Machine Learning Algorithms Combining Slope Deceleration and Fetal Heart Rate Features to Predict Acidemia
Abstract
1. Introduction
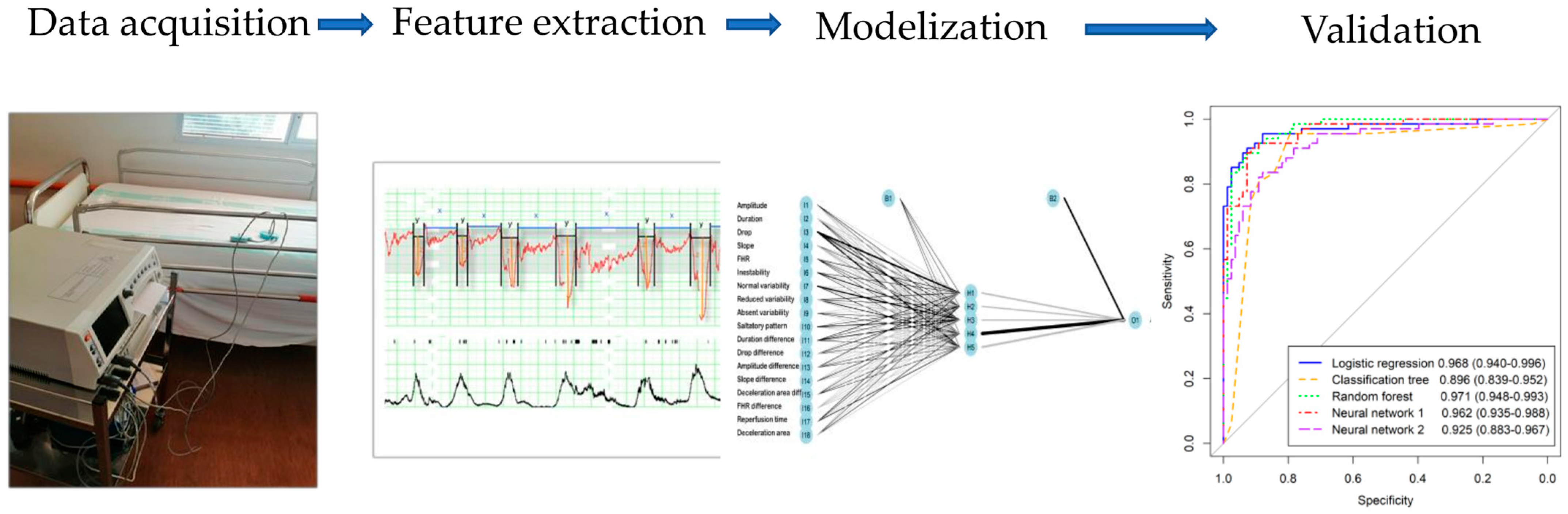
2. Material and Methods
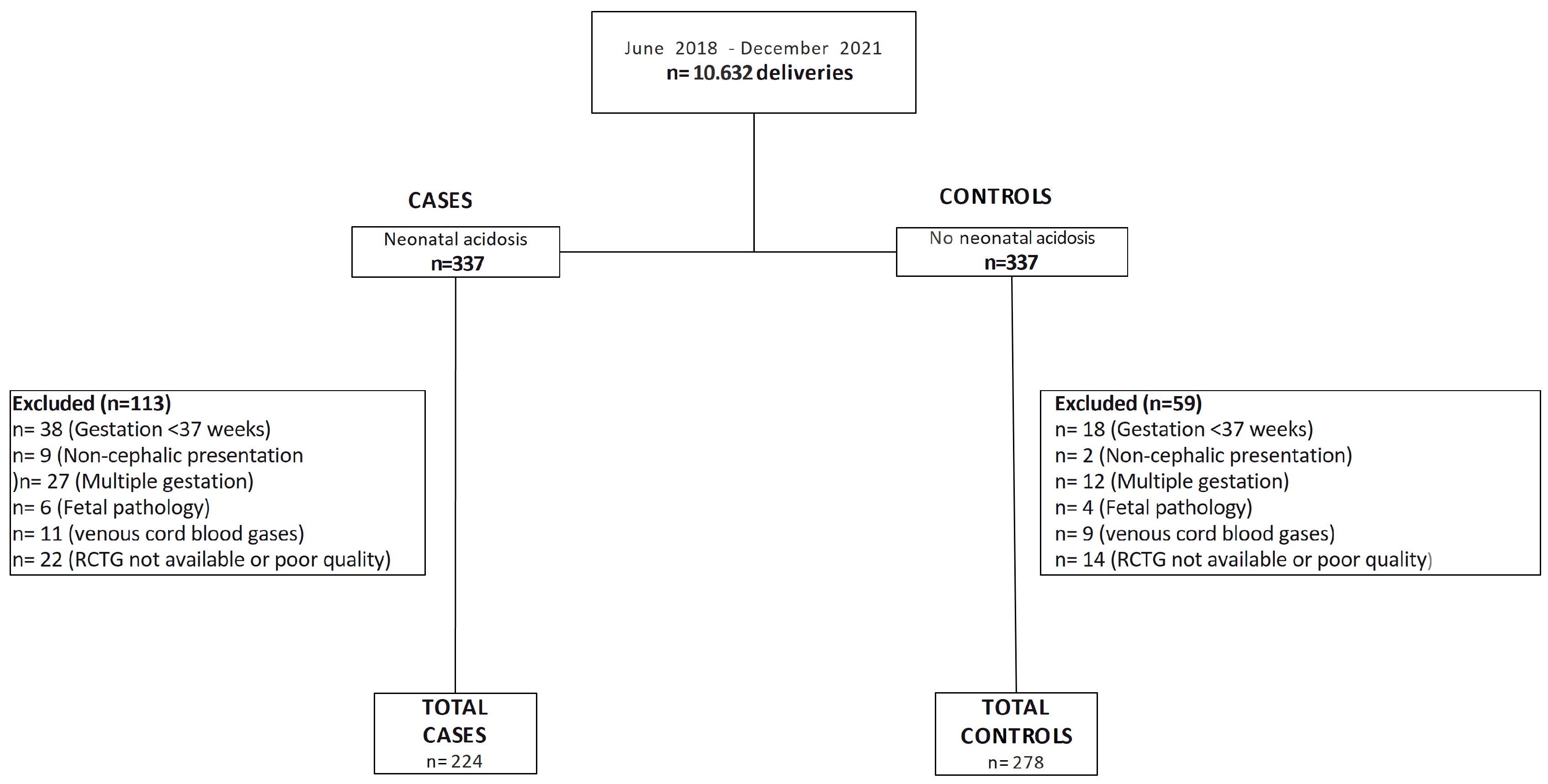
2.1. Data Recruitment
2.2. Electronic Fetal Monitoring
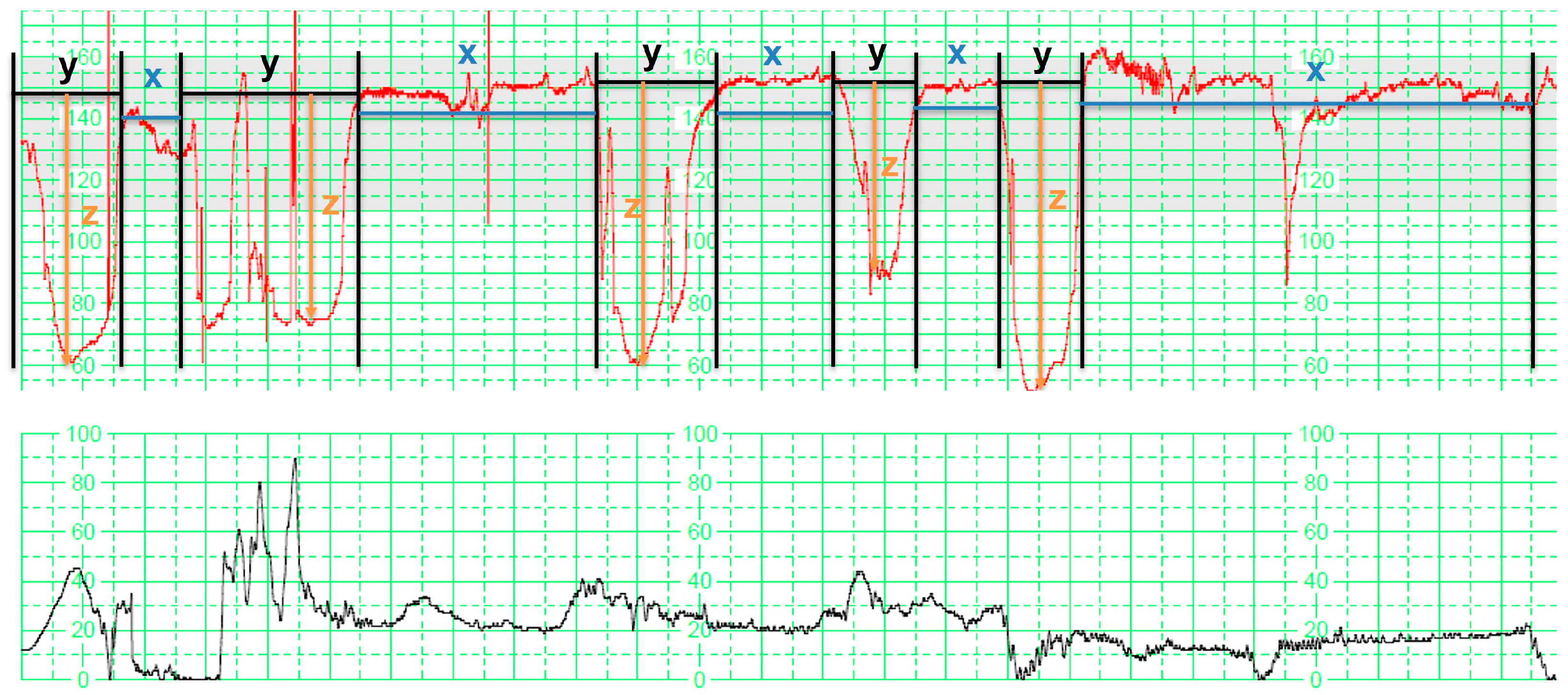
- Total reperfusion time: This parameter was calculated by adding up the duration, measured in minutes, during which the fetus maintained a baseline state without any deceleration within the last 30 min (), where is the total number of interdeceleration periods.
- Deceleration time: This parameter was determined by summing the duration, measured in minutes, of the period during which the fetus displayed decelerations within the last 30 min (), where is the total number of deceleration periods.
- Total deceleration area: This parameter was computed by summing the areas of all the decelerations. Each deceleration’s area was determined by multiplying the duration of the deceleration in seconds by its maximum depth of fall from the baseline, given in beats per minute, and dividing the result by two ().
2.3. Statistical Analysis
2.3.1. Model Building
2.3.2. Model Validation
3. Results
3.1. Descriptive Characteristics
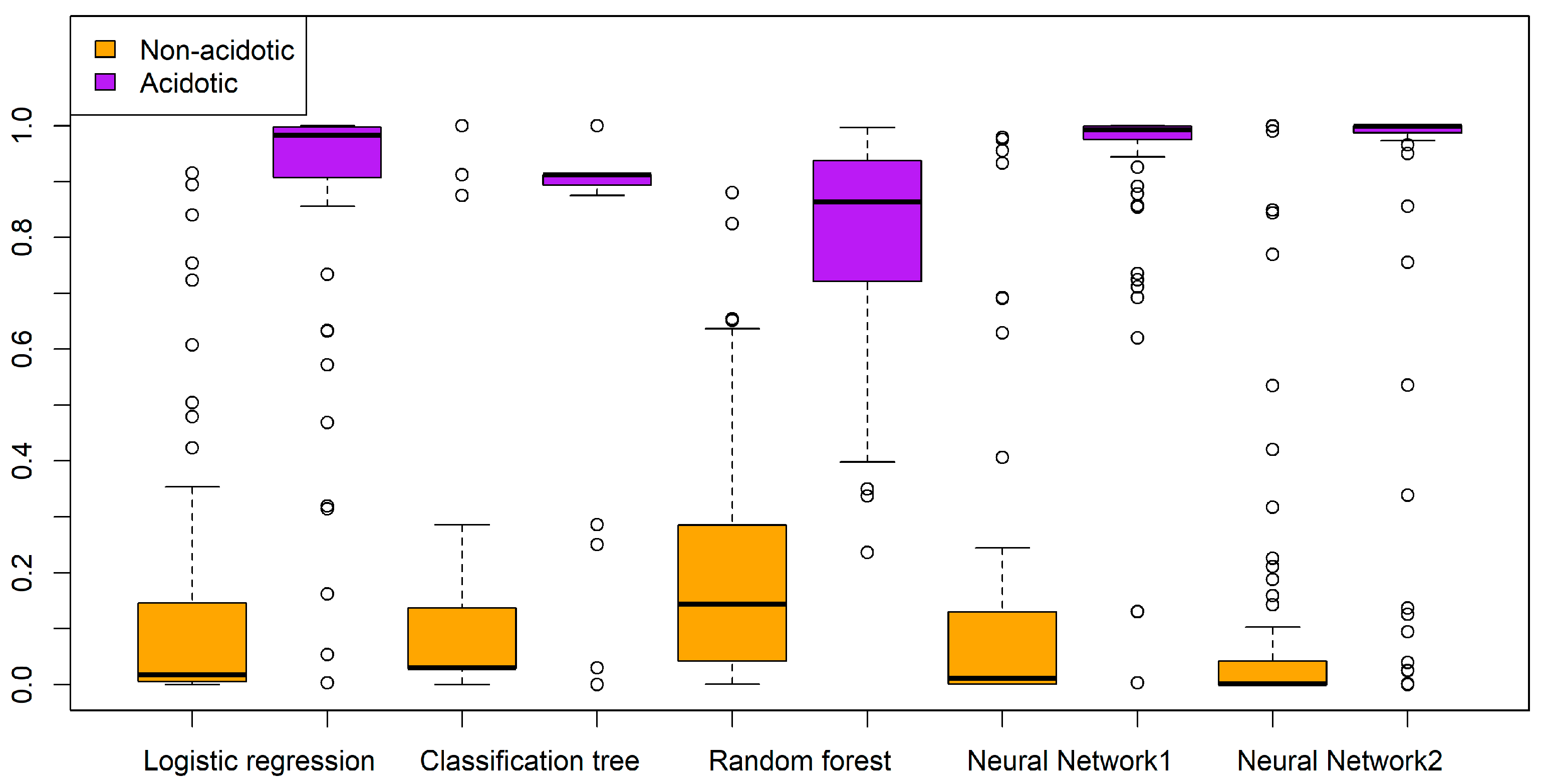
3.2. Multivariate Prediction Models
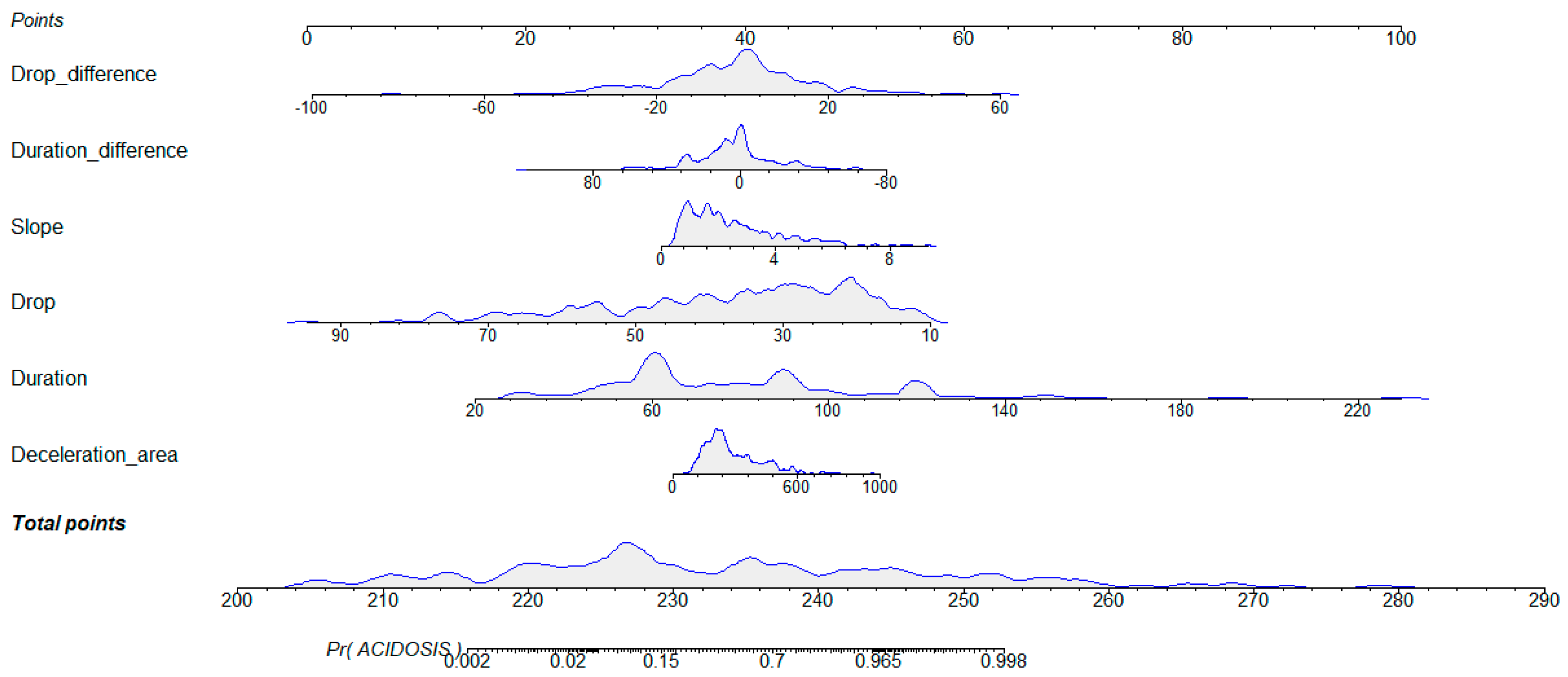
3.2.1. Logistic Regression
3.2.2. Classification Trees
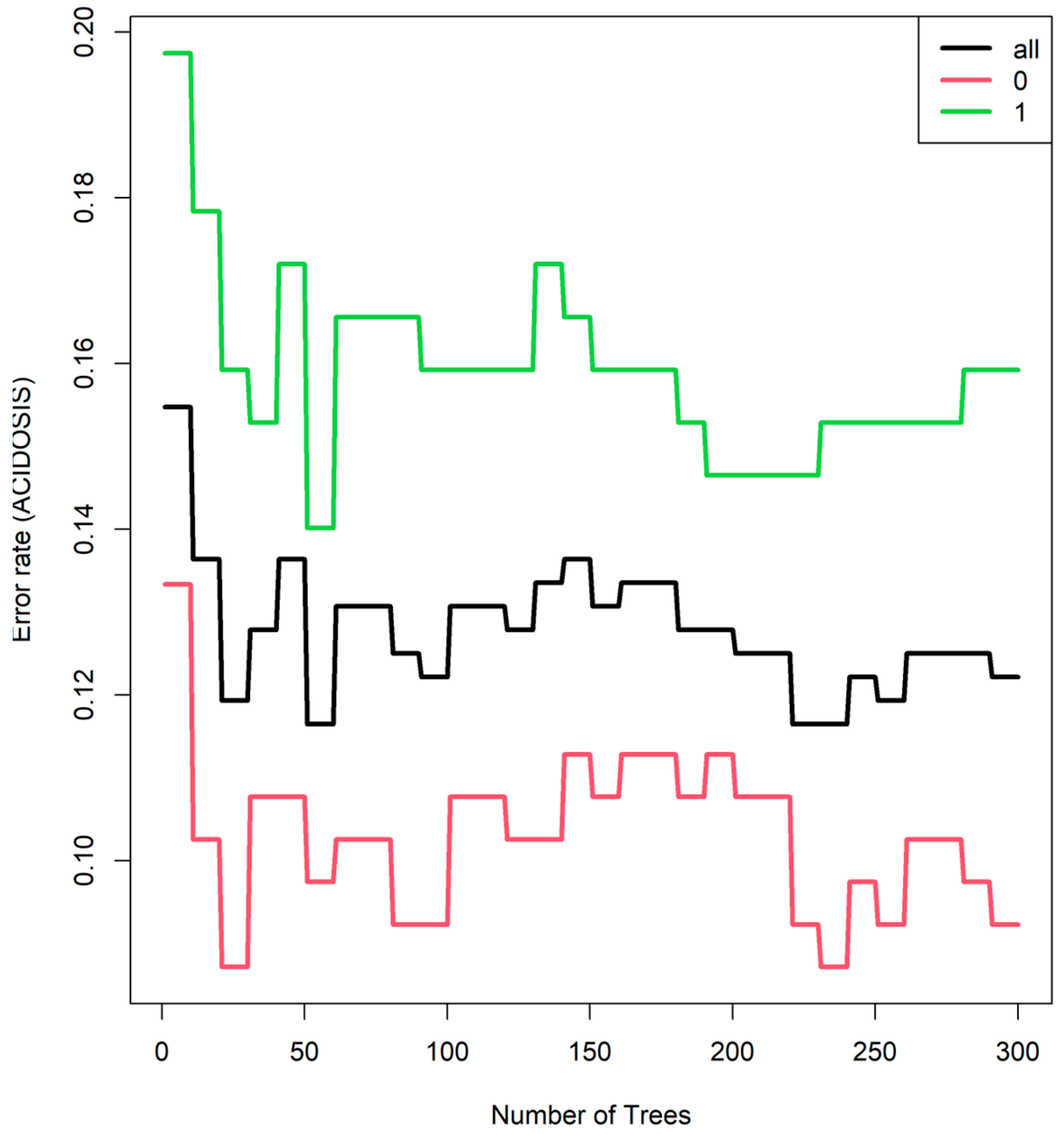
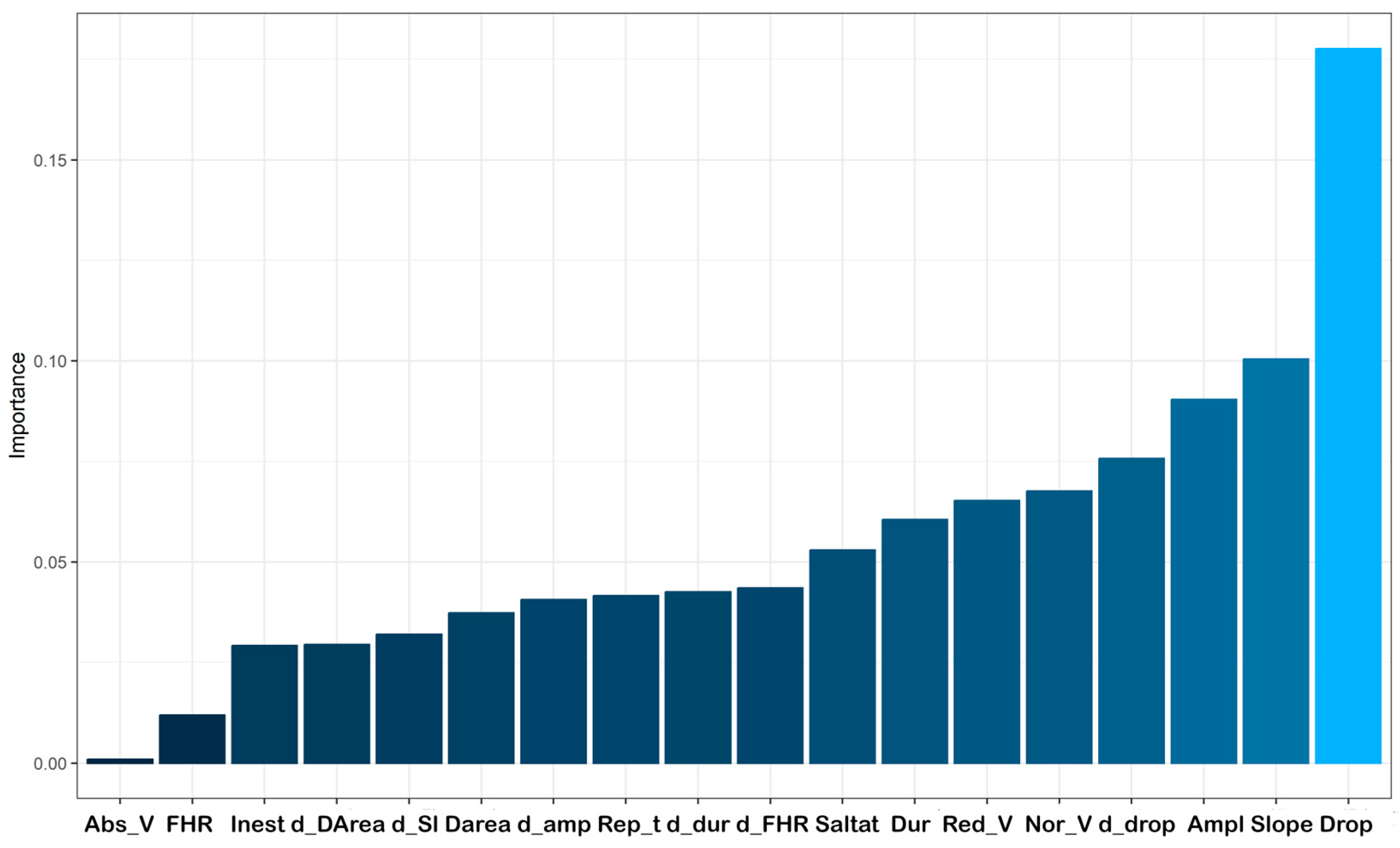
3.2.3. Random Forest
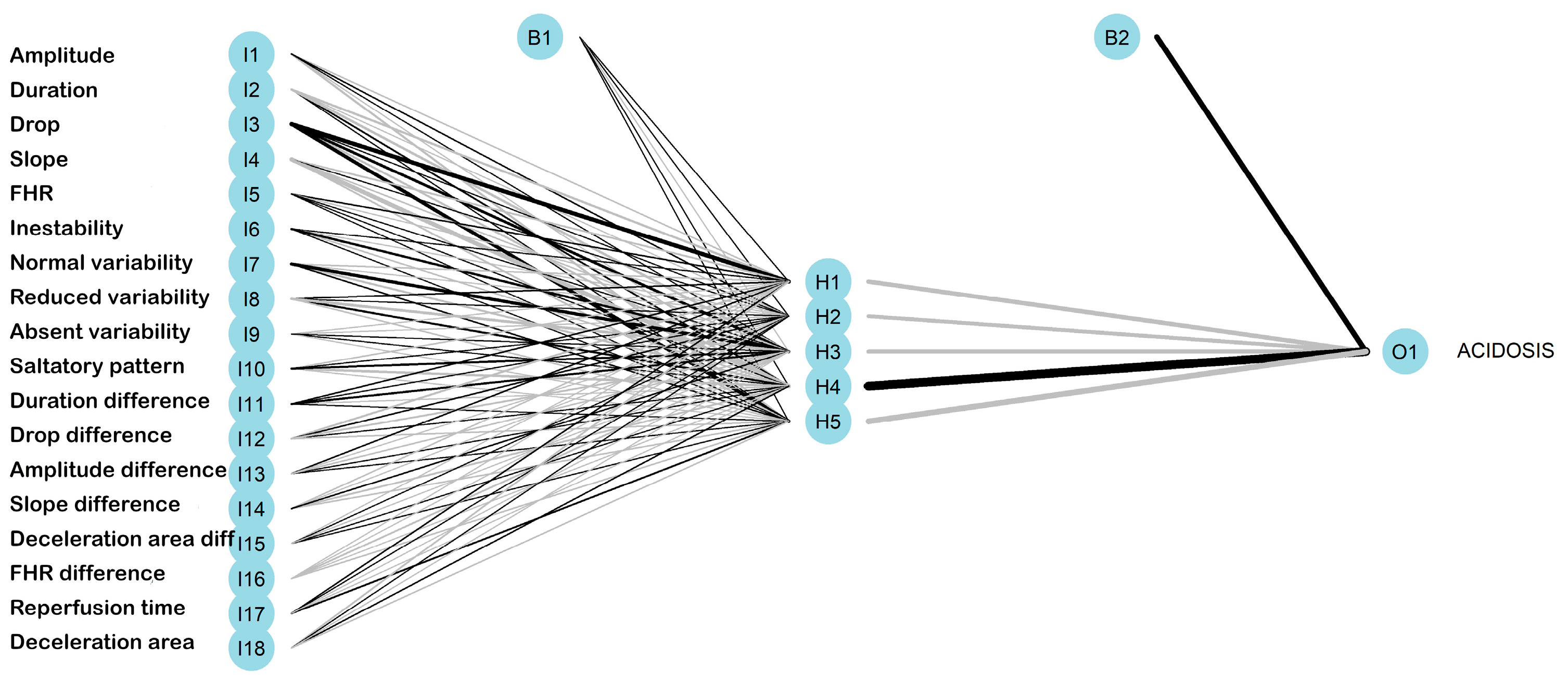
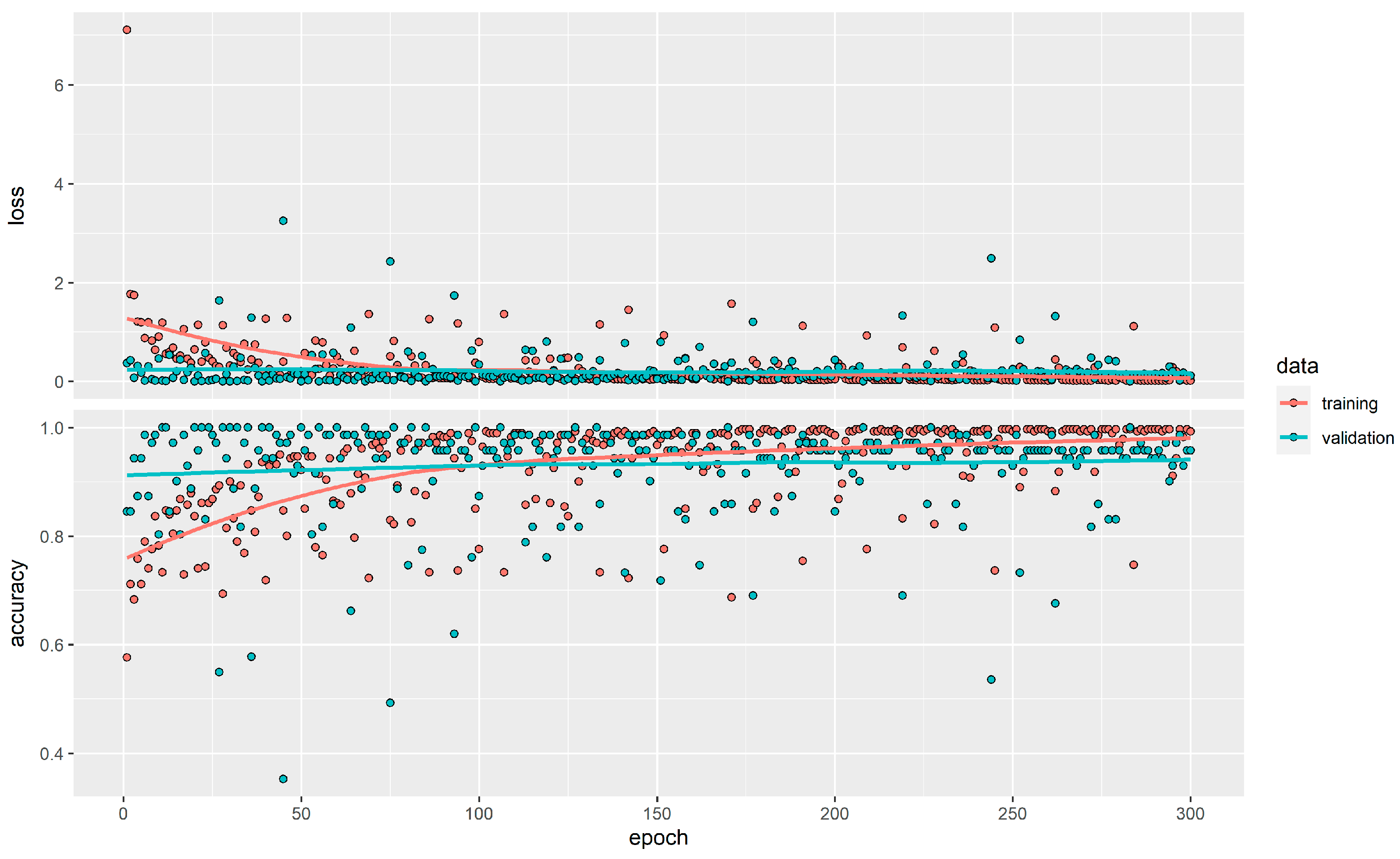
3.2.4. Neuronal Networks
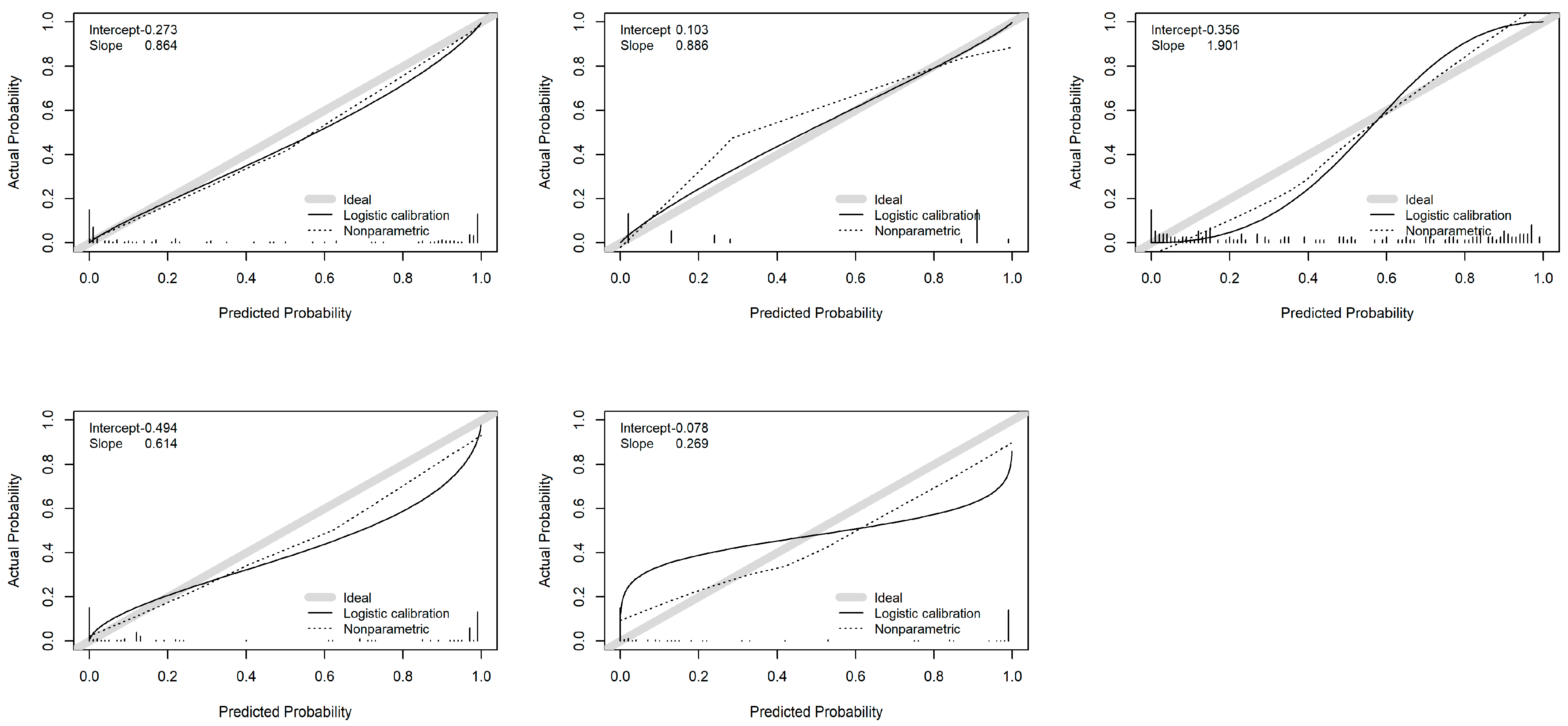
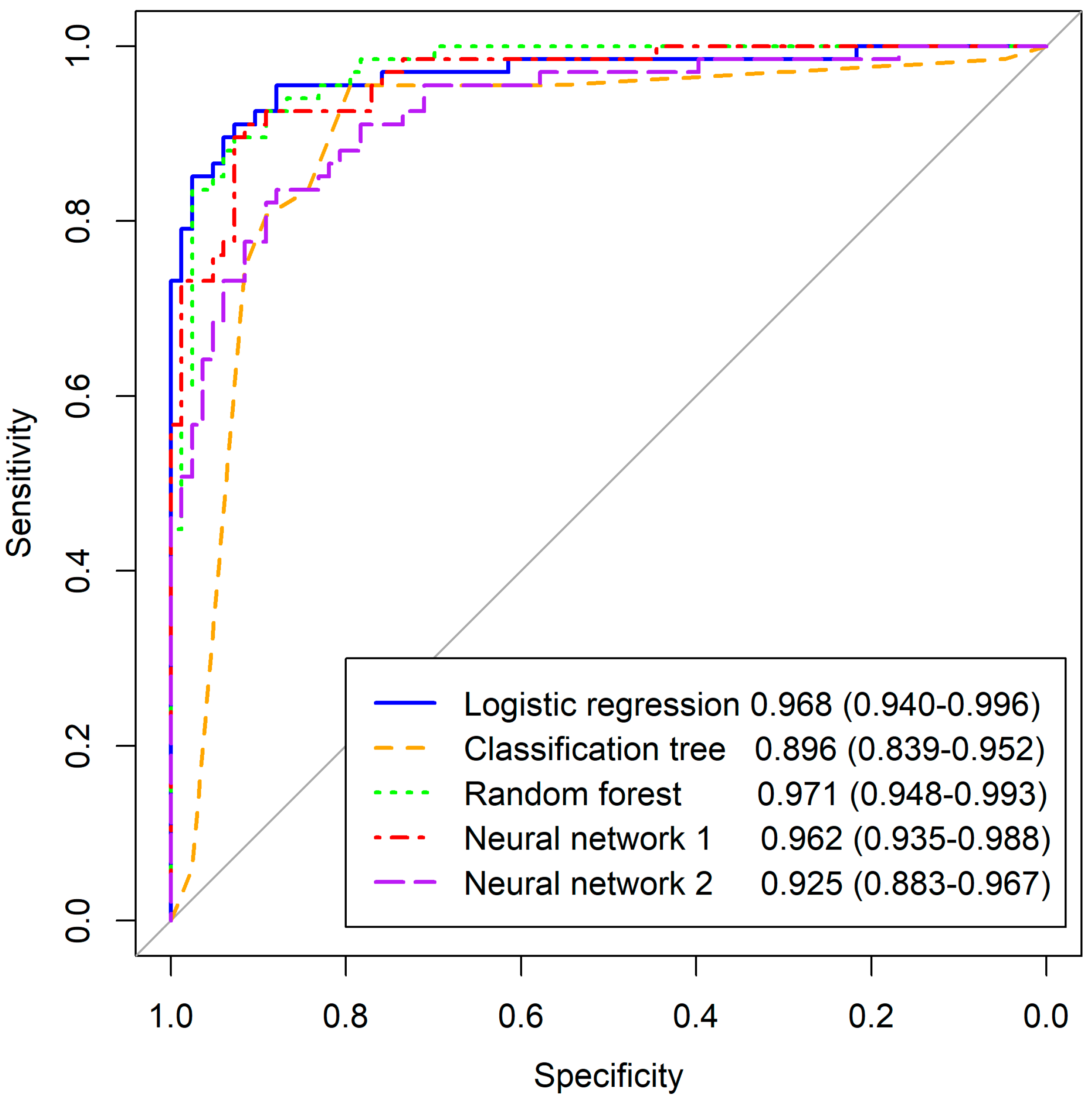
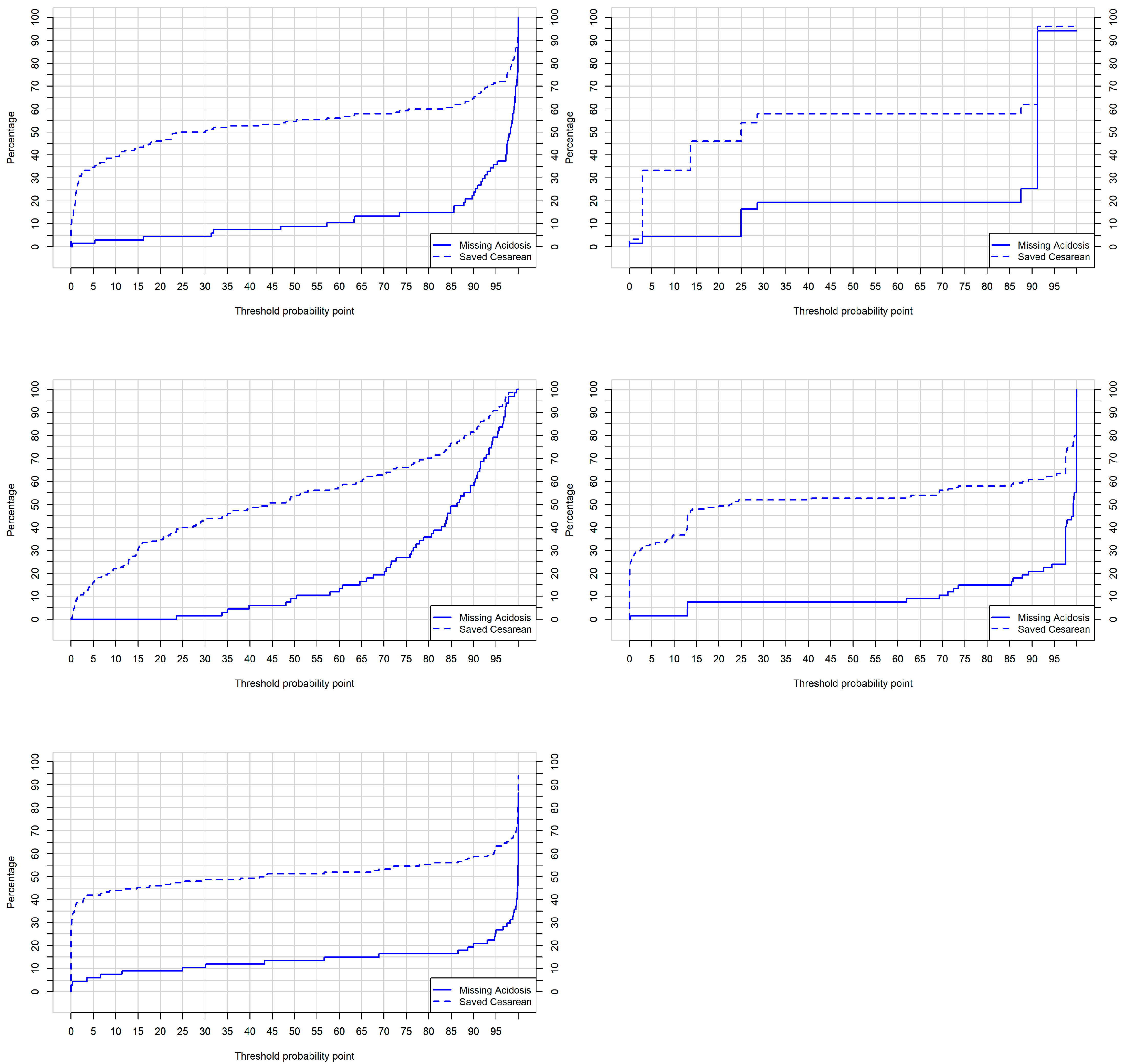
3.3. Validation of Developed Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nunes, I.; Ayres-de-Campos, D. Computer analysis of foetal monitoring signals. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 30, 68–78. [Google Scholar] [CrossRef]
- Clark, S.L.; Hamilton, E.F.; Garite, T.J.; Timmins, A.; Warrick, P.A.; Smith, S. The limits of electronic fetal heart rate monitoring in the prevention of neonatal metabolic acidemia. Am. J. Obstet. Gynecol. 2017, 216, 163.e1–163.e6. [Google Scholar] [CrossRef] [PubMed]
- American College of Obstetricians and Gynecologists. Fetal heart rate monitoring: Guidelines. ACOG Tech. Bull. 1974, 32, 1–10. [Google Scholar]
- American College of Obstetricians and Gynecologists. Practice bulletin no. 116: Management of intrapartum fetal heart rate tracings. Obstet. Gynecol. 2010, 116, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Zamora, C.; Chóliz, M.; Mejía, I.; Díaz de Terán, E.; Esteban, L.M.; Rivero, A.; Castán, B.; Andeyro, M.; Savirón, R. Diagnostic capacity and interobserver variability in FIGO, ACOG, NICE and Chandraharan cardiotocographic guidelines to predict neonatal acidemia. J. Matern. Fetal Neonatal Med. 2021, 80, 6479. [Google Scholar]
- Rei, M.; Tavares, S.; Pinto, P.; Machado, A.P.; Monteiro, S.; Costa, A.; Costa-Santos, C.; Bernardes, J.; Ayres-De-Campos, D. Interobserver agreement in CTG interpretation using the 2015 FIGO guidelines for intrapartum fetal monitoring. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 205, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Martí, S.; Lapresta, M.; Pascual, J.; Lapresta, C.; Castán, S. Deceleration area and fetal acidemia. J. Matern. Fetal Neonatal Med. 2017, 30, 2578–2584. [Google Scholar] [CrossRef]
- Cahill, A.G.; Tuuli, M.G.; Stout, M.J.; López, J.D.; Macones, G.A. A prospective cohort study of fetal heart rate monitoring: Deceleration area is predictive of fetal acidemia. Am. J. Obstet. Gynecol. 2018, 218, 523.e1–523.e12. [Google Scholar] [CrossRef]
- Chóliz, M.; Savirón, R.; Esteban, L.M.; Zamora, C.; Espiau, A.; Castán, B.; Castán Mateo, S. Total intrapartum fetal reperfusión time (fetal resilience) and neonatal acidemia. J. Matern. Fetal Neonatal Med. 2021, 91, 5977. [Google Scholar]
- Bennet, L.; Gunn, A.J. The fetal heart rate response to hypoxia: Insights from animal models. Clin. Perinatol. 2009, 36, 655–672. [Google Scholar] [CrossRef]
- Westgate, J.A.; Wibbens, B.; Bennet, L.; Wassink, G.; Parer, J.T.; Gunn, A.J. The intrapartum deceleration in center stage: A physiologic approach to the interpretation of fetal heart rate changes in labor. Am. J. Obstet. Gynecol. 2007, 197, e1–e11. [Google Scholar] [CrossRef] [PubMed]
- Sbrollini, A.; Agostinelli, A.; Marcantoni, I.; Morettini, M.; Burattini, L.; Di Nardo, F.; Fioretti, S.; Burattini, L. eCTG: An automatic procedure to extract digital cardiotocographic signals from digital images. Comput. Methods Programs Biomed. 2018, 156, 133–139. [Google Scholar] [CrossRef]
- Doret, M.; Helgason, H.; Abry, P.; Goncalves, P.; Gharib, C.; Gaucherand, P. Multifractal analysis of fetal heart rate variability in fetuses with and without severe acidosis during labor. Am. J. Perinatol. 2011, 28, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Doret, M.; Spilka, J.; Chudáček, V.; Gonçalves, P.; Abry, P. Fractal analysis and hurst parameter for intrapartum fetal heart rate variability analysis: A versatile alternative to frequency bands and LF/HF ratio. PLoS ONE 2015, 10, e0136661. [Google Scholar] [CrossRef]
- Cömert, Z.; Kocamaz, A.F. Open-access software for analysis of fetal heart rate signals. Biomed. Signal Process. Control 2018, 45, 98–108. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.; Deng, Y. A comprehensive feature analysis of the fetal heart rate signal for the intelligent assessment of fetal state. J. Clin. Med. 2018, 7, 223. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Escaño, J.; Castán, B.; Castán, S.; Chóliz-Ezquerro, M.; Asensio, C.; Laliena, A.R.; Sanz-Enguita, G.; Sanz, G.; Savirón, R. Machine learning algorithm to predict acidemia using electronic fetal monitoring recording parameters. Entropy 2022, 24, 68. [Google Scholar] [CrossRef]
- Ayres-de-Campos, D.; Nogueira-Reis, Z. Technical characteristics of current cardiotocographic monitors. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 30, 22–32. [Google Scholar] [CrossRef]
- Docker, M. Doppler ultrasound monitoring technology. BJOG Int. J. Obstet. Gynaecol. 1993, 100, 18–20. [Google Scholar] [CrossRef]
- Nunes, I.; Ayres-de-Campos, D.; Figueiredo, C.; Bernardes, J. An overview of central fetal monitoring systems in labour. J. Perinat. Med. 2013, 41, 93–99. [Google Scholar] [CrossRef]
- Da Silva Neto, M.G.; Do Vale Madeiro, J.P.; Gomes, D.G. On designing a biosignal-based fetal state assessment system: A systematic mapping study. Comput. Methods Programs Biomed. 2022, 216, 106671. [Google Scholar] [CrossRef]
- Comert, Z.; Kocamaz, A.F. A novel software for comprehensive analysis of cardiotocography signals “ctg-oas”. In Proceedings of the 2017 International Artificial Intelligence and Data Processing Symposium (IDAP), Malatya, Turkey, 16–17 September 2017; pp. 1–6. [Google Scholar]
- Anisha, M.; Kumar, S.S.; Nithila, E.E.; Benisha, M. Detection of fetal cardiac anomaly from composite abdominal electrocardiogram. Biomed. Signal Process. Control 2021, 65, 102308. [Google Scholar]
- Zhao, Z.; Zhang, Y.; Comert, Z.; Deng, Y. Computer-aided diagnosis system of fetal hypoxia incorporating recurrence plot with convolutional neural network. Front. Physiol. 2019, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Alsaggaf, W.; Cömert, Z.; Nour, M.; Polat, K.; Brdesee, H.; Toğaçar, M. Predicting fetal hypoxia using common spatial pattern and machine learning from cardiotocography signals. Appl. Acoust. 2020, 167, 107429. [Google Scholar] [CrossRef]
- Barquero-Pérez, Ó.; Santiago-Mozos, R.; Lillo-Castellano, J.M.; García-Viruete, B.; Goya-Esteban, R.; Caamaño, A.J.; Rojo-Alvarez, J.R.; Martín-Caballero, C. Fetal heart rate analysis for automatic detection of perinatal hypoxia using normalized compression distance and machine learning. Front. Physiol. 2017, 8, 113. [Google Scholar] [CrossRef]
- Cömert, Z.; Kocamaz, A.F.; Subha, V. Prognostic model based on image-based time-frequency features and genetic algorithm for fetal hypoxia assessment. Comput. Biol. Med. 2018, 99, 85–97. [Google Scholar] [CrossRef]
- Das, S.; Obaidullah, S.M.; Santosh, K.C.; Roy, K.; Saha, C.K. Cardiotocograph-based labor stage classification from uterine contraction pressure during ante-partum and intra-partum period: A fuzzy theoretic approach. Health Inf. Sci. Systems. 2020, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Deng, Y.; Zhang, Y.; Zhang, Y.; Zhang, X.; Shao, L. DeepFHR: Intelligent prediction of fetal Acidemia using fetal heart rate signals based on convolutional neural network. BMC Med. Inform. Decis. Mak. 2019, 19, 286. [Google Scholar] [CrossRef]
- Cömert, Z.; Şengür, A.; Budak, Ü.; Kocamaz, A.F. Prediction of intrapartum fetal hypoxia considering feature selection algorithms and machine learning models. Health Inf. Sci. Syst. 2019, 7, 17. [Google Scholar] [CrossRef]
- Petrozziello, A.; Redman, C.W.G.; Papageorghiou, A.T.; Jordanov, I.; Georgieva, A. Multimodal convolutional neural networks to detect fetal compromise during labor and delivery. IEEE Access 2019, 7, 112026–112036. [Google Scholar] [CrossRef]
- Cömert, Z.; Kocamaz, A.F. Fetal hypoxia detection based on deep convolutional neural network with transfer learning approach. In Software Engineering and Algorithms in Intelligent Systems, Proceedings of the CSOC2018 Computer Science Online Conference, Online, 25–28 April 2018; Silhavy, R., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 239–248. [Google Scholar]
- Feng, G.; Quirk, J.G.; Djurić, P.M. Supervised and unsupervised learning of fetal heart rate tracings with deep gaussian processes. In Proceedings of the 2018 14th Symposium on Neural Networks and Applications (NEUREL), Belgrade, Serbia, 20–21 November 2018; pp. 1–6. [Google Scholar]
- Fergus, P.; Chalmers, C.; Montanez, C.C.; Reilly, D.; Lisboa, P.; Pineles, B. Modelling segmented cardiotocography time-series signals using one-dimensional convolutional neural networks for the early detection of abnormal birth outcomes. IEEE Trans. Emerg. Top. Comput. Intell. 2020, 5, 882–892. [Google Scholar] [CrossRef]
- Gao, W.; Lu, Y. Fetal heart baseline extraction and classification based on deep learning. In Proceedings of the 2019 International Conference on Information Technology and Computer Application (ITCA), Guangzhou, China, 20–22 December 2019; pp. 211–216. [Google Scholar]
- Ma’sum, M.A.; Riskyana Dewi Intan, P.; Jatmiko, W.; Krisnadhi, A.A.; Setiawan, N.A.; Suarjaya, I.M.A.D. Improving deep learning classifier for fetus hypoxia detection in cardiotocography signal. In Proceedings of the 2019 International Workshop on Big Data and Information Security (IWBIS), Bali, Indonesia, 11 October 2019; pp. 51–56. [Google Scholar]
- Iraji, M.S. Prediction of fetal state from the cardiotocogram recordings using neural network models. Artif. Intell. Med. 2019, 96, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, T.; Li, M.; Yang, X. The design and implementation of cardiotocography signals classification algorithm based on neural network. Comput. Math. Methods Med. 2018, 2018, 8568617. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.r-project.org/ (accessed on 15 May 2023).
- Steyerberg, E.W.; Van Calster, B.; Pencina, M.J. Performance measures for prediction models and markers: Evaluation of predictions and classifications. Rev. Esp. Cardiol. 2011, 64, 788–794. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Borque-Fernando, Á.; Esteban-Escaño, L.M.; Rubio-Briones, J.; Lou-Mercadé, A.C.; García-Ruiz, R.; Tejero-Sánchez, A.; Muñoz-Rivero, M.V.; Cabañuz-Plo, T.; Alfaro-Torres, J.; Marquina-Ibáñez, I.M. A preliminary study of the ability of the 4Kscore test, the Prostate Cancer Prevention Trial-Risk Calculator and the European Research Screening Prostate-Risk Calculator for predicting high-grade prostate cancer. Actas Urológicas Españolas 2016, 40, 155–163. [Google Scholar] [CrossRef]
- Ishwaran, H.; Lu, M. Standard errors and confidence intervals for variable importance in random forest regression, classification, and survival. Stat. Med. 2019, 38, 558–582. [Google Scholar] [CrossRef] [PubMed]
- Garson, G.D. Interpreting neural network connection weights. Artif. Intell. Expert 1991, 6, 46–51. [Google Scholar]
- Shelley, T.; Tipton, R.H. Dip area. A quantitative measure of fetal heart rate patterns. BJOG Int. J. Obstet. Gynaecol. 1971, 78, 694–701. [Google Scholar] [CrossRef]
- Beguin, F.; Yeh, S.Y.; Forsythe, A.; Hon, E.H. A study of fetal heart rate deceleration areas II. Correlation between deceleration areas and fetal pH during labor. Obstet. Gynecol. 1975, 45, 292–298. [Google Scholar]
- Tranquilli, A.L.; Biagini, A.; Greco, P.; Di Tommaso, M.; Giannubilo, S.R. The correlation between fetal bradycardia area in the second stage of labor and acidemia at birth. J. Matern. Fetal Neonatal Med. 2013, 26, 1425–1429. [Google Scholar] [CrossRef]
- Hamilton, E.; Warrick, P.; O’Keeffe, D. Variable decelerations: Do size and shape matter? J. Matern. Fetal Neonatal Med. 2012, 25, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Neilson, D.; Hamilton, E. Cumulative deceleration area: A simplified predictor of metabolic acidemia. J. Matern. Neonatal. Med. 2021, 19, 3104–3111. [Google Scholar] [CrossRef] [PubMed]
- Garabedian, C.; De Jonckheere, J.; Butruille, L.; Deruelle, P.; Storme, L.; Houfflin-Debarge, V. Understanding fetal physiology and second line monitoring during labor. J. Gynecol. Obstet. Hum. Reprod. 2017, 46, 113–117. [Google Scholar] [CrossRef]
- Sartwelle, T.P.; Johnston, J.C. Continuous electronic fetal monitoring during labor: A critique and a reply to contemporary proponents. Surg. J. 2018, 4, e23–e28. [Google Scholar] [CrossRef] [PubMed]
- Balayla, J.; Shrem, G. Use of artificial intelligence (AI) in the interpretation of intrapartum fetal heart rate (FHR) tracings: A systematic review and meta-analyssis. Arch. Gynecol. Obstet. 2019, 300, 7–14. [Google Scholar] [CrossRef]




| Total Sample n = 502 | Non-Acidotic n = 278 | Acidotic n = 224 | p-Value | |
|---|---|---|---|---|
| Maternal age | 34 (30–37) | 34 (30–36) | 34 (30–37) | 0.440 |
| Nulliparity | 302 (60.16%) | 148 (53.24%) | 149 (66.52%) | <0.001 |
| Age gestational | 280.5 (274–286) | 280 (273.75–286) | 281 (274.25–286) | 0.205 |
| Sex | 0.87 | |||
| Male | 262 (52.19%) | 146 (52.51) | 116 (51.78) | |
| Female | 240 (47.81%) | 132 (47.48) | 108 (48.21) | |
| Newborn percentile weight | 3251.74 (471.07) | 3281.09 (454.45) | 3215.31 (489.34) | 0.123 |
| p < 10 | 73 (14.54) | 30 (10.79%) | 43 (15.47%) | 0.011 |
| >90 | 60 (11.95) | 33 (11.87%) | 27 (12.05%) | 0.999 |
| Birth eutocic | 303 (60.36%) | 193 (69.42%) | 110 (49.11%) | <0.001 |
| Instrumental delivery | 121 (24.1%) | 60 (21.58%) | 61 (27.23%) | 0.141 |
| Cesarean section | 78 (15.54%) | 25 (8.99%) | 53 (23.66%) | <0.001 |
| Apgar 5 m < 7 | 36 (7.17%) | 4 (1.44%) | 32 (14.29) | <0.001 |
| Umbilical arterial pH | 7.12 (7.07–7.20) | 7.18 (7.14–7.27) | 7.06 (7.01–7.09) | <0.001 |
| Umbilical venous pH | 7.18 (7.13–7.22) | 7.23 (7.18–7.26) | 7.15 (7.09–7.19) | <0.001 |
| Total n = 502 | Non Acidotic n = 278 | Acidotic n = 224 | p-Value | Odds Ratio | AUC | |
|---|---|---|---|---|---|---|
| 30 min window | ||||||
| Deceleration range | 14.16 (7.71, 22.54) | 9.71 (5.37, 15.42) | 20.93 (13.75, 28.65) | <0.001 | 1.141 (1.112, 1.171) | 0.807 |
| Reperfusion time | 19.35 (15.37, 23.20) | 21.69 (18.10, 25.12) | 17.04 (13.86, 19.10) | <0.001 | 0.822 (0.787, 0.859) | 0.750 |
| Final deceleration window | ||||||
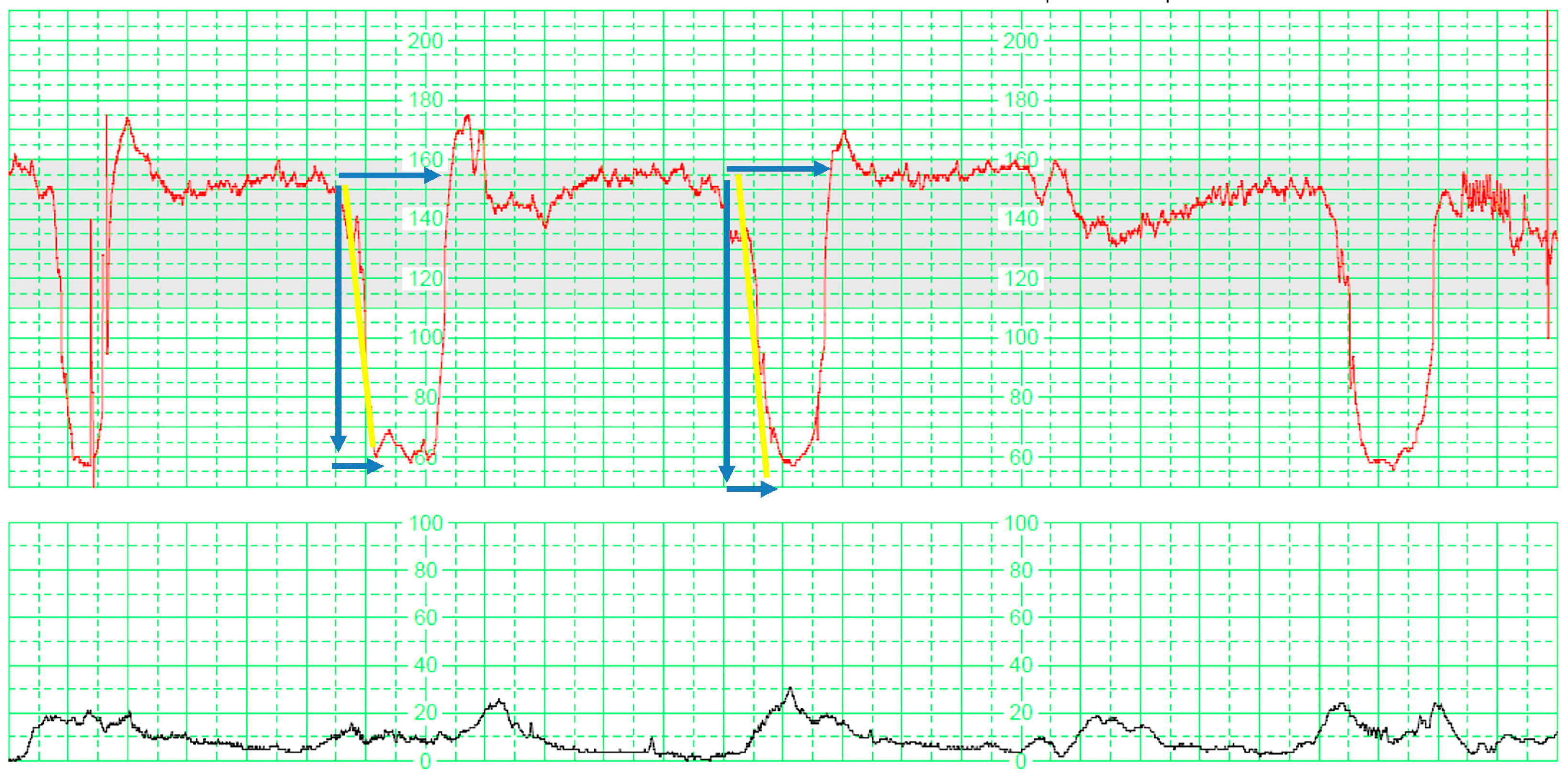
| Amplitude | 72.99 (54.94, 87.04) | 60.06 (49.96, 74.92) | 84.87 (74.18, 95.11) | <0.001 | 1.063 (1.050, 1.076) | 0.796 |
| Duration | 71.25 (59.88, 90.69) | 61.8 (55.68, 89.92) | 80.9 (64.32, 100.14) | <0.001 | 1.023 (1.015, 1.030) | 0.670 |
| Drop | 32.63 (23.26, 46.51) | 40.9 (30.79, 53.47) | 23.96 (19.18, 32.32) | <0.001 | 0.933 (0.918, 0.947) | 0.792 |
| Slope | 2.08 (1.31, 3.47) | 1.57 (1.01, 2.05) | 3.51 (2.49, 4.69) | <0.001 | 3.331 (2.681, 4.138) | 0.853 |
| Area | 2.51 (1.89, 3.71) | 2.15 (1.6, 2.72) | 3.55 (2.45, 4.69) | <0.001 | 2.470 (2.048, 2.979) | 0.781 |
| FHR (bpm) | 155 (145, 165) | 150 (140, 160) | 160 (150, 170) | <0.001 | 1.043 (1.029, 1.057) | 0.664 |
| Overshoot | 67 (13.35%) | 24 (8.63%) | 43 (19.19%) | 0.001 | 2.514 (1.473, 4.291) | 0.552 |
| Inestability | 188 (37.45%) | 50 (17.98%) | 138 (61.61%) | <0.001 | 7.317 (4.867, 11.000) | 0.718 |
| Reduced variability | 110 (21.9%) | 38 (13.67%) | 72 (32.14%) | <0.001 | 2.992 (1.922, 4.656) | 0.592 |
| Initial window | ||||||
| Amplitude (bpm) | 6.12 (−1.97, 19.63) | 4.16 (−4.11, 13.11) | 12.32 (1.32, 24.38) | <0.001 | 1.023 (1.013, 1.034) | 0.631 |
| Duration (s) | 2.15 (−7.31, 14.44) | 1.31 (−4.71, 13.45) | 3.90 (−8.76, 23.55) | 0.136 | 1.008 (1.001, 1.015) | 0.538 |
| Drop (sg) | −1.41 (−10.84, 6.24) | −1.62 (−12.47, 8.11) | −0.78 (−8.72, 5.04) | 0.611 | 1.001(0.991, 1.012) | 0.513 |
| Slope (bpm/sg) | 0.29 (−0.24, 1.01) | 0.24 (−0.22, 0.59) | 0.64 (−0.29, 1.70) | <0.001 | 1.325 (1.154, 1.520) | 0.599 |
| Area (mm2) | 32.41 (−19.11, 99.12) | 17.20 (−25.10, 54.57) | 70.28 (−1.05, 163.74) | <0.001 | 1.004 (1.003, 1.006) | 0.657 |
| FHR (bpm) | 0 (0, 10) | 0 (0, 5) | 5 (0, 15) | <0.001 | 1.006 (1.004, 1.008) | 0.651 |
| Saltatory Pattern | 88 (17.52%) | 32 (11.51%) | 56 (23.14%) | <0.001 | 2.643 (1.506. 4.638) | 0.572 |
| Odds Ratio (95% CI) | p-Value | |
|---|---|---|
| 30 min window | ||
| Deceleration area | 1.121 (1.064, 1.189) | <0.001 |
| Final deceleration window | ||
| Duration (s) | 1.157 (1.117, 1.209) | <0.001 |
| Drop (s) | 0.809 (0.743, 0.871) | <0.001 |
| Slope (bpm/sg) | 2.814 (1.541, 5.523) | 0.001 |
| Difference between Final and Initial deceleration window | ||
| Duration (s) | 0.950 (0.925, 0.974) | <0.001 |
| (s) | 1.133 (1.088, 1.188) | <0.001 |
| Specificities | Logistic Regression | Classification Tree | Random Forest | Neural Network 1 | Neural Network 2 |
|---|---|---|---|---|---|
| 0.80 | 0.976 | 0.894 | 0.976 | 0.928 | 0.892 |
| 0.85 | 0.976 | 0.837 | 0.952 | 0.928 | 0.831 |
| 0.90 | 0.928 | 0.817 | 0.892 | 0.916 | 0.783 |
| 0.95 | 0.879 | 0.797 | 0.831 | 0.771 | 0.710 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteban, L.M.; Castán, B.; Esteban-Escaño, J.; Sanz-Enguita, G.; Laliena, A.R.; Lou-Mercadé, A.C.; Chóliz-Ezquerro, M.; Castán, S.; Savirón-Cornudella, R. Machine Learning Algorithms Combining Slope Deceleration and Fetal Heart Rate Features to Predict Acidemia. Appl. Sci. 2023, 13, 7478. https://doi.org/10.3390/app13137478
Esteban LM, Castán B, Esteban-Escaño J, Sanz-Enguita G, Laliena AR, Lou-Mercadé AC, Chóliz-Ezquerro M, Castán S, Savirón-Cornudella R. Machine Learning Algorithms Combining Slope Deceleration and Fetal Heart Rate Features to Predict Acidemia. Applied Sciences. 2023; 13(13):7478. https://doi.org/10.3390/app13137478
Chicago/Turabian StyleEsteban, Luis Mariano, Berta Castán, Javier Esteban-Escaño, Gerardo Sanz-Enguita, Antonio R. Laliena, Ana Cristina Lou-Mercadé, Marta Chóliz-Ezquerro, Sergio Castán, and Ricardo Savirón-Cornudella. 2023. "Machine Learning Algorithms Combining Slope Deceleration and Fetal Heart Rate Features to Predict Acidemia" Applied Sciences 13, no. 13: 7478. https://doi.org/10.3390/app13137478
APA StyleEsteban, L. M., Castán, B., Esteban-Escaño, J., Sanz-Enguita, G., Laliena, A. R., Lou-Mercadé, A. C., Chóliz-Ezquerro, M., Castán, S., & Savirón-Cornudella, R. (2023). Machine Learning Algorithms Combining Slope Deceleration and Fetal Heart Rate Features to Predict Acidemia. Applied Sciences, 13(13), 7478. https://doi.org/10.3390/app13137478







