MIDOM—A DICOM-Based Medical Image Communication System
Abstract
1. Introduction
- A framework for exchange of DICOM-based medical images between remote medical professionals in heterogeneous environments with closed and isolated PACS systems and varying connectivity;
- Reduction of transmission latency and cost through integration of our customized lossless image compression method (CBPC) into the system in question;
- A system for the increase in the availability of medical care in underdeveloped parts of the world, dependent on low-grade mobile networks.
2. Background
2.1. DICOM
- Universal standard of digital medicine—for its data and image transmission and storage.
- High image quality—up to 16 bits monochrome images.
- Support for various image-acquisition parameters—much information about the image is also stored, such as size, orientation, and number of layers.
- Encoding of medical data—it defines more than 2000 standardized attributes, such as patients name, age, or diagnosis.
- Well described imaging devices—it precisely describes the device and its functionality.
2.2. CBPC
- Predictive model,
- Entropy coder.
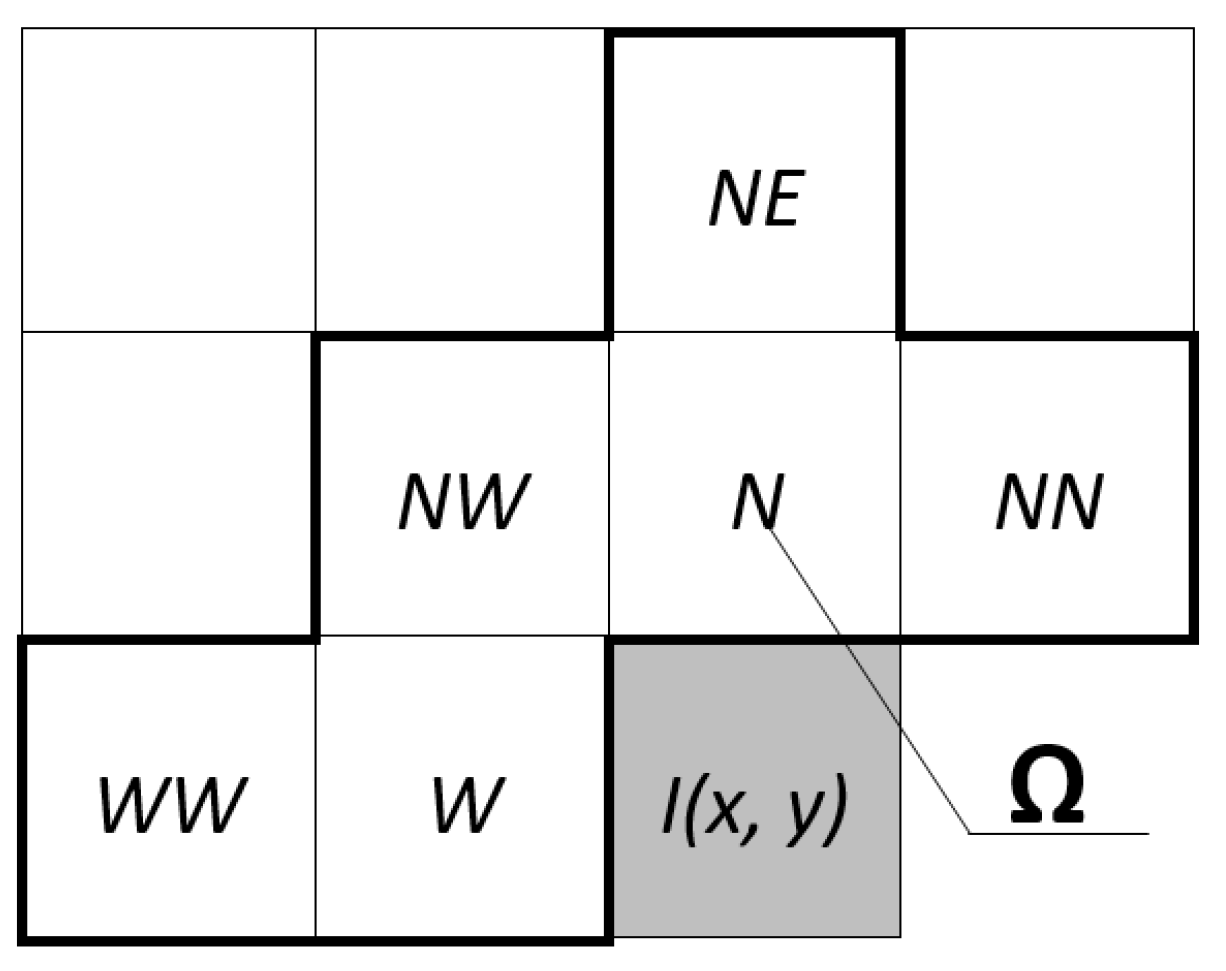
2.2.1. Predictive Model
- N—returns value of the image element positioned north (up) from the current element.
- W—returns value of the image element positioned west (left) from the current element.
- NW—returns value of the image element positioned northwest (up-left) from the current element.
- NE—returns value of the image element positioned northeast (up-right) from the current element.
- GradW—returns result of calculating .
- GradN—returns result of calculating .
- L-JPEG4—returns result of calculating .
- MED—returns result according to the function displayed in Equation (1).
- GAP—chooses one of the previously mentioned functions based on the horizontal and vertical gradient around the current element.
- FPG—combines neighbouring elements based on the approximation of the gradients from the context.
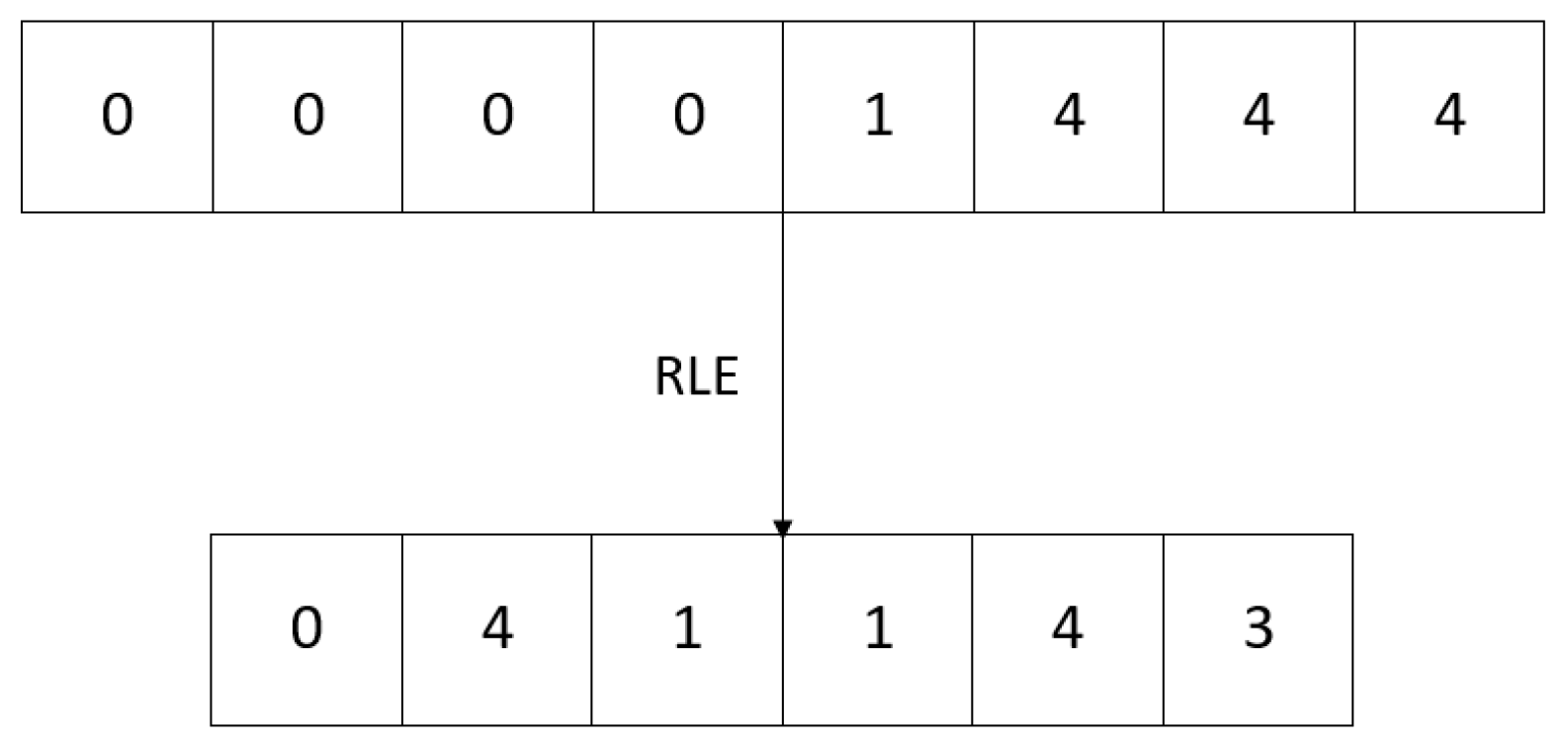
2.2.2. Entropy Coder
2.3. CBPC2
2.4. Related Work
3. MIDOM
3.1. Overview
- Study—A basic set of diagnostic data created with the purpose of exchange in the system;
- Consultation Request (CR)—A request for medical advice, opinion, or feedback for the existing study;
- Administrator—A system user tasked with user administration, management of system settings, and application-specific tasks;
- Study Provider (SP)—A physician or other medical professional who is in charge of creating medical studies and issuing consultation requests for medical specialists;
- Medical Specialist (MS)—A physician or domain expert who can accept or reject consultation requests and provide feedback to the requesting study provider.
- On-demand server-side compression of medical images of ultra-high-resolution—Algorithms for lossless compression of one or series of images comprising a study, adaptation of such algorithms for multicore and many-core systems, the performance of such algorithms for different compression levels;
- Client-side decompression of medical images of ultra-high-resolution—Capabilities of Android mobile platform for high storage and computing requirements posed by the application;
- Adaptation to current environmental characteristics—Strategies and techniques for different communication channel throughput and client platform capabilities. Adaptation mechanisms as a specific feature of MIDOM consider three main challenges, user experience, data transmission cost (in particular for clients connected through mobile networks), and throughput capabilities of client devices.
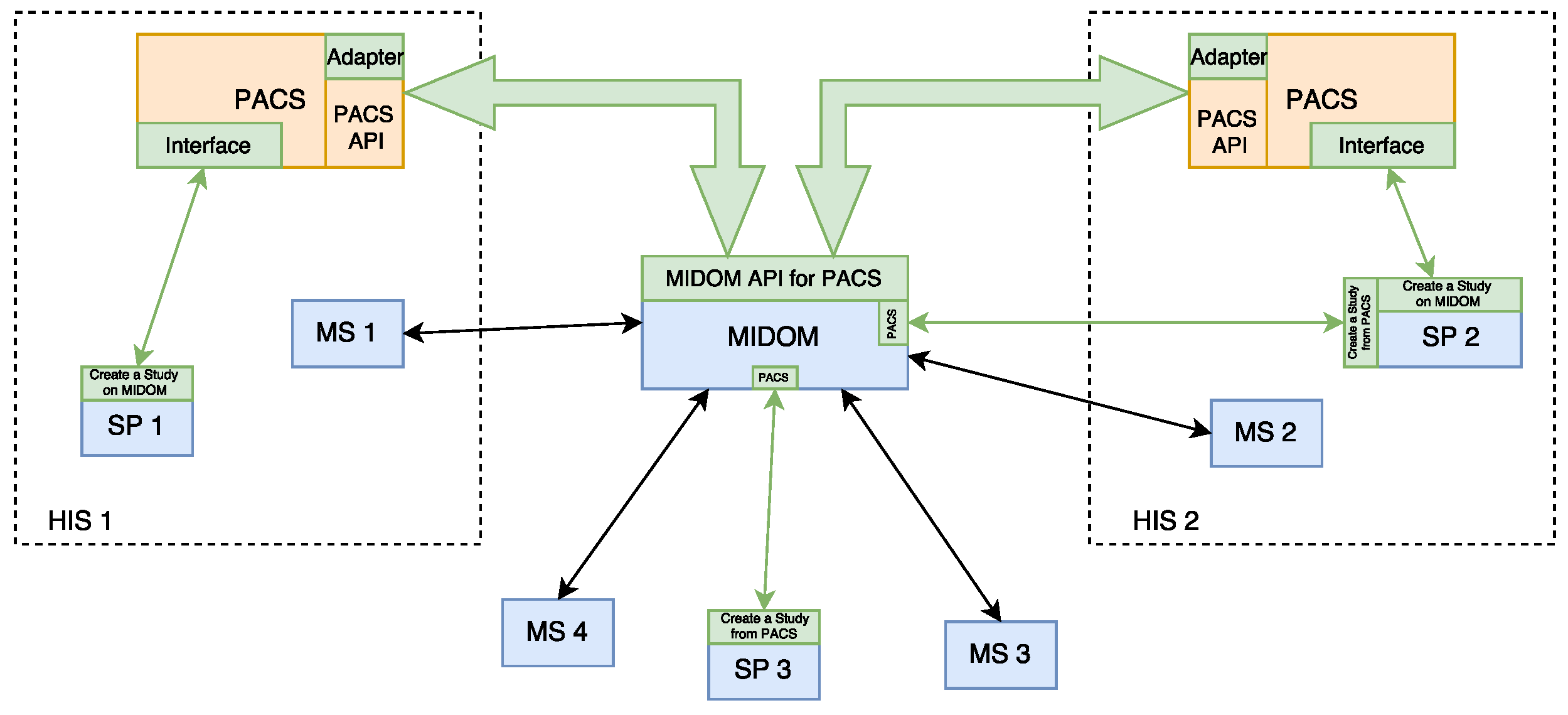
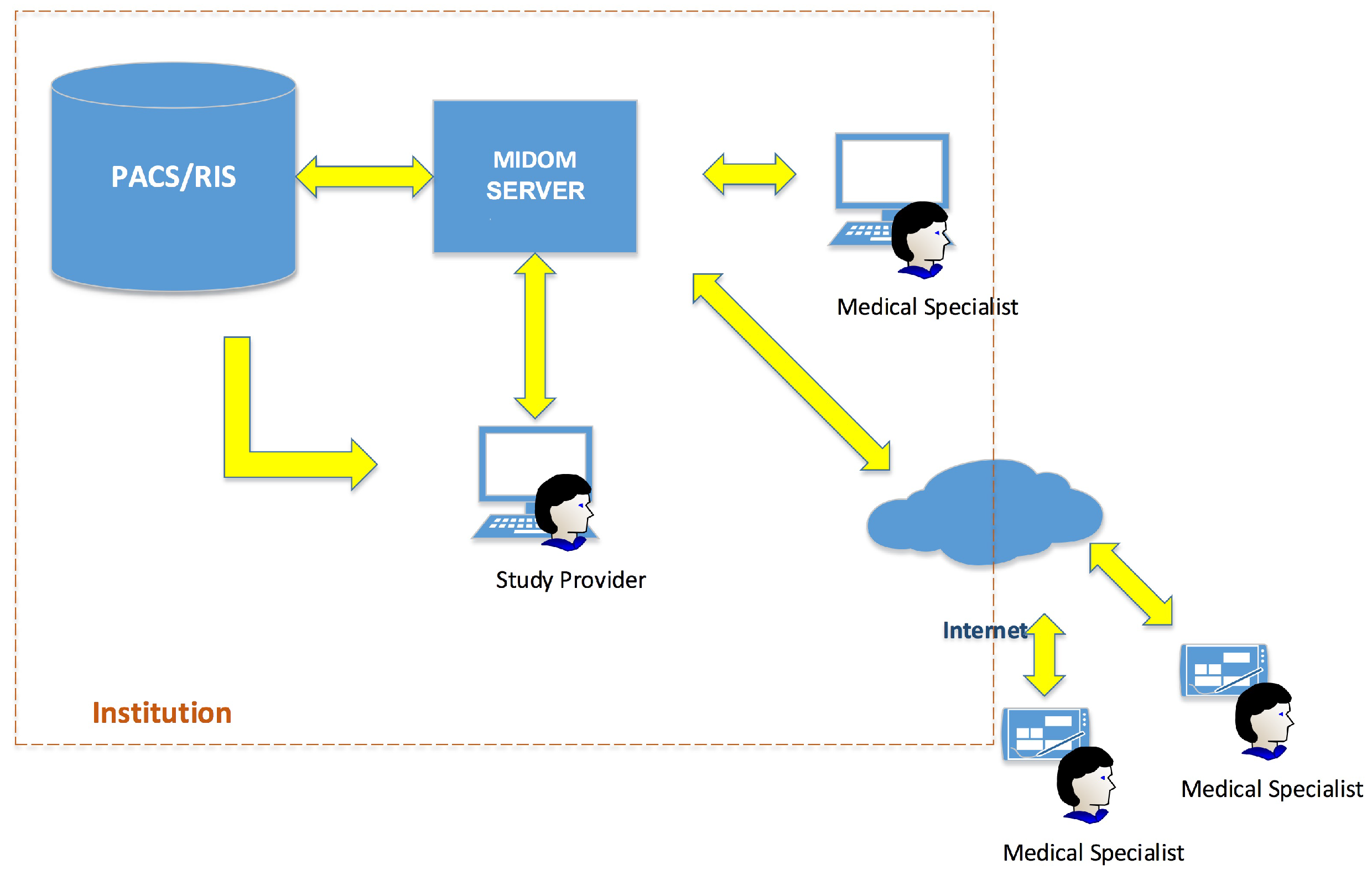
3.2. MIDOM Components
- MIDOM server—The main component which facilitates the collaboration of medical specialists on sets of diagnostic procedure results, i.e., studies. This component hosts studies and related multimedia data, mainly in high-resolution images, and keeps track of the statuses of each study and consultation request. It also provides methods for system administration and performance monitoring, and performs computationally and storage-intensive operations in order to adjust to communication and processing power characteristics of the medical specialist and devices transparently to the system users;
- PACS server—This component, originally not an integrated part of MIDOM, is used for storage purposes, for efficient medical data accessing and fetching. By attaching PACS component with MIDOM, fast access to a large number of medical images and their import for usage was enabled;
- Study Provider—A component used by medical professionals seeking consultations with other colleagues. It allows inspection, selection, and extraction of diagnostic imaging data from the PACS server to form a study. It also allows for requests for consultations and the collection of feedback returned by medical specialists. The study provider component can manage the study and associated consultation requests’ lifecycle. The main goals of this work have been developed through this component by enabling the study provider to access the PACS server and allowing it to create a study using image data from that server;
- Medical Provider—This component is usually implemented as a mobile part of the system. A medical specialist offers consultation services to the study provider in a standard flow. The study provider assigns consultation requests to a medical specialist then they seek further consultations about a particular study. A single study usually consists of medical images paired with their description and further explanation of the problem. A medical specialist can accept or reject a consultation request. If the consultation request is accepted, a medical specialist can inspect image data from the study for which the consultation has been requested and provide either textual or audio feedback. Medical specialists’ end devices can be connected through various types of networks, which can cause significant delays in terms of latency or time needed to pass studies through network channels. This aspect is thoroughly addressed in MIDOM by appropriate tooling and required logic for compressing exchanged imaging data to optimize user experience, throughput, and data communication costs associated with mobile networks.
4. Evaluation
4.1. Benchmarks and Methodology
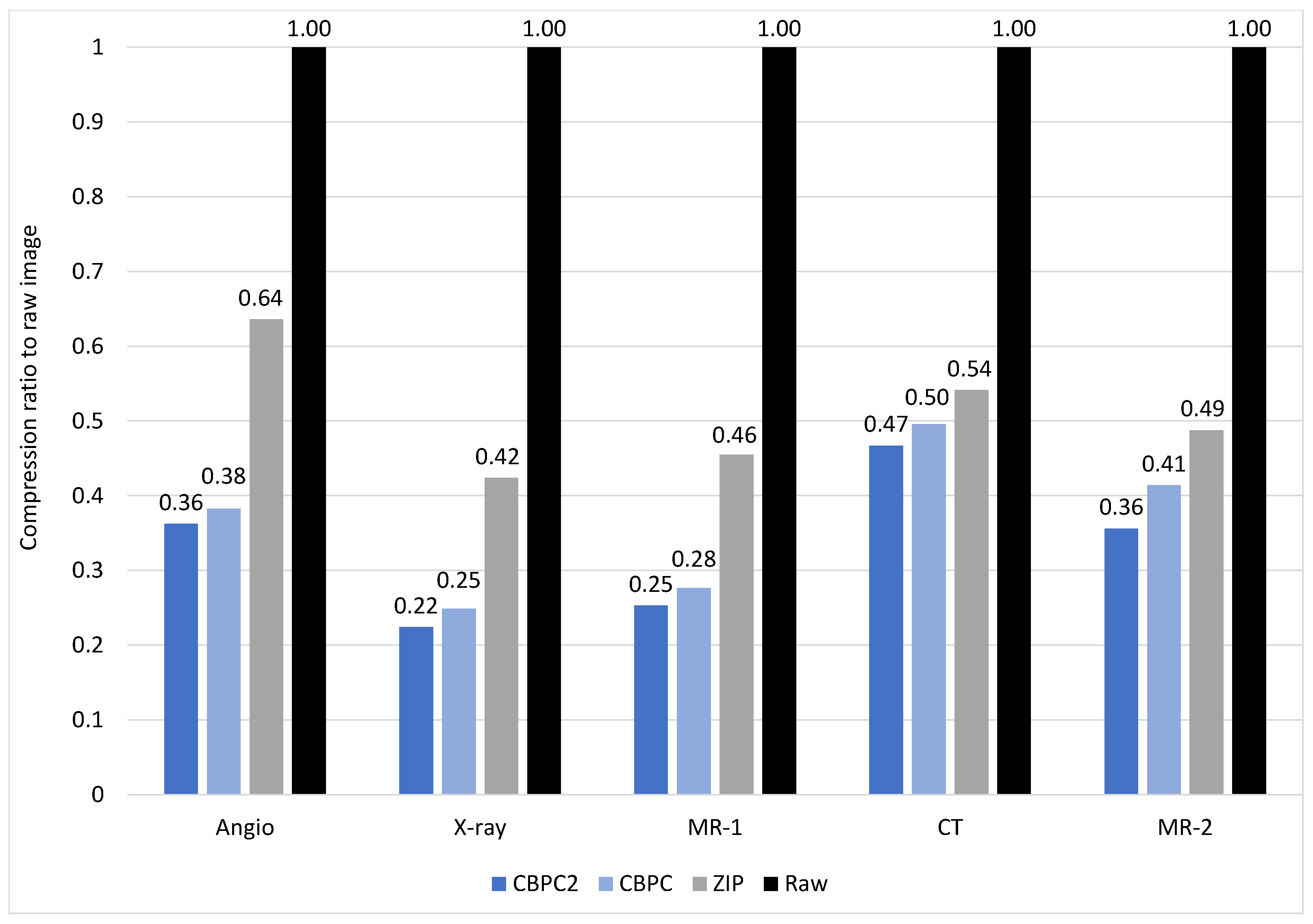
- Angio—One image, DICOM modality: XA, pixels in size, MB large;
- X-ray—Two images, DICOM modality: PX, and pixels in size, total of MB large;
- MR-1—383 images, DICOM modality: MR, pixels in size, total of MB large;
- CT—307 images, DICOM modality: CT, pixels in size, total of MB large;
- MR-2—2534 images, DICOM modality: MR, pixels in size, total of MB large.
4.2. Compression Ratio
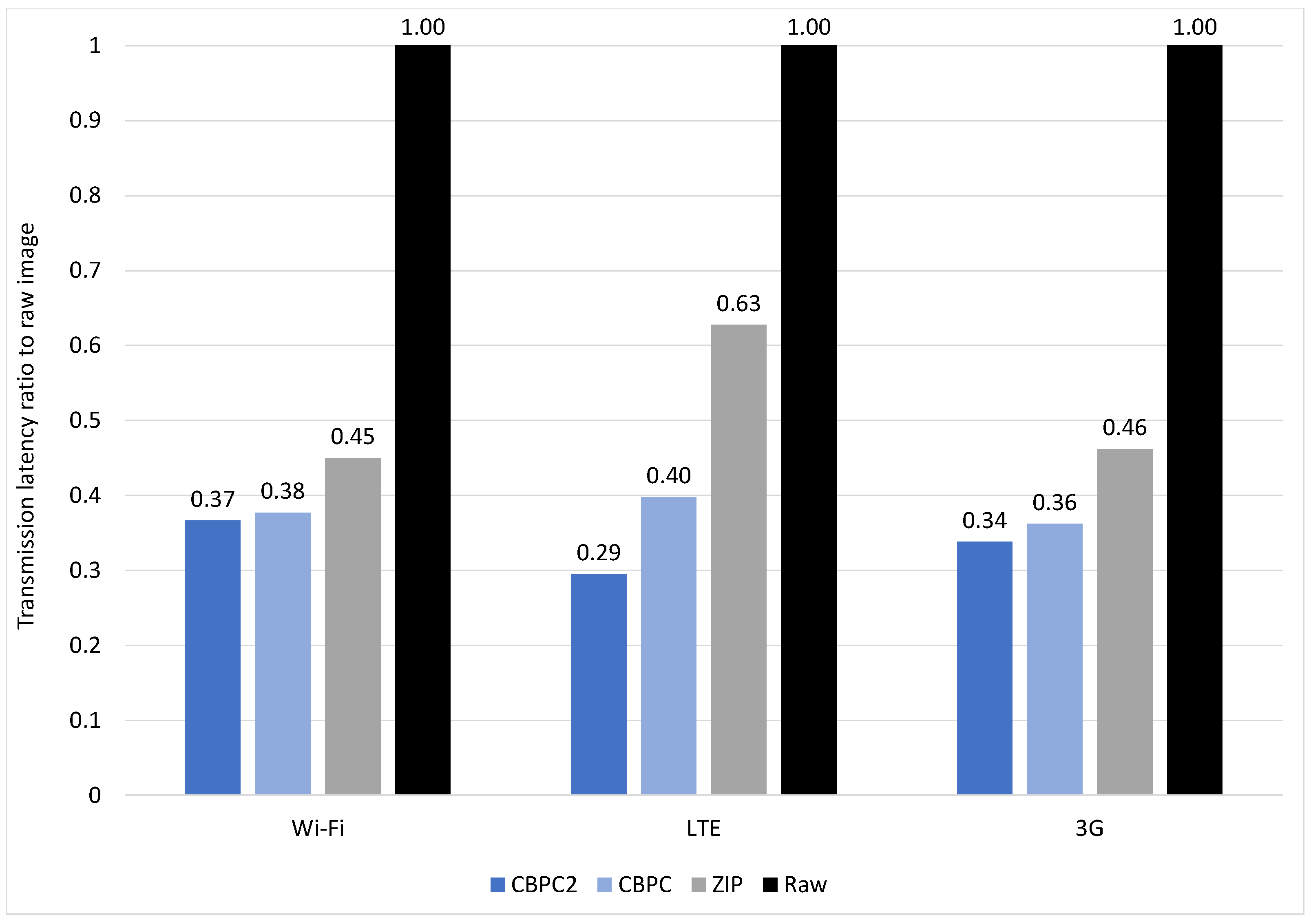
4.3. Latency Reduction
4.3.1. Angio
4.3.2. X-ray
4.3.3. MR-1
4.3.4. CT
4.3.5. MR-2
4.4. Discussion
5. Conclusions
5.1. Summary
5.2. Limitations
5.3. Suggestions for Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application Programming Interface |
| CBPC | Classification and Blending Predictor Coder |
| CR | Consultation Request |
| CT | Computed Tomography |
| DICOM | Digital Imaging and Communications in Medicine |
| GDPR | General Data Protection Regulation |
| HIPAA | Health Insurance Portability and Accountability Act |
| HIS | Hospital Information System |
| IOD | Information Object Definitions |
| MIDOM | Medical Imaging and Diagnostics on the Move |
| MR | Magnetic Resonance |
| MS | Medical Specialist |
| NEMA | National Electrical Manufacturers Association |
| OPT_JPEG_XT | Optimized JPEG-XT |
| PACS | Picture Archiving and Communication System |
| RLE | Run-Length Encoding |
| SP | Study Provider |
| UHL | University Hospital Limerick |
| VPN | Virtual Private Network |
References
- Li, H.; Yu, L.; He, W. The Impact of GDPR on Global Technology Development. J. Glob. Inf. Technol. Manag. 2019, 22, 1–6. [Google Scholar] [CrossRef]
- Jeong, Y.U.; Yoo, S.; Kim, Y.H.; Shim, W.H. De-identification of facial features in magnetic resonance images: Software development using deep learning technology. J. Med. Internet Res. 2020, 22, e22739. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Scott, R.E.; Mars, M. Whatsapp in clinical practice—The challenges of record keeping and storage. A scoping review. Int. J. Environ. Res. Public Health 2021, 18, 13426. [Google Scholar] [CrossRef]
- Ncube, B.; Mars, M.; Scott, R.E. The need for a telemedicine strategy for Botswana? A scoping review and situational assessment. BMC Health Serv. Res. 2020, 20, 794. [Google Scholar] [CrossRef]
- BBC News. WhatsApp Issued Second-Largest GDPR Fine of €225m. Available online: https://www.bbc.com/news/technology-58422465 (accessed on 8 May 2023).
- Mars, M.; Scott, R.E. Being Spontaneous: The Future of Telehealth Implementation? Telemed. J. Health Off. J. Am. Telemed. Assoc. 2017, 23, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Knezovic, J. Kompresija Slika Bez Gubitaka Metodom Predvidjanja; Faculty of Electrical Engineering and Computing, University of Zagreb: Zagreb, Croatia, 2005. [Google Scholar]
- Knezovic, J.; Kovac, M.; Mlinaric, H. Classification and Blending Prediction for Lossless Image Compression. In Proceedings of the MELECON 2006—2006 IEEE Mediterranean Electrotechnical Conference, Malaga, Spain, 16–19 May 2006; IEEE: Piscataway, NJ, USA, 2006; Volume 2006, pp. 486–489. [Google Scholar] [CrossRef]
- Strizic, L.; Knezovic, J. Optimization of losless image compression method for GPGPU. In Proceedings of the 2016 18th Mediterranean Electrotechnical Conference (MELECON), Lemesos, Cyprus, 18–20 April 2016; IEEE: Piscataway, NJ, USA, 2016. number April. pp. 1–6. [Google Scholar] [CrossRef]
- Liu, F.; Hernandez-Cabronero, M.; Sanchez, V.; Marcellin, M.W.; Bilgin, A. The current role of image compression standards in medical imaging. Information 2017, 8, 131. [Google Scholar] [CrossRef]
- van Ooijen, P.M.; Aryanto, K.Y.; Broekema, A.; Horii, S. DICOM data migration for PACS transition: Procedure and pitfalls. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1055–1064. [Google Scholar] [CrossRef]
- García-Vílchez, F.; Muñoz-Marí, J.; Zortea, M.; Blanes, I.; González-Ruiz, V.; Camps-Valls, G.; Plaza, A.; Serra-Sagristà, J. On the impact of lossy compression on hyperspectral image classification and unmixing. IEEE Geosci. Remote Sens. Lett. 2011, 8, 253–257. [Google Scholar] [CrossRef]
- Li, Z.; Ramos, A.; Li, Z.; Osborn, M.L.; Li, X.; Li, Y.; Yao, S.; Xu, J. An optimized JPEG-XT-based algorithm for the lossy and lossless compression of 16-bit depth medical image. Biomed. Signal Process. Control 2021, 64, 102306. [Google Scholar] [CrossRef]
- Li, Z.; Ramos, A.; Li, Z.; Osborn, M.L.; Zaid, W.; Li, X.; Li, Y.; Xu, J. Nearly-lossless-to-lossy medical image compression by the optimized JPEGXT and JPEG algorithms through the anatomical regions of interest. Biomed. Signal Process. Control 2023, 83, 104711. [Google Scholar] [CrossRef]
- Nassef, M.; Alkinani, M.H. A Novel Multilevel Lossy Compression Algorithm for Grayscale Images Inspired by the Synthesization of Biological Protein Sequences. IEEE Access 2021, 9, 149657–149680. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, Z.; Xiong, G. A New Image Compression Algorithm Based on Non-Uniform Partition and U-System. IEEE Trans. Multimed. 2021, 23, 1069–1082. [Google Scholar] [CrossRef]
- Kumar, S.N.; Fred, A.L.; Kumar, H.A.; Varghese, P.S. Lossless Compression of CT Images by an Improved Prediction Scheme Using Least Square Algorithm. Circuits Syst. Signal Process. 2020, 39, 522–542. [Google Scholar] [CrossRef]
- Kumar, S.N.; Ahilan, A.; Haridhas, A.K.; Sebastian, J. Gaussian Hermite polynomial based lossless medical image compression. Multimed. Syst. 2021, 27, 15–31. [Google Scholar] [CrossRef]
- Aldemir, E.; Dueñas, O.A.T.; Kavur, A.E.; Tohumoglu, G.; Sánchez-Cruz, H.; Selver, M.A. Chain code strategy for lossless storage and transfer of segmented binary medical data. Expert Syst. Appl. 2023, 216, 119449. [Google Scholar] [CrossRef]
- Latha, H.R.; Ramaprasath, A. HWCD: A hybrid approach for image compression using wavelet, encryption using confusion, and decryption using diffusion scheme. J. Intell. Syst. 2023, 32, 20229056. [Google Scholar] [CrossRef]
- de Aguiar, E.J.; dos Santos, A.J.; Meneguette, R.I.; Grande, R.E.D.; Ueyama, J. A blockchain-based protocol for tracking user access to shared medical imaging. Future Gener. Comput. Syst. 2022, 134, 348–360. [Google Scholar] [CrossRef]
- Lewis, T.; Berkowitz, S.; Weinstein, J. Abstract No. 565 Smartphone and web-based interventional radiology case logging system to facilitate recording of procedures performed by interventional radiology residents. J. Vasc. Interv. Radiol. 2021, 32, S157. [Google Scholar] [CrossRef]
- O’Sullivan, D.M.; O’Sullivan, E.; O’Connor, M.; Lyons, D.; McManus, J. WhatsApp Doc? BMJ Innov. 2017, 3, 238–239. [Google Scholar] [CrossRef]
- Giansanti, D.; Cosentino, L. WhatsApp in mHealth: Design and evaluation of an mHealth tool to share dynamic images in hemodynamics. mHealth 2021, 7, 1–7. [Google Scholar] [CrossRef]
- Pongkunakorn, A.; Aksornthung, C.; Sritumpinit, N. Accuracy of a New Digital Templating Method for Total Hip Arthroplasty Using Picture Archiving and Communication System (PACS) and iPhone Technology: Comparison with Acetate Templating on Digital Radiography. J. Arthroplast. 2021, 36, 2204–2210. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; von Wangenheim, A.; von Wangenheim, C.; Inácio, A.; de Macedo, D. Usability Engineering of a Radiological Workstation for Mobile Devices: An Experience Report. Soc. Bras. Comput. SBC 2016, 7, 2471–2480. [Google Scholar] [CrossRef]
- Wattanapisit, A.; Amaek, W.; Wattanapisit, S.; Tuangratananon, T.; Wongsiri, S.; Pengkaew, P. Challenges of implementing an mhealth application for personalized physical activity counselling in primary health care: A qualitative study. Int. J. Gen. Med. 2021, 14, 3821–3831. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, E.; Urban, T.; Brown, D.; Petts, J.; Pieper, S.D.; Lewis, R.; Hafey, C.; Harris, G.J. Open Health Imaging Foundation Viewer: An Extensible Open-Source Framework for Building Web-Based Imaging Applications to Support Cancer Research. JCO Clin. Cancer Inform. 2020, 4, 336–345. [Google Scholar] [CrossRef]
- Bai, S.; Ma, C.; Wang, X.; Zhou, S.; Jiang, H.; Ma, L.; Jiang, H. Application of Medical Image 3D Visualization Web Platform in Auxiliary Diagnosis and Preoperative Planning. J. Image Graph. 2023, 11, 32–39. [Google Scholar] [CrossRef]
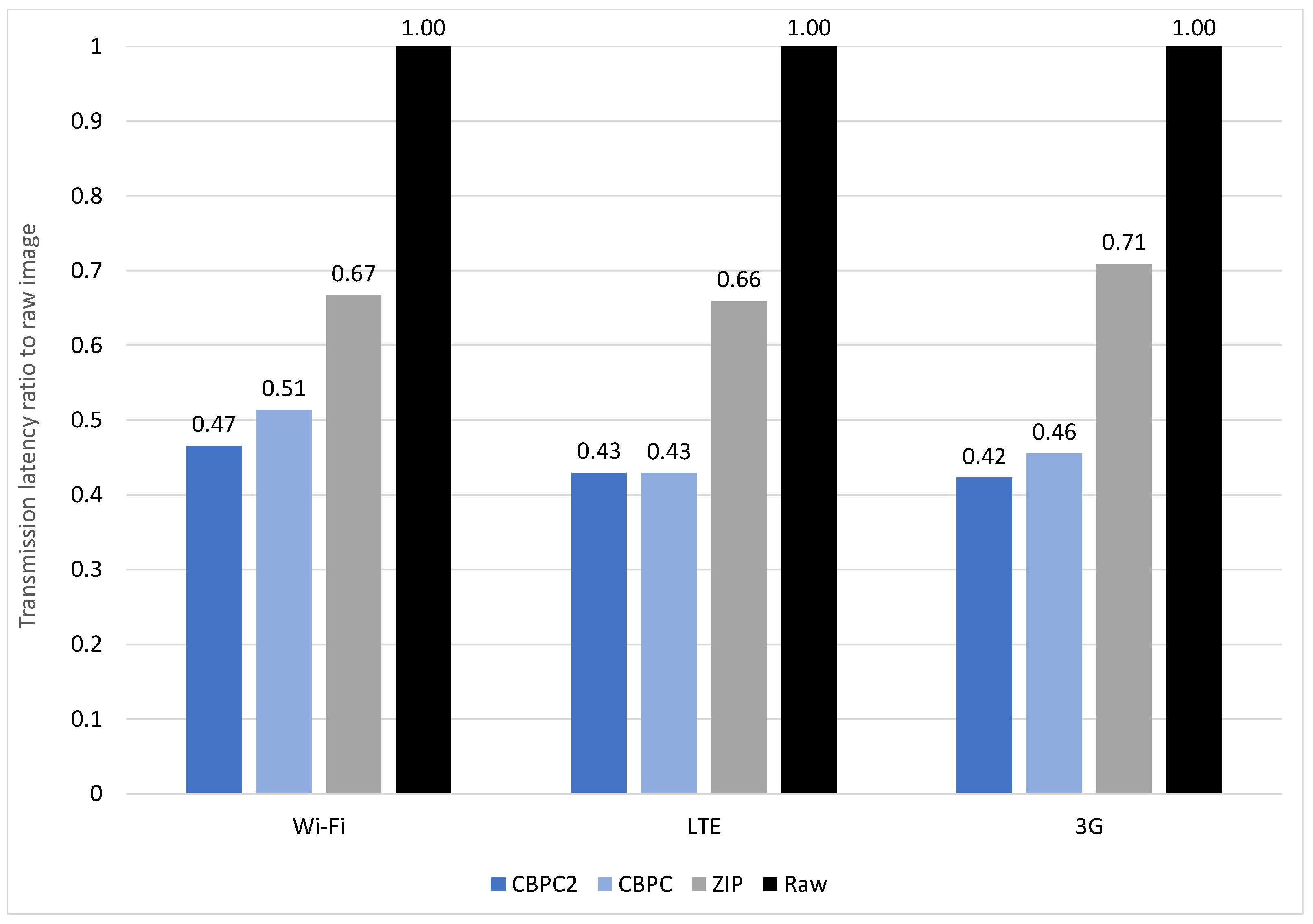
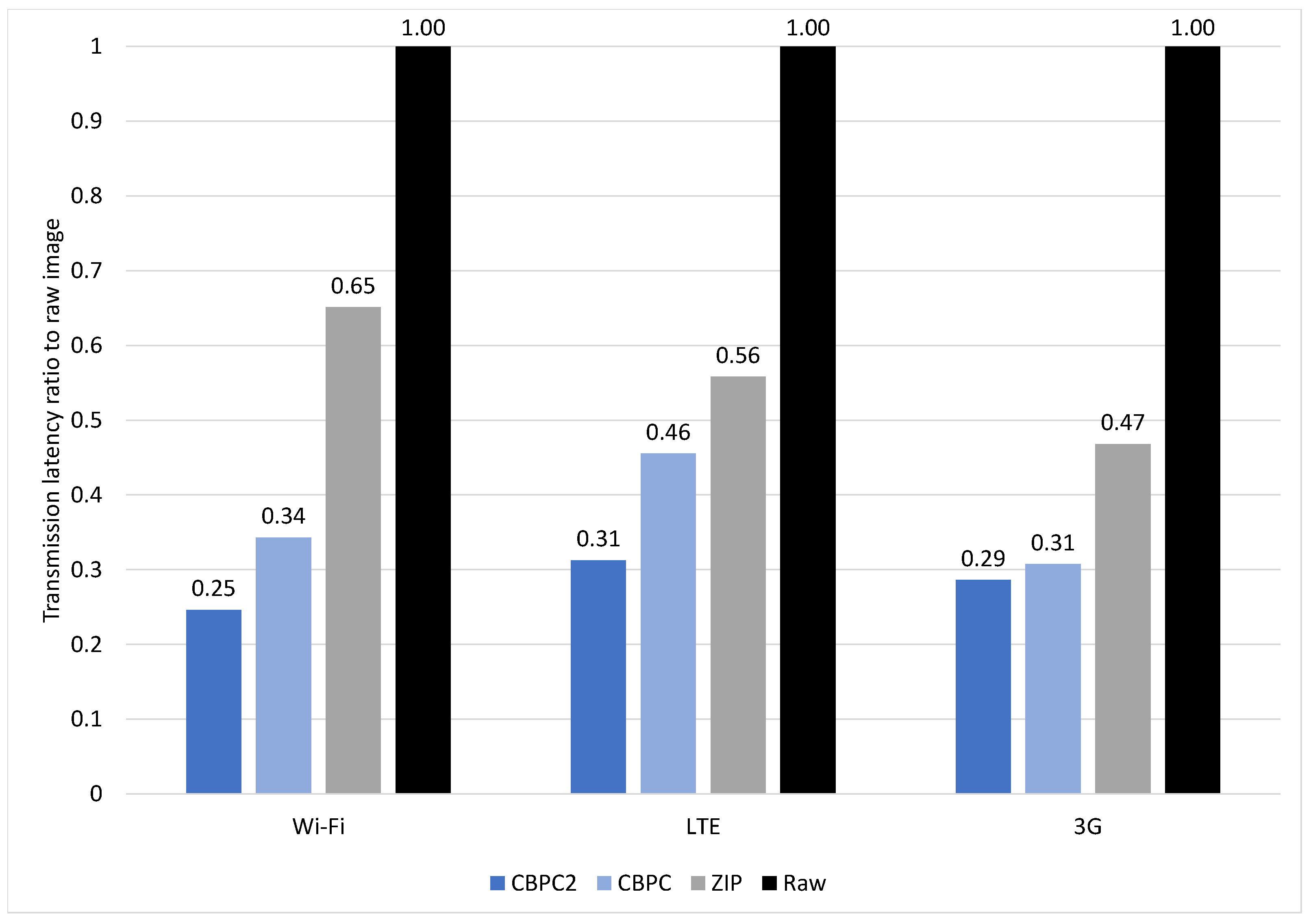


| Profile | Bandwidth | Delay | ||
|---|---|---|---|---|
| Download | Upload | Download | Upload | |
| 3G | 780 Kbps | 330 Kbps | 100 ms | 100 ms |
| Wi-Fi | 40 Mbps | 30 Mbps | 1 ms | 1 ms |
| LTE | 50 Mbps | 10 Mbps | 50 ms | 50 ms |
| Data Size [MB] | ||||
|---|---|---|---|---|
| Original | CBPC2 | CBPC | ZIP | |
| Angio | 1.43 | 0.52 | 0.55 | 0.91 |
| X-ray | 11.88 | 2.66 | 2.95 | 5.03 |
| MR-1 | 95.82 | 24.23 | 26.48 | 43.62 |
| CT | 76.81 | 35.87 | 38.07 | 41.56 |
| MR-2 | 486.83 | 173.41 | 201.52 | 237.37 |
| Latency [s] | ||||||
|---|---|---|---|---|---|---|
| Angio | X-ray | MR-1 | CT | MR-2 | ||
| 3G | Original | 28.41 | 146.57 | 1108.39 | 879.66 | 6197.09 |
| CBPC2 | 12.01 | 41.97 | 286.48 | 415.87 | 2095.19 | |
| CBPC | 12.94 | 45.06 | 312.58 | 443.33 | 2242.86 | |
| ZIP | 20.15 | 68.61 | 506.64 | 483.02 | 2861.06 | |
| LTE | Original | 3.28 | 10.46 | 58.29 | 43.58 | 532.27 |
| CBPC2 | 1.41 | 3.27 | 14.78 | 21.41 | 156.95 | |
| CBPC | 1.41 | 4.76 | 18.00 | 23.57 | 211.59 | |
| ZIP | 2.17 | 5.84 | 30.32 | 26.73 | 334.01 | |
| Wi-Fi | Original | 1.06 | 5.94 | 36.24 | 28.58 | 224.94 |
| CBPC2 | 0.49 | 1.46 | 8.89 | 14.75 | 82.40 | |
| CBPC | 0.55 | 2.04 | 12.73 | 17.17 | 84.78 | |
| ZIP | 0.71 | 3.87 | 18.77 | 19.87 | 101.17 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pervan, B.; Tomic, S.; Ivandic, H.; Knezovic, J. MIDOM—A DICOM-Based Medical Image Communication System. Appl. Sci. 2023, 13, 6075. https://doi.org/10.3390/app13106075
Pervan B, Tomic S, Ivandic H, Knezovic J. MIDOM—A DICOM-Based Medical Image Communication System. Applied Sciences. 2023; 13(10):6075. https://doi.org/10.3390/app13106075
Chicago/Turabian StylePervan, Branimir, Sinisa Tomic, Hana Ivandic, and Josip Knezovic. 2023. "MIDOM—A DICOM-Based Medical Image Communication System" Applied Sciences 13, no. 10: 6075. https://doi.org/10.3390/app13106075
APA StylePervan, B., Tomic, S., Ivandic, H., & Knezovic, J. (2023). MIDOM—A DICOM-Based Medical Image Communication System. Applied Sciences, 13(10), 6075. https://doi.org/10.3390/app13106075








