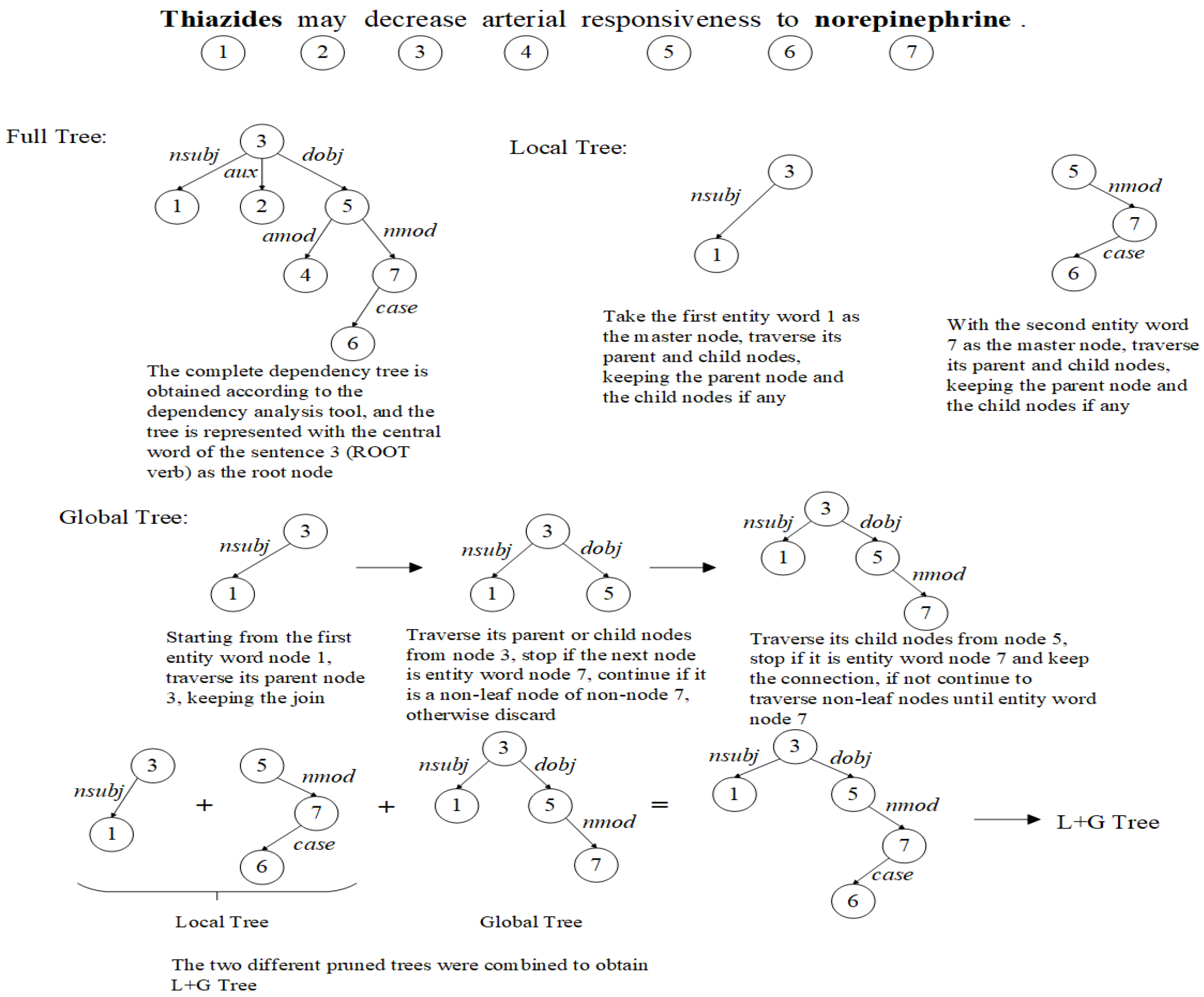
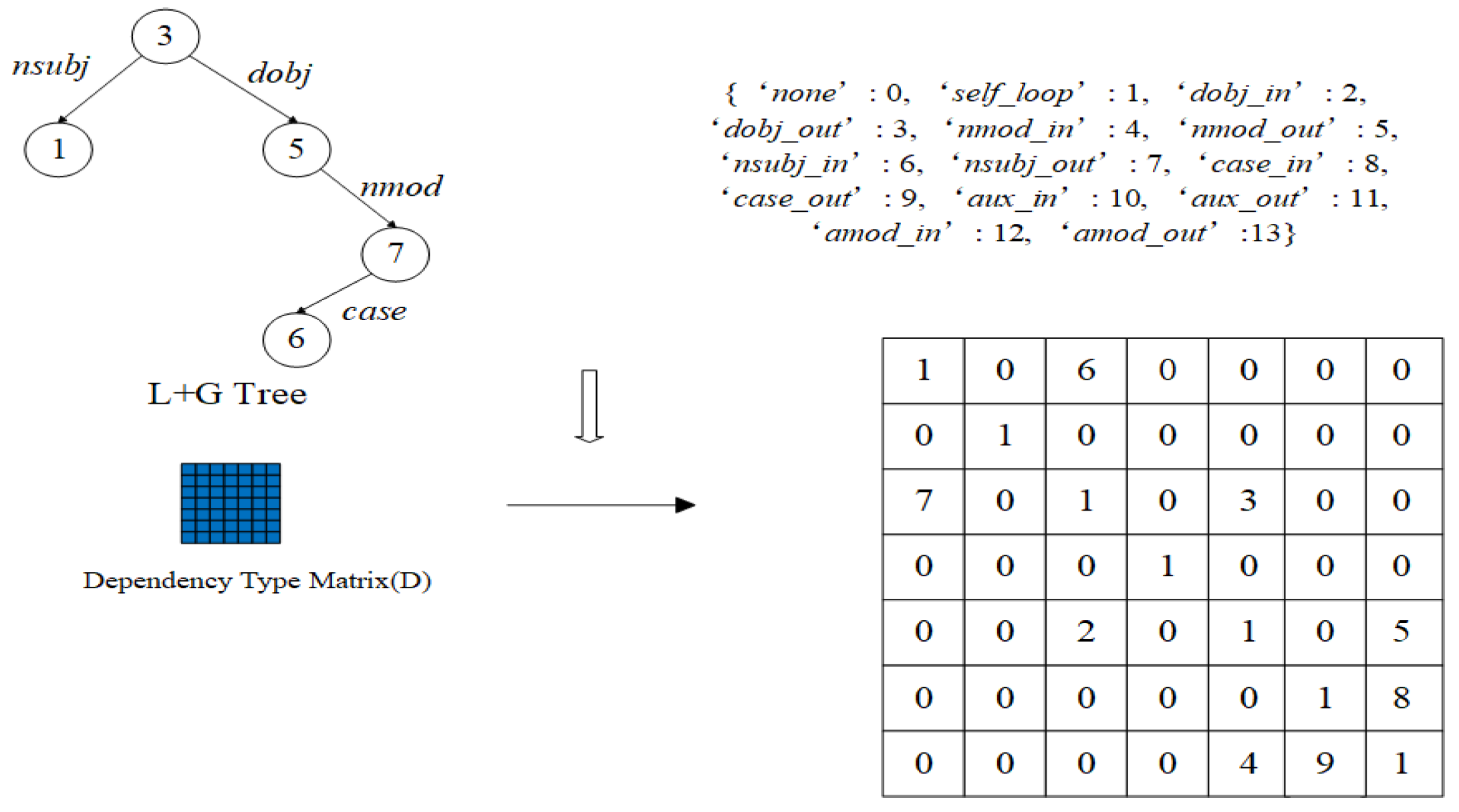
3.1. Dependency Tree Pruning Strategy
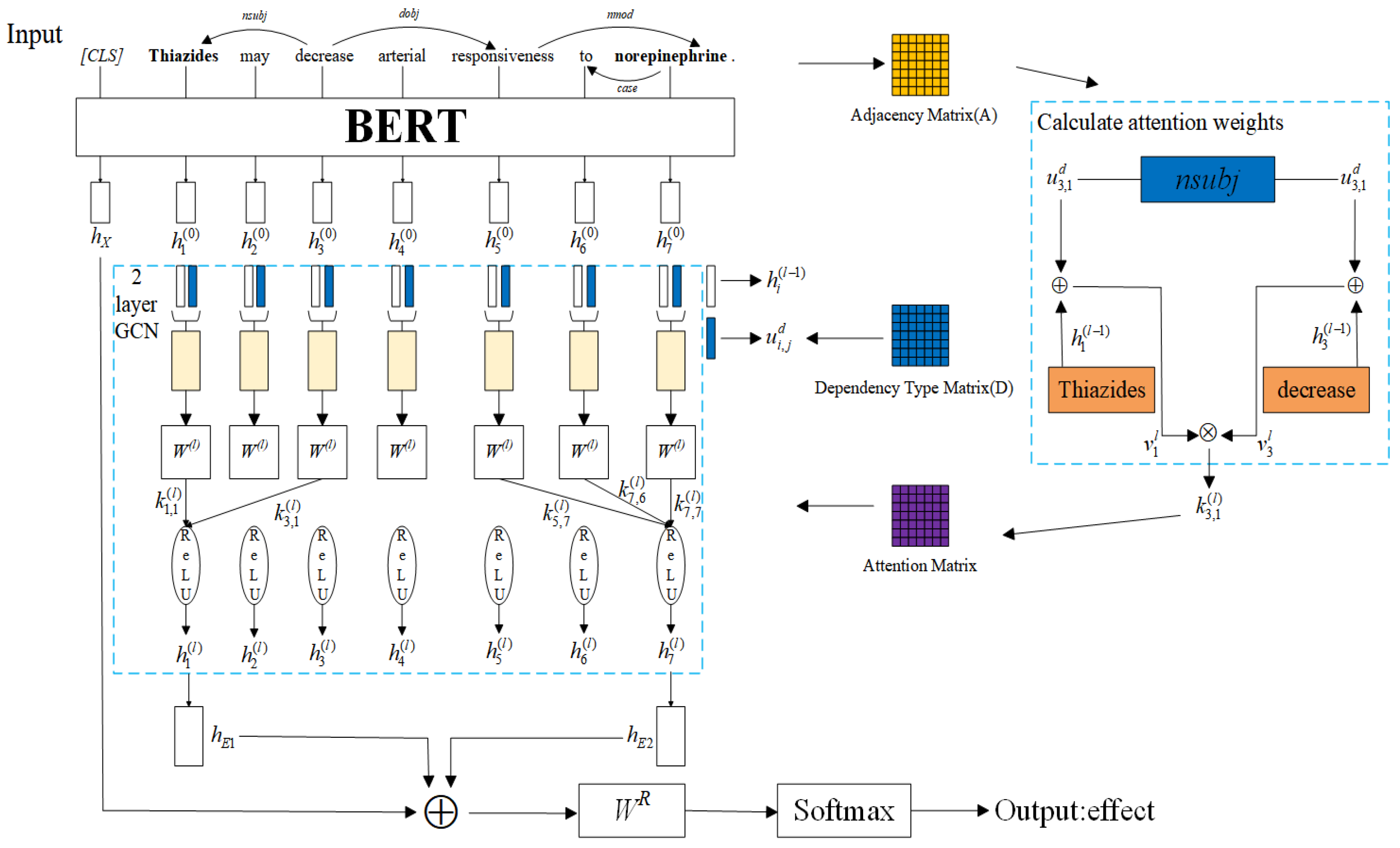
3.2. Dependency Information Fusion Attention Graph Convolutional Network
3.2.1. Input Layer
3.2.2. BERT Encoding Layer
3.2.3. Graph Convolution Layer
3.2.4. Classification Layer
4. Experiment and Analysis
4.1. Experiment Data
4.2. Experiment Setup and Results
4.2.1. Verification of Pruning Strategy
4.2.2. Verification of GCN Layers
4.2.3. Model Comparison
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, Y.; Li, J.; Lian, Z.; Jiao, C.; Hu, X. Supporting Medical Relation Extraction via Causality-Pruned Semantic Dependency Forest. In Proceedings of the 29th International Conference on Computational Linguistics, Gyeongju, Republic of Korea, 12–17 October 2022; pp. 2450–2460. [Google Scholar]
- Liu, K. A survey on neural relation extraction. Sci. China Technol. Sci. 2020, 63, 1971–1989. [Google Scholar] [CrossRef]
- Kambhatla, N. Combining lexical, syntactic, and semantic features with maximum entropy models for information extraction. In Proceedings of the ACL Interactive Poster and Demonstration Sessions, Barcelona, Spain, 21–26 July 2004; pp. 178–181. [Google Scholar]
- Bunescu, R.; Mooney, R. A shortest path dependency kernel for relation extraction. In Proceedings of the Human Language Technology Conference and Conference on Empirical Methods in Natural Language Processing, Vancouver, BC, Canada, 6–8 October 2005; pp. 724–731. [Google Scholar]
- Qian, M.; Wang, J.; Lin, H.; Zhao, D.; Zhang, Y.; Tang, W.; Yang, Z. Auto-learning convolution-based graph convolutional network for medical relation extraction. In Proceedings of the Information Retrieval: 27th China Conference, CCIR 2021, Dalian, China, 29–31 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 195–207. [Google Scholar]
- Zeng, D.; Liu, K.; Lai, S.; Zhou, G.; Zhao, J. Relation classification via convolutional deep neural network. In Proceedings of the COLING 2014, the 25th International Conference On Computational Linguistics: Technical Papers, Dublin, Ireland, 23–29 August 2014; pp. 2335–2344. [Google Scholar]
- dos Santos, C.; Xiang, B.; Zhou, B. Classifying Relations by Ranking with Convolutional Neural Networks. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Beijing, China, 26–31 July 2015. [Google Scholar]
- Socher, R.; Huval, B.; Manning, C.D.; Ng, A.Y. Semantic compositionality through recursive matrix-vector spaces. In Proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, Jeju Island, Republic of Korea, 12–14 July 2012; pp. 1201–1211. [Google Scholar]
- Suárez-Paniagua, V.; Segura-Bedmar, I. Extraction of drug-drug interactions by recursive matrix-vector spaces. In Proceedings of the 6th International Workshop on Combinations of Intelligent Methods and Applications (CIMA 2016), The Hague, Holland, 30 August 2016; Volume 2016, p. 65. [Google Scholar]
- Zhang, S.; Zheng, D.; Hu, X.; Yang, M. Bidirectional long short-term memory networks for relation classification. In Proceedings of the 29th Pacific Asia Conference on Language, Information and Computation, Shanghai, China, 30 October–1 November 2015; pp. 73–78. [Google Scholar]
- Song, L.; Zhang, Y.; Wang, Z.; Gildea, D. N-ary relation extraction using graph state LSTM. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018. [Google Scholar]
- Vu, N.T.; Adel, H.; Gupta, P.; Schütze, H. Combining Recurrent and Convolutional Neural Networks for Relation Classification. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, CA, USA, 12–17 June 2016; pp. 534–539. [Google Scholar]
- Zhou, P.; Shi, W.; Tian, J.; Qi, Z.; Li, B.; Hao, H.; Xu, B. Attention-based bidirectional long short-term memory networks for relation classification. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Berlin/Heidelberg, Germany, 7–12 August 2016; pp. 207–212. [Google Scholar]
- Lin, Y.; Shen, S.; Liu, Z.; Luan, H.; Sun, M. Neural relation extraction with selective attention over instances. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin/Heidelberg, Germany, 7–12 August 2016; pp. 2124–2133. [Google Scholar]
- Wu, S.; He, Y. Enriching pre-trained language model with entity information for relation classification. In Proceedings of the 28th ACM International Conference on Information and Knowledge Management, Beijing China, 3–7 November 2019; pp. 2361–2364. [Google Scholar]
- Miwa, M.; Bansal, M. End-to-End Relation Extraction using LSTMs on Sequences and Tree Structures. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin/Heidelberg, Germany, 7–12 August 2016; pp. 1105–1116. [Google Scholar]
- Xu, Y.; Mou, L.; Li, G.; Chen, Y.; Peng, H.; Jin, Z. Classifying relations via long short term memory networks along shortest dependency paths. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015; pp. 1785–1794. [Google Scholar]
- Zhang, Y.; Qi, P.; Manning, C.D. Graph Convolution over Pruned Dependency Trees Improves Relation Extraction. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018; pp. 2205–2215. [Google Scholar]
- Li, F.; Zhang, M.; Fu, G.; Ji, D. A neural joint model for entity and relation extraction from biomedical text. BMC Bioinform. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kenton, J.D.M.W.C.; Toutanova, L.K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the naacL-HLT, Minneapolis, MN, USA, 2–7 June 2019; Volume 1, p. 2. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2016. [Google Scholar]
- Herrero-Zazo, M.; Segura-Bedmar, I.; Martínez, P.; Declerck, T. The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions. J. Biomed. Inform. 2013, 46, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Spies, N.C.; Krysiak, K.; McMichael, J.F.; Coffman, A.C.; Danos, A.M.; Ainscough, B.J.; Ramirez, C.A.; Rieke, D.T.; Kujan, L.; et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 2017, 49, 170–174. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Z.; Luo, L.; Lin, H.; Wang, J. Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics 2016, 32, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Hua, L.; Sun, X.; Bai, W. Multichannel convolutional neural network for biological relation extraction. Biomed Res. Int. 2016, 2016, 1850404. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Anand, A. Drug-drug interaction extraction from biomedical texts using long short-term memory network. J. Biomed. Inform. 2018, 86, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gao, W.; Wong, K.F. Rumor Detection on Twitter with Tree-structured Recursive Neural Networks. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia, 15–20 July 2018. [Google Scholar]
- Zhang, Y.; Zhong, V.; Chen, D.; Angeli, G.; Manning, C.D. Position-aware attention and supervised data improve slot filling. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, 7–11 September 2017. [Google Scholar]
- Hong, L.; Lin, J.; Li, S.; Wan, F.; Yang, H.; Jiang, T.; Zhao, D.; Zeng, J. A novel machine learning framework for automated biomedical relation extraction from large-scale literature repositories. Nat. Mach. Intell. 2020, 2, 347–355. [Google Scholar] [CrossRef]
- Peng, N.; Poon, H.; Quirk, C.; Toutanova, K.; Yih, W.T. Cross-sentence n-ary relation extraction with graph lstms. Trans. Assoc. Comput. Linguist. 2017, 5, 101–115. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Y.; Lu, W. Attention Guided Graph Convolutional Networks for Relation Extraction. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; pp. 241–251. [Google Scholar]

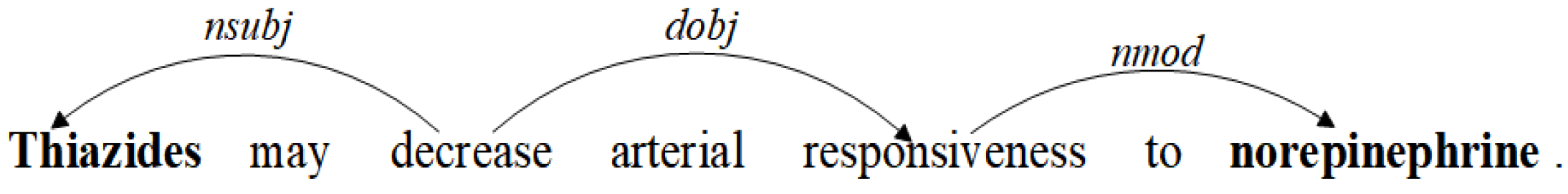
| Parameter Name | Parameter Value |
|---|---|
| Epoch | 100 |
| BatchSize | 16 |
| Learning_rate | 3 × 10−5 |
| Warmup_rate | 0.06 |
| Random_seed | 42 |
| Max_seq_length | 128 |
| Dropout | 0.2 |
| DDI | CIVIC | |
|---|---|---|
| Full | 73.3 | 74.2 |
| SDP | 74.7 | 76.1 |
| L+G(ours) | 77.1 | 77.9 |
| DDI Sentence | In a small combination study of ARAVA with methotrexate, a 2- to 3-fold elevation in liver enzymes was seen in 5 of 30 patients. |
| True relation | e1: ARAVA, e2: methotrexate, RE: effect |
| Full | e1: ARAVA, e2: methotrexate, RE: mechanism |
| SDP | e1: ARAVA, e2: methotrexate, RE: mechanism |
| L+G(ours) | e1: ARAVA, e2: methotrexate, RE: effect |
| DDI | CIVIC | |
|---|---|---|
| 1-layer GCN | 74.0 | 75.8 |
| 2-layer GCN | 77.1 | 77.9 |
| 3-layer GCN | 73.5 | 75.1 |
| Models | F1(%) |
|---|---|
| MV-RNN | 50.0 |
| SCNN | 67.0 |
| CNN-bioWE | 69.8 |
| MCCNN | 70.2 |
| Joint AB-LSTM | 71.5 |
| RvNN | 71.7 |
| Position-aware LSTM | 73.0 |
| BERE | 73.9 |
| DIF-A-GCN(ours) | 77.1 |
| Models | F1(%) |
|---|---|
| BiLSTM-Shortest-Path | 70.2 |
| GRN | 71.7 |
| CNN | 73.0 |
| BiLSTM | 73.9 |
| Tree LSTM | 75.9 |
| Graph LSTM-FULL | 76.7 |
| AGGCN | 77.4 |
| DIF-A-GCN(ours) | 77.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Xing, L.; Zhang, L.; Cai, H.; Guo, M. A Biomedical Relation Extraction Method Based on Graph Convolutional Network with Dependency Information Fusion. Appl. Sci. 2023, 13, 10055. https://doi.org/10.3390/app131810055
Yang W, Xing L, Zhang L, Cai H, Guo M. A Biomedical Relation Extraction Method Based on Graph Convolutional Network with Dependency Information Fusion. Applied Sciences. 2023; 13(18):10055. https://doi.org/10.3390/app131810055
Chicago/Turabian StyleYang, Wanli, Linlin Xing, Longbo Zhang, Hongzhen Cai, and Maozu Guo. 2023. "A Biomedical Relation Extraction Method Based on Graph Convolutional Network with Dependency Information Fusion" Applied Sciences 13, no. 18: 10055. https://doi.org/10.3390/app131810055
APA StyleYang, W., Xing, L., Zhang, L., Cai, H., & Guo, M. (2023). A Biomedical Relation Extraction Method Based on Graph Convolutional Network with Dependency Information Fusion. Applied Sciences, 13(18), 10055. https://doi.org/10.3390/app131810055






