1. Introduction
The hippocampus is a portion of the brain located between the cerebral thalamus and the medial temporal lobe; it is a crucial organ that is responsible for storing and organizing memory [
1]. Research on the main function and basic structure of the hippocampus is essential for understanding the working principles of the brain and the pathogenesis of neurodegenerative diseases and developing treatment methods. Many studies have shown that the shape and texture of the hippocampus are related to neurodegenerative diseases such as alzheimer’s disease (AD), epilepsy, etc. To some extent, atrophy of the hippocampus can reflect the condition of these diseases [
2,
3]. Magnetic resonance imaging (MRI) is a new medical imaging examination technique that creates high-definition images of organs and tissues by using powerful magnetic fields and harmless radio waves [
4]. MRI has become increasingly crucial in disease diagnosis and research due to the rapid development of neuroimaging technologies [
5]. This technology can not only help clinicians detect lesions but also provide more accurate information on the location and size of lesions, providing significant assistance in disease diagnosis. Accurately segmenting the hippocampus from brain MRI images and measuring its volume and morphological characteristics can provide an essential foundation for early diagnosis, progress monitoring, and treatment evaluation of these diseases. Thus, clinicians usually observe the shape of the hippocampus to diagnose neurodegenerative diseases and conduct surgical planning and treatment evaluation. As a result, precisely segmenting the hippocampus from brain MRIs and observing its shape are critical for disease diagnosis.
Segmenting the hippocampus from MRI images is a challenging task, as the quality of the images may vary, the shape of the hippocampus is irregular, and the hippocampus boundary is not distinct. Furthermore, manually segmenting the hippocampus from brain MRIs is a professional task that needs to be carried out by experienced experts or clinicians. Thus, accurately and automatically segmenting the hippocampus from brain MRI images rather than manually segmenting has recently drawn a lot of attention.
MRI images are 3D data that contain more information than 2D images. Typically, networks comprise numerous parameters and a high level of computational complexity when processing 3D data such as MRI images, which may consume considerable computational and storage resources. A number of lightweight networks have been proposed to reduce the network’s scale but may also limit their performance [
6]. Thus, a 3D U-Net-based segmentation model named Coordinate Attention and Dynamic Convolution U-Net (CADyUNet) is introduced, which significantly improves the network’s performance without expanding its scale by combining coordinate attention mechanisms and dynamic convolution operations. We applied the CADyUNet to hippocampus segmentation tasks and confirmed its effectiveness. The major contributions of this study are listed as follows:
To maintain a balance between the model’s performance and scale, a dynamic convolution block named dy-block is designed, which introduces new dynamic convolution operations to substitute the normal convolution operations and spatial dropout blocks to reduce the risk of overfitting. The dy-block can segment the hippocampus, especially its boundary, more precisely and quickly without increasing the depth of the network, which is defined as the number of hidden layers, or the width of the network, which is defined as the number of channels in each hidden layer;
To improve the segmentation performance, an improved coordinate attention mechanism is utilized in 3D U-Net. The enhanced attention mechanism expands the 2D-suitable structure to a 3D-suitable structure and uses larger convolutional kernels to extract spatial features, which can extract more spatial information compared to the original mechanism;
To preserve more important textural information and key background features, the soft pooling method is introduced to replace normal pooling methods such average pooling, max pooling, etc.
2. Related Work
In recent years, many researchers have segmented the hippocampus from brain MRI images using machine learning algorithms such as k-means clustering, the watershed algorithm, and the subtractive clustering algorithm [
7,
8,
9]. These machine learning algorithms can segment the hippocampus with more accuracy than manual segmentation. However, the segmentation accuracy of machine learning algorithms is limited by image noise and complex brain structure, which makes the segmentation performance very unstable. Recently, deep learning algorithms typified by convolutional neural networks (CNNs), which can automatically capture features, have demonstrated better advantages than machine learning methods in the image processing field. Many studies have shown that CNNs outperform typical semantic segmentation methods [
10]. Thus, a range of deep learning technologies are applied to the medical segmentation area, such as in retinal blood vessel segmentation [
11], brain tumor segmentation [
12,
13,
14,
15], breast cancer segmentation [
16], etc. These deep learning technologies can achieve excellent accuracy in hippocampus segmentation tasks through large-scale dataset training.
U-Net is a broadly employed deep learning medical image segmentation algorithm. It can integrate both global and local contextual features via the encoder and decoder, then compensate for feature loss resulting from downsampling via skip connections [
17]. Owing to U-Net’s simple structure and perfect performance, researchers have proposed various variant networks based on U-Net for different application scenarios in recent years. Zhou [
18] proposed UNet++ based on the UNet structure. UNet++ has more distinct scale skip connections and improved feature concatenation methods than U-Net, enabling it to capture targets of various scales and shapes. R2U-Net [
19] is also an extension of U-Net that introduces cyclic and residual connections to improve the network’s expression capabilities. Other similar models based on U-Net include Res-UNet [
20], MultiResUNet [
21], etc. These variant algorithms based on U-Net typically introduce attention mechanisms [
22], residual connections [
20], or other new network structures to improve the algorithm’s segmentation accuracy and robustness on different tasks and datasets. These algorithms provide important tools for the development of medical image segmentation and make its accuracy more accurate, faster, and more reliable.
U-Net is suitable for processing 2D but not 3D data. However, many 3D medical images collected by electronic computed tomography (ECT), MRI, ultrasound, and other medical imaging equipment contain more important spatial information and can provide more comprehensive lesion information compared to 2D medical images. These 3D images must be sliced into 2D images before being processed using U-Net, which may result in the loss of key anatomical structure information. To address this issue, a 3D U-Net model is designed, which is similar to U-Net in architecture [
23]. The 3D U-Net, which substitutes the 2D convolutional operations of U-Net with 3D volume convolutional operations, is commonly used in the 3D medical image segmentation area. For instance, Mehta R et al. [
24] showed that segmenting the brain tumor using 3D U-Net can enable accurate identification and segmentation of the brain tumor region, which contributes to the advancement of brain tumor diagnosis. V-Net [
25], UNETR [
26], Swin UNETR [
27], and other algorithms have also been designed for 3D image segmentation. These 3D segmentation networks can effectively utilize the three-dimensional information of medical images to achieve more accurate segmentation results, helping doctors to comprehensively understand the spatial distribution and morphological features of lesions to arrange the best treatment plans.
To segment targets with excellent precision in medical image segmentation tasks, a network must focus on specific target information while ignoring other unimportant information. The attention mechanism can solve this problem. There are three commonly used types of attention: spatial attention, channel attention, and mixed attention. The convolutional block attention modulus (CBAM) suggested by Woo S. et al. is a representative of mixed attention mechanisms; it infers attention maps in both channel domains and spatial domains [
28]. However, CBAM’s channel attention mechanism ignores feature map positional information, and the convolution operations used in CBAM’s spatial attention can only capture local features but not long-distance information. Thus, Qi Bin Hou et al. suggested coordinate attention (CA), which integrates coordinate features into channel attention [
29]. To obtain long-distance information, CA captures it along one dimension while retaining accurate positional information along another dimension. Attention mechanisms can significantly increase the model’s performance. For example, the Attention U-Net [
22] introduces an attention-gating module that sets high weights for segmentation targets and low weights for other background positions. The attention-gating module significantly improves the performance of 3D U-Net while maintaining computational efficiency. Other networks with attention mechanisms include SA-UNet [
30] and RA-UNet [
31].
3. Methodology
3.1. Improved Coordinate Attention Mechanism
The CA mechanism is added to 3D U-Net to achieve high segmentation accuracy in this work. A diagram of the CA is displayed in
Figure 1. First, the input images are divided into a one-dimensional aggregated feature on the width dimension and a one-dimensional aggregated feature on the height dimension by the average pooling method. Then, the two aggregated features are concatenated together and processed by a 1 × 1 convolution block to fully learn channel-domain information. Next, the concatenated features are split into two one-dimensional features followed by a 1 × 1 convolution block to learn the weight of each pixel of the two aggregated features. Then, the two aggregated features are multiplied by one another to obtain the final attention weight. Finally, to assign each input element a different weight value, the attention weight element is multiplied by the correlating input element.
Many datasets for medical image segmentation tasks comprise 3D data, providing additional depth-dimension information compared to 2D medical images. The depth dimension can capture contextual information about the segmentation target along the depth direction, which is crucial for accurately locating and segmenting targets, as it provides the relative position and relationship between targets and surrounding structures [
32]. However, the CA mechanism is designed for 2D data [
29]. Therefore, 3D images must be sliced into 2D images before being processed by the CA mechanism. Thus, an additional depth dimension structure is added to the CA structure, and the original 2D convolution operations are replaced by 3D convolution operations in the CA structure to process 3D images. Furthermore, convolution operations with a kernel size of 1 × 1 can only extract channel-domain information and ignore adjacent features in the spatial domain. Therefore, convolution operations with kernel sizes of 1 × 1 × 3, 1 × 3 × 1, and 3 × 1 × 1 are substitutes for the 1 × 1 convolution layers to extract spatial domain features and channel features simultaneously. The average pooling operation also loses important texture features. Inspired by the structure of CBAM [
28], the combination of average pooling and max pooling operations is used to preserve important texture features.
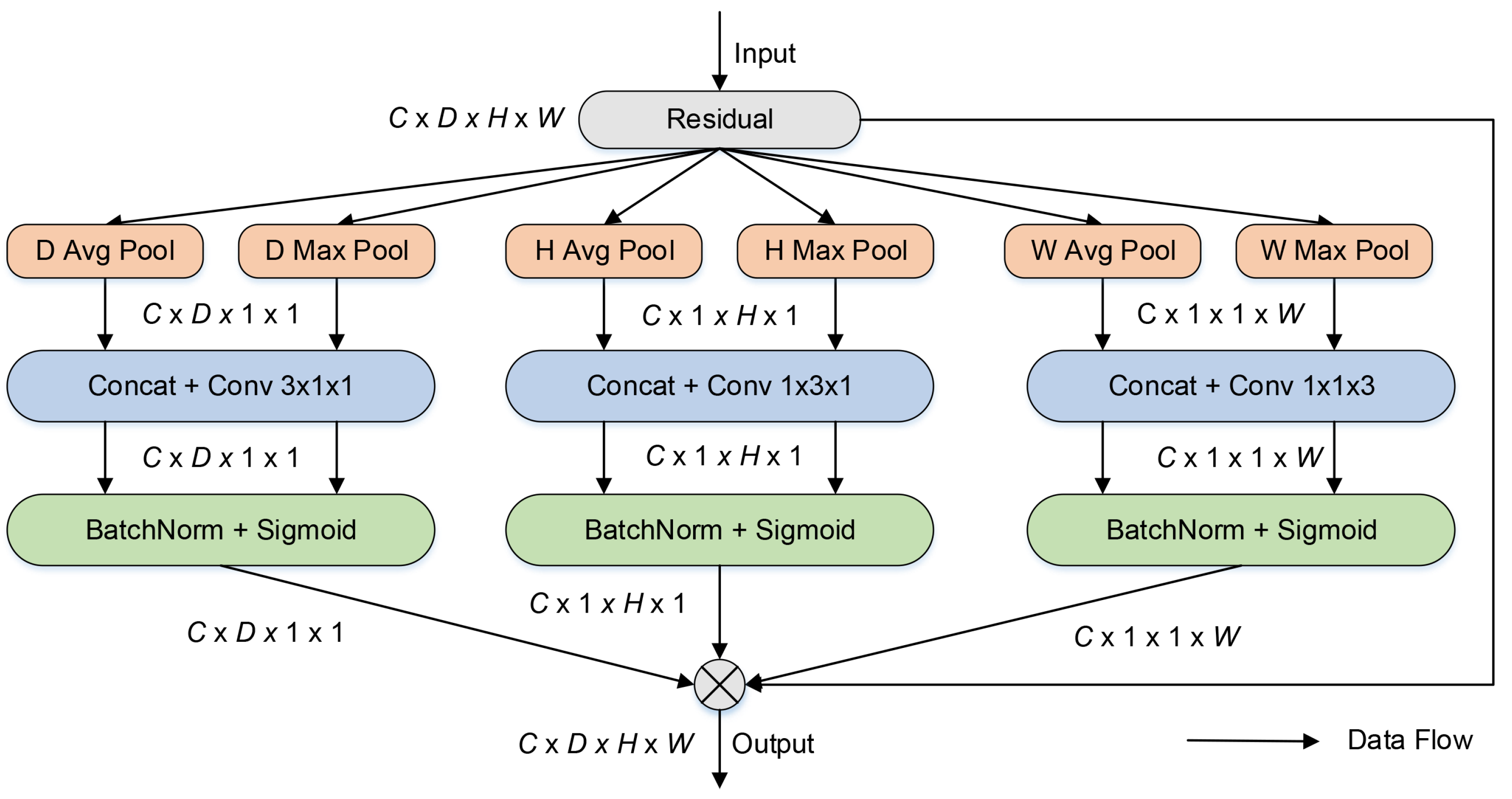
Figure 2 shows the framework of the improved CA mechanism.
As shown in
Figure 2, the input images are pooled into six aggregated features: three different dimensions of aggregated features by the average pooling method and three different dimensions of aggregated features by the max pooling method. Similar to CA, the two aggregated features from the same dimension are concatenated together. Then, channel- and spatial-domain features are extracted using convolutional operations with kernel sizes of 1 × 1 × 3, 1 × 3 × 1, and 3 × 1 × 1. Finally, to adaptively refine features, these aggregated characteristics are multiplied by the input data. In the structure of the improved CA, the combination of average pooling operations and max pooling operations can reduce the loss of important texture features and background information. The larger kernel of the convolution can extract more adjacent features of the spatial domain than convolution operations with a kernel size of 1. The improved CA mechanism is introduced in the skip connection of 3D U-Net to improve segmentation accuracy in this research.
3.2. Soft Pool
Pooling operations are applied to capture the most important characteristics and reduce feature map dimensionality after convolutional operations [
33]. Specifically, the pooling operation splits the input feature map into many regions that are not overlapping, with each region taking a representative value (such as maximum, average, etc.) as the output feature value. The pooling operation can decrease the model’s computational complexity while retaining important information and improving its robustness. Max pooling and average pooling are the main pooling operations in convolutional networks [
34].
The max pooling method takes the maximum pixel value of the pooling region as the output feature value, preserving the texture features of the input images but may lose some useful background information [
34]. The average pooling method computes the mean value of pixels in each pooling region as the output value, preserving the overall information of the image, but is more sensitive to noise than other pooling methods [
34]. Thus, Stergiou A. et al. designed the soft pooling method, which calculates the weight of each pixel in the pooling region, then multiplies each pixel by its corresponding weight, and sums them up [
35]. Soft pooling does not simply calculate the maximum or average value of pixels in the pooling region as the representative feature but calculates the representative feature based on the softmax weighting method [
35]. Soft pooling balances the effects of average pooling and max pooling while utilizing the beneficial characteristics of both. In this study, to reduce the loss of important texture features in the downsampling step, the soft pooling methods are a substitute for the max pooling methods in the downsampling of 3D UNet.
3.3. Dynamic Convolution Block
Over the years, CNN-based algorithms have made significant progress in the image processing area. However, convolution operations use the same convolution kernel weights for all inputs, which limits the representational capacity of the model. Thus, to increase the complexity of the network, researchers extend the width or depth of the network, which consumes considerable computational resources [
36]. Therefore, Chen Y et al. proposed dynamic convolution, which can considerably increase the network complexity without expanding the model’s scale [
37]. Standard convolutional operations use the same convolutional kernel weight for all input images, which may lead to weak representational ability and poor prediction for some complex input images [
38]. Dynamic convolutional networks can dynamically calculate the parameters of convolutional kernels based on input images, thereby enabling better feature representational abilities of the model [
36]. Compared with standard convolution operations, dynamic convolution can utilize prior knowledge of input images to dynamically adjust convolutional kernel weights to enhance feature representation capabilities and thereby improve model performance [
38]. The calculation process of dynamic convolution is displayed in
Figure 3. In contrast to ordinary convolution operations, dynamic convolution involves dynamically calculating the attention weights of multiple parallel convolution kernels, then aggregating the attention weights of these kernels to obtain the final kernel weights [
37]. The inputs vary, and the dynamic convolution kernel weights also change accordingly.
An overfitting problem occurs if the network structure is too complex or if the training data are too large. The phenomenon of overfitting means that the network performs perfectly on the training datasets but terribly on the test datasets [
39]. In order to avoid overfitting, some regularization methods are usually added in the training phase of a network, such as early stopping, batch normalization, dropout, etc. To decrease the possibility of overfitting, the “dropout” method reduces information transmission between neural nodes by randomly inactivating some neurons in the network during training. This method is usually used as a regularization method for fully connected neural networks (FCNs) [
40]. The datasets used in this research comprise 3D MRI images with strong spatial correlation. A standard dropout strategy cannot effectively reduce overfitting, as the information can still be transmitted through adjacent pixels in 3D space once a pixel is inactive [
41]. Compared to standard dropout, spatial dropout deactivates some channels of the 3D image randomly, which can effectively prevent the transmission of information in the channels and thereby reduce the possibility of overfitting. Thus, spatial dropout, as opposed to standard dropout, is chosen as the regulation method in this work.
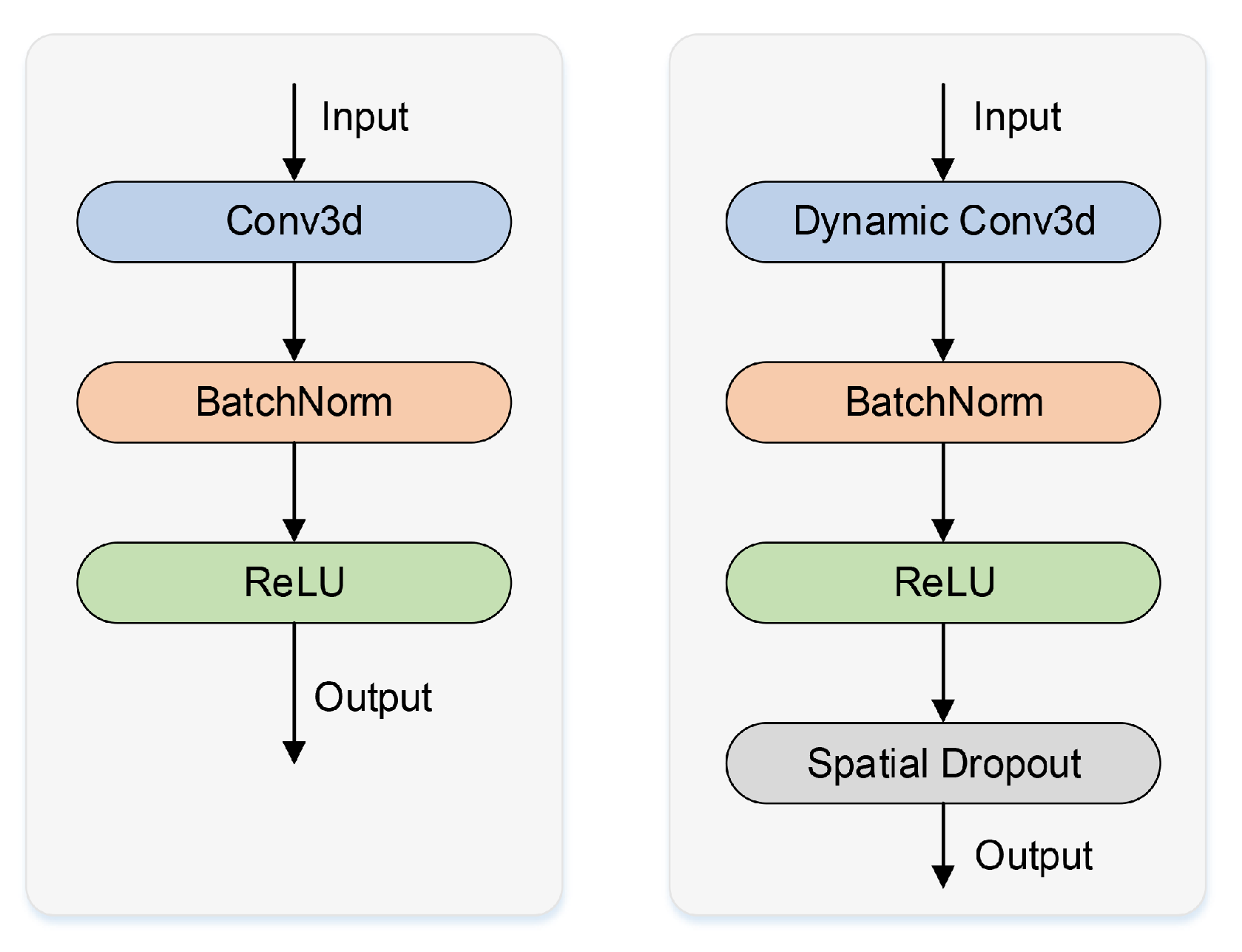
In this paper, a dynamic convolutional block named dy-block is designed as a substitute for the original 3D U-Net convolutional block (conv-block) to increase the model’s representational ability. Dynamic convolution is a new form of convolution that can dynamically calculate the weights of convolution kernels based on the characteristics of inputs. Compared with standard convolutional kernels, dynamic convolutional kernels have prior knowledge of inputs and can extract features with stronger ability. The dy-block designed in this work has better feature representational capabilities compared to the conv-block of 3D U-Net. The framework diagrams of the conv-block and the dy-block are shown in
Figure 4. The dy-block includes a dynamic convolutional layer to extract features, a batch normalization layer to speed up the convergence of the 3D U-Net, a ReLU layer to enhance the nonlinear representation ability, and a spatial dropout layer to reduce the risk of overfitting. Compared to the framework of the original conv-block, the dy-block can significantly increase target segmentation accuracy without expanding the model’s depth or width.
3.4. CADyUNet Architecture
The proposed CADyUNet consists of three separate components: the encoder, the decoder, and the skip connections. The framework of CADyUNet is displayed in
Figure 5.
The CADyUNet encoder, which contains four encryption blocks, is used to capture image features. Every encryption block contains a dy-block and a conv-block to increase the representational capacity of the proposed CADyUNet. Each encryption block is followed by a downsampling layer, which uses the soft pool method to preserve critical information, with the exception of the last encryption block.
The decoder of CADyUNet is used to recover image pixels, including three decryption blocks, each consisting of a conv-block and a dy-block. The images processed by a decryption block are transmitted to an upsampling layer to restore the image pixels. Then, through the skip connection structure, the recovered images are concatenated with images of corresponding sizes coded by the encoder stage.
The skip-connection structure of CADyUNet is combined with an improved CA mechanism. The shallow features captured by the encoder are recoded through the improved CA block before being transmitted to the decoder in the skip connection. The improved CA mechanism recodes the data and sets different location pixels to different weights. The pixels at the location of the hippocampus are set to high weights, and the other background location pixels are set to low weights. The improved CA mechanism proposed in this work can significantly increase hippocampus segmentation accuracy.
The last layer of CADyUNet is a convolutional operation with a kernel size of 1 × 1 × 1, which restores the image’s number of channels to 1. Additionally, to conserve computing resources, the number of channels in CADyUNet is decreased by four times compared to the number of channels in 3D U-Net in this study.
The designed CADyUNet is an automatic hippocampus segmentation network similar in architecture to 3D U-Net. In the structure of CADyUNet, dynamic convolution operations with stronger feature extraction capabilities are introduced in the encoding and decoding steps. The introduction of dynamic convolution greatly increases the network’s performance without increasing its depth or width. In addition, enhanced CA mechanisms are introduced in each skip connection so that shallow features are recoded with different weights. Finally, soft pooling methods are used in each downsampling layer of CADyUNet, which can greatly reduce the loss of important information.
4. Experiment and Analysis
4.1. Datasets
In our work, two datasets are used: the MICCAI 2013 SATA Challenge (MICCAI) dataset and the Harmonized Protocol initiative of the Alzheimer’s Disease Neuroimaging Initiative (HarP) [
42]. The MICCAI contains 35 groups of T1-weighted images in the training set and 12 groups in the testing set; every training image has its own corresponding multi-atlas label. Every image in this dataset is in NIFTI format, and both images and labels are 256 × 256 × 287 pixels, with a voxel spacing of 1 × 1 × 1 pixels. The MICCAI dataset can be accessed publicly at
https://my.vanderbilt.edu/masi/workshops/ (accessed on 15 April 2023). The HarP contains 135 groups of T1-weighted MRI images and their corresponding hippocampus labels. All of the images and labels have a voxel size of 1 × 1 × 1 pixels and a resolution of 197 × 233 × 189 pixels. The HarP dataset can be accessed publicly at
http://www.hippocampal-protocol.net (accessed on 23 March 2023).
For convenience of display, the segmentation labels are mapped in the original images with the same resolution between the raw MRI image and its hippocampus segmentation label in the HarP and the MICCAI datasets. We set the corresponding hippocampus label pixel in the original MRI image to a specific value to represent the hippocampus, and other non-hippocampus pixels were kept unchanged to distinguish them from the hippocampus. The visualization segmentation results of the HarP and MICCAI datasets are displayed in
Figure 6 and
Figure 7, respectively. The area with a red pixel represents where the hippocampus is located, and other pixel values are non-hippocampus areas.
4.2. Evaluation Indicators
Comparison of each element of the output results with the corresponding label’s element shows that if a positive element of the segmentation is correctly predicted as a positive element, then the element is classified in the true-positive (TP) category. A negative element is classified in the false-positive (FP) category when it is falsely predicted as a positive element. The opposite is the case for elements that are divided into the false-negative (FN) and true-negative (TN) categories. The dice, the mIoU, and the F1 are then calculated according to the four variables to measure the model’s effectiveness in this study. The formulas of these indicators are displayed below; among them, the F1 is determined by precision and recall.
4.3. Implementation Details
The segmentation labels of the MICCAI dataset are multi-atlas, including 15 different labels, such as amygdala, caudate, hippocampus, etc. Howerver, in this work, only the labels of the hippocampus are useful. Thus, the segmentation labels of the hippocampus are first separated from the multi-atlas MRI images of the MICCAI. The processed MICCAI labels are displayed in
Figure 8.
To reduce computation and conserve resources, the MICCAI dataset and the HarP dataset are cropped to 64 × 64 × 96 pixels with the hippocampus preserved. Because the datasets are too small, some commonly used data augmentation strategies that do not cause MRI resolution change or MRI distortion, such as random flipping and random rotation, are used to expand the two datasets. Random flipping makes the model learn hippocampus features in a broader direction, and random rotation improves the recognition ability of the model for the hippocampus at different angles. The HarP dataset is expanded from 135 groups to 540 groups. Among them, 400 groups are used for training, and 140 groups are used for validation. The MICCAI dataset is expanded from 35 groups to 140 groups. Among them, 100 groups are used for training, and 40 groups are used for validation.
In this research, the dice loss and the binary cross-entropy loss are combined for the loss function.The Adam optimizer with a weight decay of 0.0001 is utilized. The model is trained multiple times with different values of hyperparameters, and the results of each training step are recorded. Finally, the hyperparameter settings with the best performance are obtained. The spatial dropout rate is 0.1, and the early stopping epoch is set to 20. The hyperparameters used in the HarP experiment include a learning rate of 0.01, a batch size of 16, and a total of 50 epochs. The learning rate for the MICCAI dataset is set to 0.005, the batch size is set to 4, and the number of epochs is set to 50. The proposed CADyUNet is based on Pytorch, and all experiments in this research were performed on two NVIDIA Tesla GPUs, each with 14.8 GB of memory.
4.4. Experimental Results
To demonstrate the efficacy of CADyUNet on hippocampus segmentation tasks, some commonly used medical image segmentation models are selected to conduct comparison experiments on the HarP and MICCAI datasets, including 3D U-Net, Attention U-Net, UNETR, Swin UNETR, and our proposed model. The dice, the mIoU, and the F1 were used to analyze the experimental results.
Table 1 and
Table 2 display the results of the contrastive experiment on the HarP and MICCAI datasets, respectively.
As demonstrated by
Table 1 and
Table 2, CADyUNet segments the hippocampus more accurately in the hippocampus segmentation task compared to other models. Compared to the baseline, the dice on the HarP dataset rose by 3.52%, the mIoU rose by 2.65%, and the F1 rose by 3.38%. On the MICCAI dataset, the dice, mIoU, and F1 score rose by 1.13%, 0.85%, and 1.08%, respectively. The results illustrate the efficacy of CADyUNet in hippocampus segmentation tasks.
To show the hippocampus segmentation results of these algorithms more conveniently, the sagittal section, coronal section, and axial section segmentation results are provided in
Figure 9,
Figure 10, and
Figure 11, respectively.
The first column shows sections of the three dimensions of the input MRI image (axial, sagittal, and coronal), along with the corresponding hippocampus segmentation labels. The 3D U-Net segmentation results are displayed in the second column. The segmentation results obtained by the Attention U-Net, UNETR, Swin UNETR, and CADyUNet models are shown in the third, fourth, fifth, and last columns, respectively. As illustrated in these figures, in contrast to the outputs of other algorithms, the outputs of the CADyUNet model are closer to the standard segmentation labels (particularly for marginal hippocampus segmentation), which proves the efficacy of CADyUNet in hippocampus segmentation tasks.
To show the model’s efficacy more comprehensively, the Params, GFLOPs (giga floating-point operations), and FPS (frames per second) are also used to evaluate the model’s performance. The experimental results are presented in
Table 3. As shown in
Table 3, the proposed CADyUNet significantly reduces the model’s memory usage and computation usage while greatly increasing its inference speed compared with other models. The results presented in
Table 1,
Table 2 and
Table 3 show that CADyUNet has better segmentation accuracy and uses the fewest computing resources on hippocampus segmentation tasks, which proves the superiority of our model.
To identify the efficacy of the designed dy-block, the improved CA, and the introduction of the soft pool method in hippocampus segmentation tasks, five models are chosen to conduct ablation experiments on the HarP dataset and the MICCAI dataset: 3D U-Net, 3D U-Net + CA, 3D U-Net + improved CA, 3D U-Net + dy-lock, 3D U-Net + softpool, and CADyUNet. To preserve computing resources, the number of channels is reduced by four times based on the number of channels in the 3D U-Net model. Furthermore, the results of the ablation experiment are presented in
Table 4 and
Table 5. Several useful conclusions can be drawn from the results.
All of the indicators increase as a result of the improved CA mechanism being used in 3D U-Net’s skip connection. Furthermore, introducing the improved CA mechanism in 3D U-Net results in better performance than introducing the CA mechanism, demonstrating that the improved CA mechanism extracts more texture and background information through larger convolution kernels, as well as the mix of max-pool and average pool methods compared to the CA mechanism.
The introduction of the CA mechanism into the skip connection of 3D U-Net resulted in almost no increase in any of the indicators, indicating that the CA mechanism extracts many useless and redundant features to combine with the deep information result from the decoder, with a negative influence on the hippocampus segmentation accuracy in this work.
The introduction of the proposed dy-block in 3D U-Net leads to all the indicators significantly increasing because the use of dynamic convolution operations in the dy-block strengthens its representational ability compared to standard convolutional operations in the conv-block. In addition, the introduction of the softpool method greatly improves the model’s segmentation performance because the softpool causes less information loss in the downsampling steps compared to other commonly used pooling methods.
5. Conclusions
The hippocampus can reflect neurodegenerative conditions such as AD. However, the volume of the hippocampus is too small to segment accurately using U-Net. To deal with this problem, CADyUNet, a hippocampus segmentation model based on coordinate attention and dynamic convolution, is recommended. An improved coordinate attention mechanism is designed to reduce information loss and retain more critical texture details. The improved coordinate attention mechanism is introduced into 3D U-Net so that the network focuses on important features and suppresses redundant features. Additionally, a dynamic convolution block called dy-block is introduced to replace the ordinary convolutional block in 3D U-Net, which greatly increases the representational capability without expanding the network’s width and depth. Furthermore, the soft pooling method is used instead of max pooling to reduce information loss during downsampling. The experimental results obtained on the HarP and MICCAI datasets show that CADyUNet outperforms all other models on all metrics in comparison to the baseline, demonstrating the superiority of CADyUNet in hippocampus segmentation tasks.
6. Discussion
Compared with existing medical image segmentation algorithms, our model achieves higher accuracy and faster inference speed and uses fewer computational resources in hippocampus segmentation tasks, which indicates that it has potential clinical value in the medical imaging field. For example, it can be used to assist clinicians in the diagnosis and evaluation of hippocampus lesions, as well as the quantitative analysis of hippocampus structure in neuroscience research. However, there are some shortcomings associated with our research, one of which is the inadequacy of the datasets. Due to the limitations of MRI image collection and hippocampus labeling, we can only use a limited number of MRI images for training and evaluation, which may limit the model’s generalizability to broader datasets. The second limitation is the fixed size of the training data. Fixed-size training data may not fully cover hippocampus of different sizes and shapes. Thus, we propose some possible methods for future work. First, more hippocampus images can be collected labeled accurately to expand the dataset. Second, for hippocampus with different sizes, the introduction of adaptive segmentation methods should be considered so that the model can adapt to different sizes of images. In future work, we intend to solve these problems then apply the method in practice.