Study of the Influence of the Dielectrophoretic Force on the Preferential Growth of Bacterial Biofilms in 3D Printed Microfluidic Devices
Abstract
1. Introduction
2. Materials and Methods
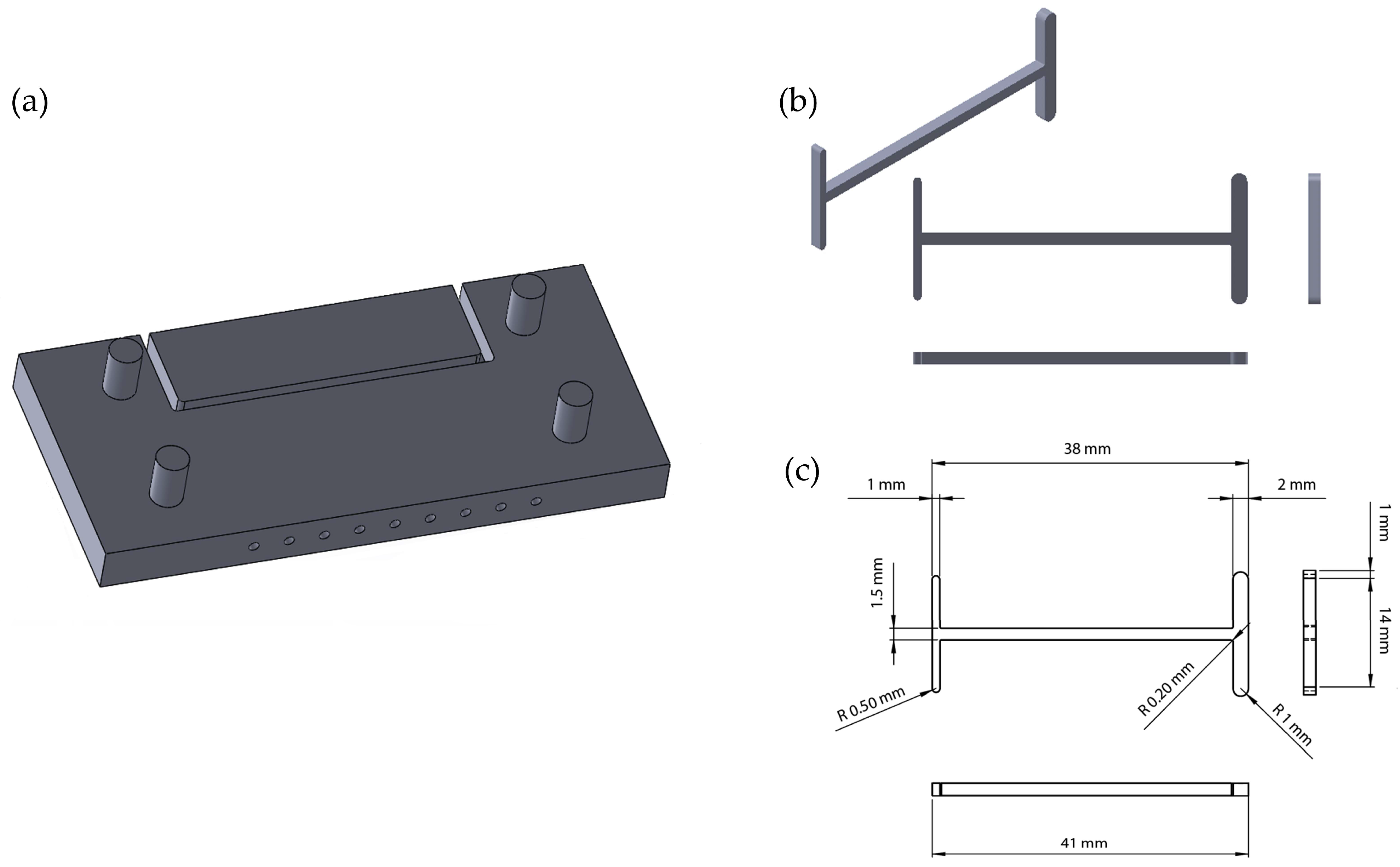
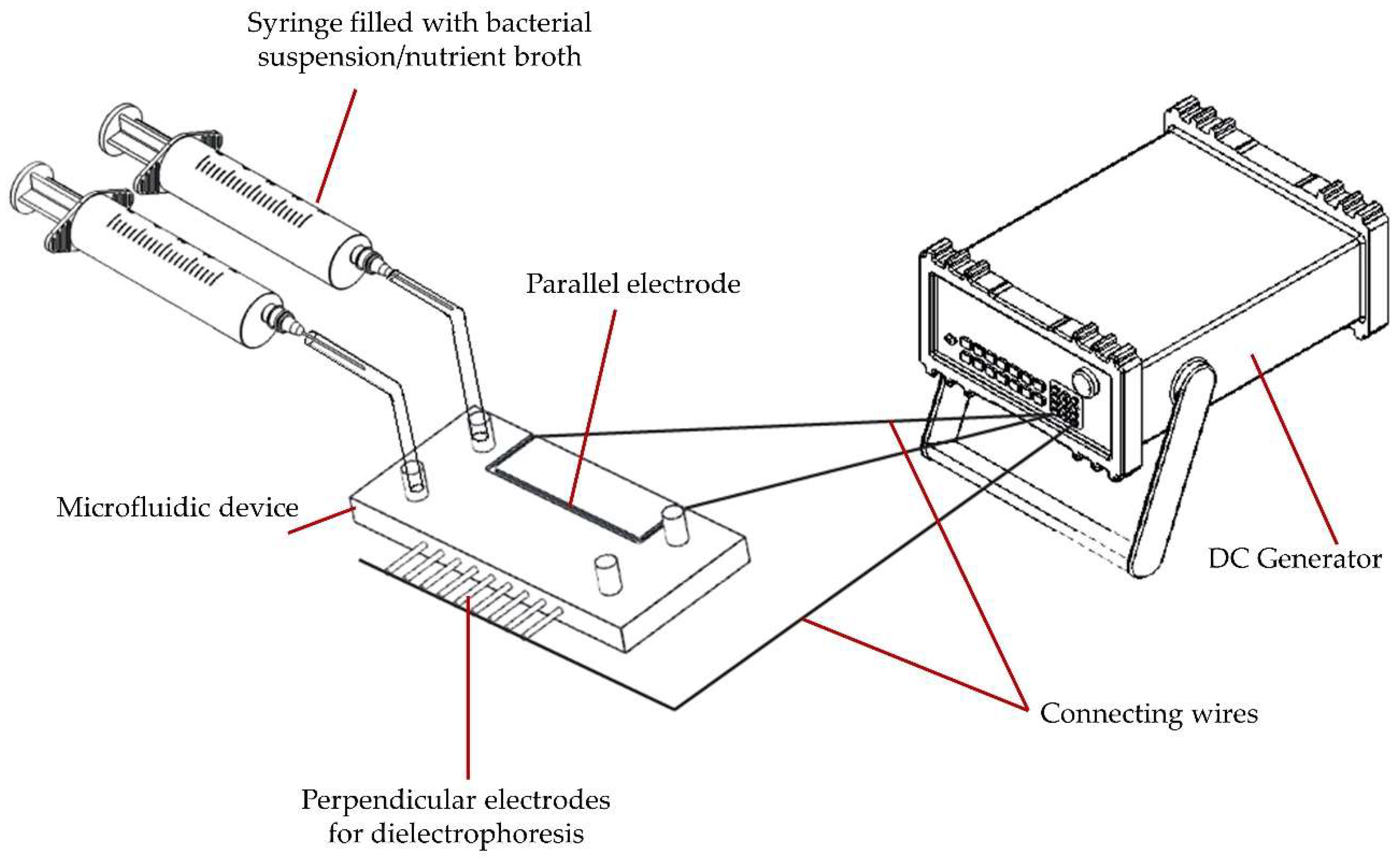
2.1. Device Setup
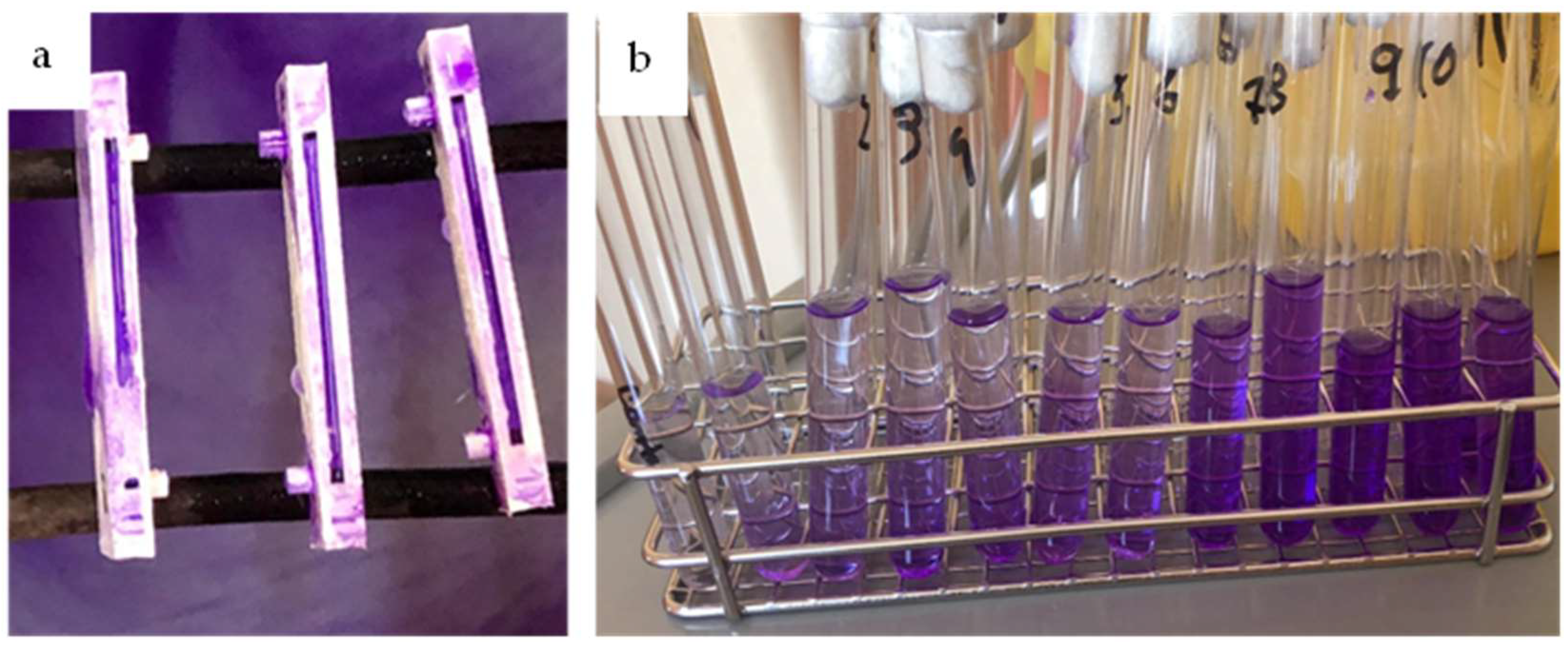
2.2. Biofilm Cultivation, Formation, and Growth
2.3. Analysis Methods
3. Results and Discussions
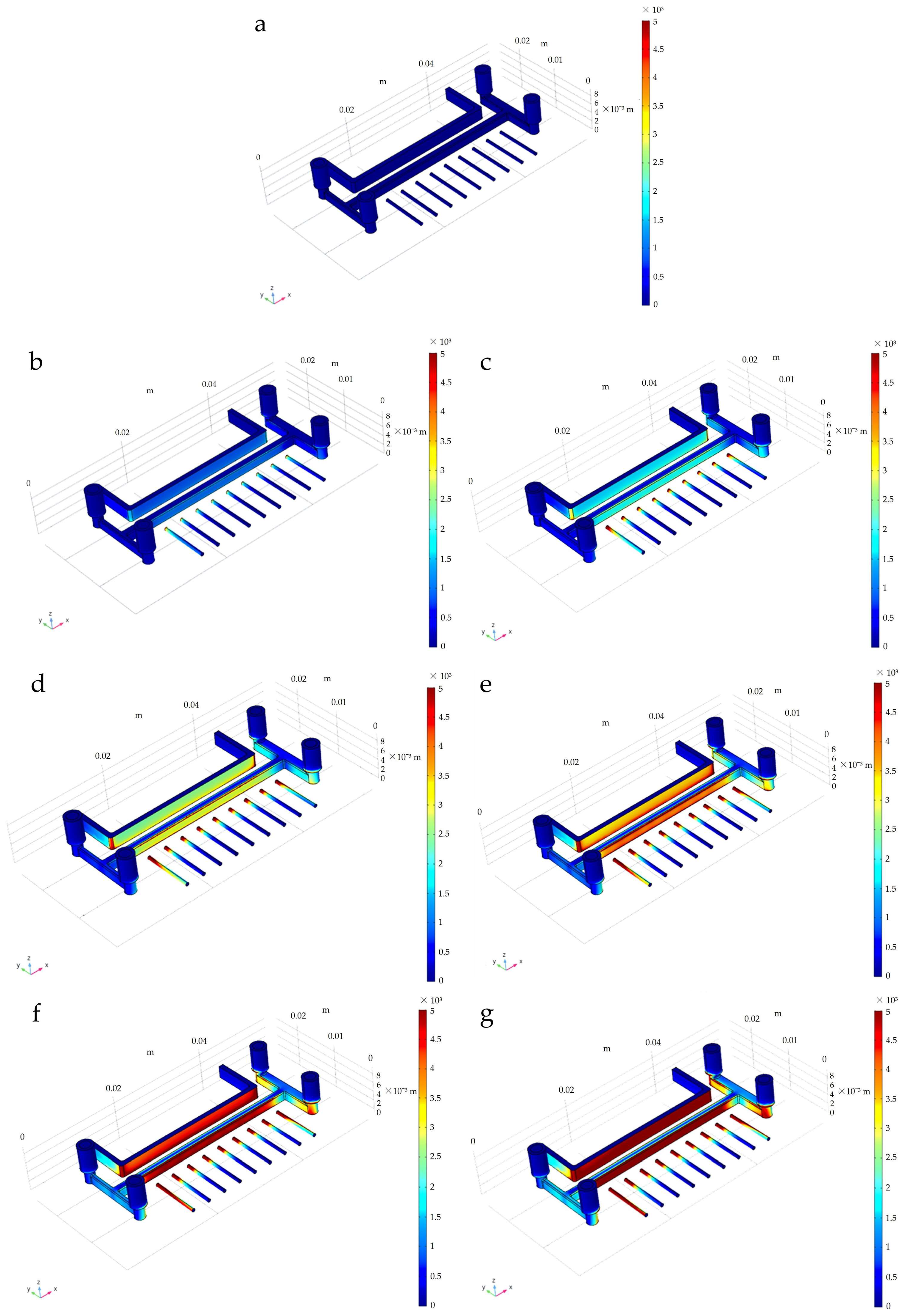
3.1. Distribution of the Electric Field
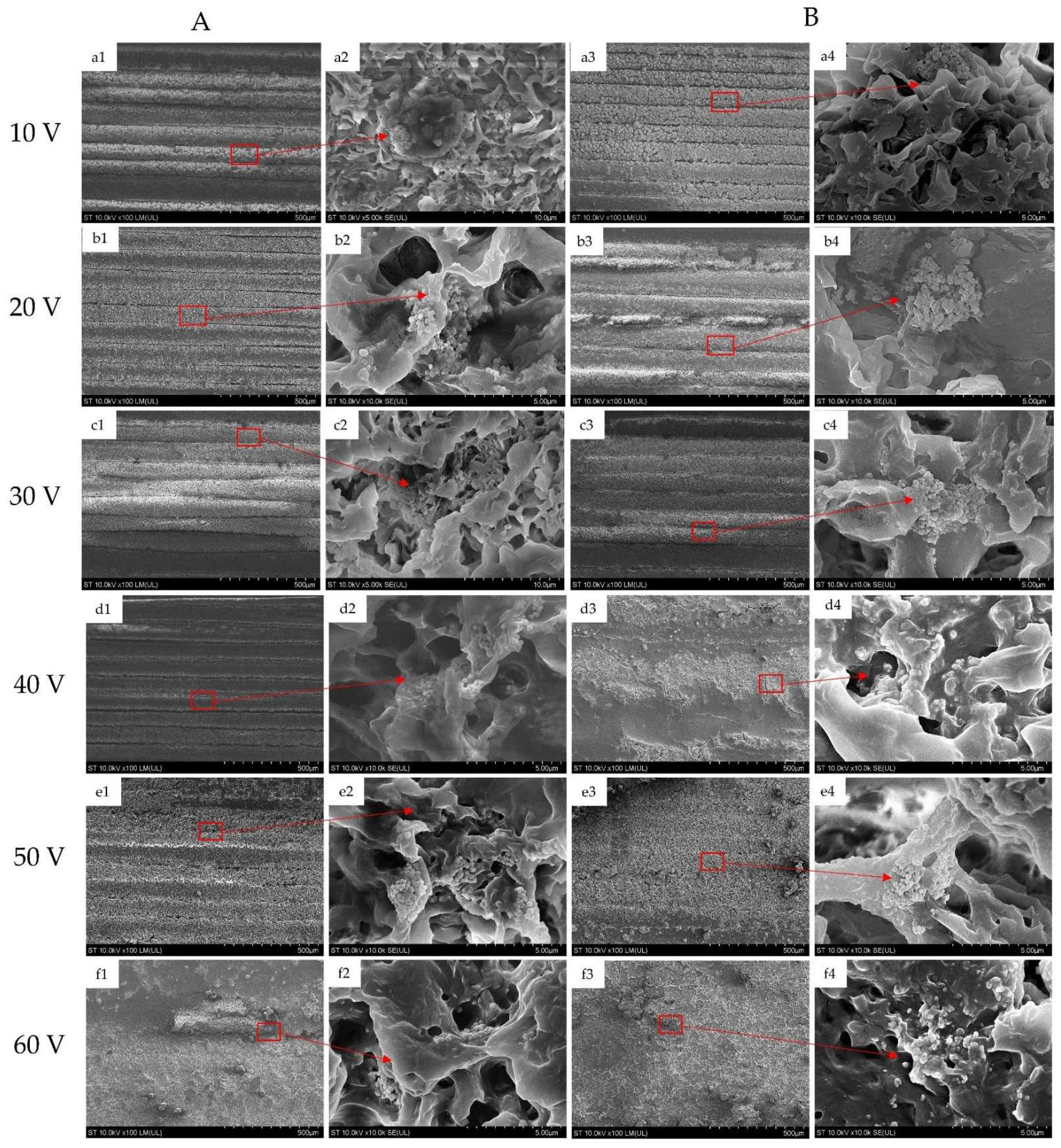
3.2. Scanning Electron Microscopy (SEM)
3.3. Quantitative Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasiri, R.; Shamloo, A.; Ahadian, S.; Amirifar, L.; Akbari, J.; Goudie, M.J.; Lee, K.; Ashammakhi, N.; Dokmeci, M.R.; Di Carlo, D.; et al. Microfluidic-Based Approaches in Targeted Cell/Particle Separation Based on Physical Properties: Fundamentals and Applications. Small 2020, 16, e2000171. [Google Scholar] [CrossRef] [PubMed]
- Cottet, J.; Renaud, P. Introduction to Microfluidics; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Breslauer, D.N.; Lee, P.J.; Lee, L.P. Microfluidics-based systems biology. Mol. Biosyst. 2006, 2, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.-D.; Chung, S. Microfluidic Approaches to Bacterial Biofilm Formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef]
- Mehta, V.; Rath, S.N. 3D printed microfluidic devices: A review focused on four fundamental manufacturing approaches and implications on the field of healthcare. Bio-Design Manuf. 2021, 4, 311–343. [Google Scholar] [CrossRef]
- Nelson, M.D.; Ramkumar, N.; Gale, B.K. Flexible, transparent, sub-100 µm microfluidic channels with fused deposition modeling 3D-printed thermoplastic polyurethane. J. Micromech. Microeng. 2019, 29, 1–9. [Google Scholar] [CrossRef]
- Romanov, V.; Samuel, R.; Chaharlang, M.; Jafek, A.R.; Frost, A.; Gale, B.K. FDM 3D Printing of High-Pressure, Heat-Resistant, Transparent Microfluidic Devices. Anal. Chem. 2018, 90, 10450–10456. [Google Scholar] [CrossRef] [PubMed]
- Gaal, G.; Mendes, M.; de Almeida, T.P.; Piazzetta, M.H.; Gobbi, Â.L.; Riul, A.; Rodrigues, V. Simplified fabrication of integrated microfluidic devices using fused deposition modeling 3D printing. Sens. Actuators B Chem. 2017, 242, 35–40. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, K.; Feyeux, M.; Gurchenkov, B.; Delgado, C.; Trushko, A.; Krause, K.-H.; Vignjević, D.; Nassoy, P.; Roux, A. A 3D printed microfluidic device for production of functionalized hydrogel microcapsules for culture and differentiation of human Neuronal Stem Cells (hNSC). Lab Chip 2016, 16, 1593–1604. [Google Scholar] [CrossRef]
- Ong, L.J.Y.; Islam, A.; DasGupta, R.; Iyer, N.G.; Leo, H.L.; Toh, Y.-C. A 3D printed microfluidic perfusion device for multicellular spheroid cultures. Biofabrication 2017, 9, 045005. [Google Scholar] [CrossRef]
- Lerman, M.J.; Lembong, J.; Gillen, G.; Fisher, J.P. 3D printing in cell culture systems and medical applications. Appl. Phys. Rev. 2018, 5, 041109. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Zlatanovic, S.; Ozkan, M.; Birkbeck, A.L.; Esener, S.C. Manipulation of microspheres and biological cells with multiple agile VCSEL traps. Sens. Actuators B Chem. 2006, 113, 866–874. [Google Scholar] [CrossRef]
- Huang, S.-B.; Wu, M.-H.; Lin, Y.-H.; Hsieh, C.-H.; Yang, C.-L.; Lin, H.-C.; Tseng, C.-P.; Lee, G.-B. High-purity and label-free isolation of circulating tumor cells (CTCs) in a microfluidic platform by using optically-induced-dielectrophoretic (ODEP) force. Lab Chip 2013, 13, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Wilkins, E. Electrochemical Biosensors for Detection of Biological Warfare Agents. Electroanalysis 2003, 15, 157–167. [Google Scholar] [CrossRef]
- Lewpiriyawong, N.; Yang, C. Dielectrophoresis Field-Flow Fractionation for Continuous-Flow Separation of Particles and Cells in Microfluidic Devices. In Advances in Transport Phenomena; Wang, L., Ed.; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Suehiro, J.; Zhou, G.; Member, S.; Imamura, M.; Hara, M. Dielectrophoretic Filter for Separation and Recovery of Biological Cells in Water. IEEE Trans. Ind. Appl. 2003, 39, 1514–1521. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Shen, S.; Lee, S.; Dou, H. Field-flow fractionation: A gentle separation and characterization technique in biomedicine. TrAC Trends Anal. Chem. 2018, 108, 231–238. [Google Scholar] [CrossRef]
- Lilliehorn, T.; Simu, U.; Nilsson, M.; Almqvist, M.; Stepinski, T.; Laurell, T.; Nilsson, J.; Johansson, S. Trapping of microparticles in the near field of an ultrasonic transducer. Ultrasonics 2005, 43, 293–303. [Google Scholar] [CrossRef]
- Yun, H.; Kim, K.; Lee, W.G. Cell manipulation in microfluidics. Biofabrication 2013, 5, 022001. [Google Scholar] [CrossRef]
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for Bioparticle Manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309. [Google Scholar] [CrossRef]
- Kharboutly, M.; Gauthier, M.; Chaillet, N. Modeling the trajectory of a microparticle in a dielectrophoresis device. J. Appl. Phys. 2009, 106, 114312. [Google Scholar] [CrossRef]
- Vert, M.; Doi, Y.; Hellwich, K.-H.; Hess, M.; Hodge, P.; Kubisa, P.; Rinaudo, M.; Schué, F. Terminology for Biorelated Polymers and Applications (IUPAC Recommendations 2012). Chem. Int. 2012, 34, 25. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Koo, H.; Yamada, K.M. Dynamic cell–matrix interactions modulate microbial biofilm and tissue 3D microenvironments. Curr. Opin. Cell Biol. 2016, 42, 102–112. [Google Scholar] [CrossRef]
- Hobley, L.; Harkins, C.; MacPhee, C.; Stanley-Wall, N.R. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Hengge, R.; Stahlhut, S.G.; Chattopadhyay, S.; Kisiela, D.I.; Hvidtfeldt, K.; Clegg, S.; Struve, C.; Sokurenko, E.V.; et al. Cellulose as an Architectural Element in Spatially Structured Escherichia coli Biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Csapai, A.; Ţoc, D.-A.; Paşcalău, V.; Toşa, V.; Opruţa, D.; Popa, F.; Popa, C. Study on the Effect of in Situ Surface Treatments of Medical Microfluidic Systems Obtained Through Additive Manufacturing. Arch. Metall. Mater. 2022, 67, 1071–1078. [Google Scholar] [CrossRef]
- Toyofuku, M.; Inaba, T.; Kiyokawa, T.; Obana, N.; Yawata, Y.; Nomura, N. Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 2016, 80, 7–12. [Google Scholar] [CrossRef]
- Fang, H.; Toyofuku, M.; Kiyokawa, T.; Ichihashi, A.; Tateda, K.; Nomura, N. The Impact of Anaerobiosis on Strain-Dependent Phenotypic Variations in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 2013, 77, 1747–1752. [Google Scholar] [CrossRef]
- Park, A.; Jeong, H.-H.; Lee, J.; Kim, K.P.; Lee, C.-S. Effect of shear stress on the formation of bacterial biofilm in a microfluidic channel. BioChip J. 2011, 5, 236–241. [Google Scholar] [CrossRef]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2011, 10, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J. Microbial Biofilms; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Jacobsen, S.M.; Shirtliff, M.E. Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence 2011, 2, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Boland, T.; Latour, R.A.; Stutzenberger, F.J. Molecular Basis of Bacterial Adhesion. In Handbook of Bacterial Adhesiomn; An, Y.H., Friedman, R.J., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 29–41. [Google Scholar]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The Intercellular Adhesion (ica) Locus Is Present in Staphylococcus aureus and Is Required for Biofilm Formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Tsaloglou, M.-N.; Parry-Jones, L.; Jacobs, A.; Watson, R.; Turner, C.; Amos, R.; Hadwen, B.; Buse, J.; et al. Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab Chip 2015, 15, 3065–3075. [Google Scholar] [CrossRef]
- Szita, N.; Boccazzi, P.; Zhang, Z.; Boyle, P.; Sinskey, A.J.; Jensen, K.F. Development of a multiplexed microbioreactor system for high-throughput bioprocessing. Lab Chip 2005, 5, 819–826. [Google Scholar] [CrossRef]
- Matlock-Colangelo, L.; Coon, B.; Pitner, C.L.; Frey, M.W.; Baeumner, A.J. Functionalized electrospun poly(vinyl alcohol) nanofibers for on-chip concentration of E. coli cells. Anal. Bioanal. Chem. 2015, 408, 1327–1334. [Google Scholar] [CrossRef]
- Xu, P.; Xie, R.; Liu, Y.; Luo, G.; Ding, M.; Liang, Q. Bioinspired Microfibers with Embedded Perfusable Helical Channels. Adv. Mater. 2017, 29, 1–7. [Google Scholar] [CrossRef]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a Paper-Based Analytical Device for Colorimetric Detection of Select Foodborne Pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef]
- Li, C.-Z.; Vandenberg, K.; Prabhulkar, S.; Zhu, X.; Schneper, L.; Methee, K.; Rosser, C.J.; Almeide, E. Paper based point-of-care testing disc for multiplex whole cell bacteria analysis. Biosens. Bioelectron. 2011, 26, 4342–4348. [Google Scholar] [CrossRef]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Csapai, A.; Toc, D.A.; Popa, F.; Tosa, N.; Pascalau, V.; Costache, C.; Botan, A.; Popa, C.O. 3D Printed Microfluidic Bioreactors Used for the Preferential Growth of Bacterial Biofilms through Dielectrophoresis. Micromachines 2022, 13, 1377. [Google Scholar] [CrossRef] [PubMed]
- Toc, D.; Csapai, A.; Popa, F.; Popa, C.; Pascalau, V.; Tosa, N.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; et al. Easy and Affordable: A New Method for the Studying of Bacterial Biofilm Formation. Cells 2022, 11, 4119. [Google Scholar] [CrossRef]
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus aureus Biofilms Promote Horizontal Transfer of Antibiotic Resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970. [Google Scholar] [CrossRef] [PubMed]
- Tai, J.-S.B.; Mukherjee, S.; Nero, T.; Olson, R.; Tithof, J.; Nadell, C.D.; Yan, J. Social evolution of shared biofilm matrix components. Proc. Natl. Acad. Sci. USA 2022, 119, e2123469119. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 2015, 23, 233–242. [Google Scholar] [CrossRef]
- Geraci, J.; Neubauer, S.; Pöllath, C.; Hansen, U.; Rizzo, F.; Krafft, C.; Westermann, M.; Hussain, M.; Peters, G.; Pletz, M.W.; et al. The Staphylococcus aureus extracellular matrix protein (Emp) has a fibrous structure and binds to different extracellular matrices. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Wilking, J.N.; Zaburdaev, V.; De Volder, M.; Losick, R.; Brenner, M.P.; Weitz, D.A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2012, 110, 848–852. [Google Scholar] [CrossRef]
- Patel, R. Biofilms and Antimicrobial Resistance. Clin. Orthop. Relat. Res. 2005, 437, 41–47. [Google Scholar] [CrossRef]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csapai, A.; Toc, D.A.; Pascalau, V.; Tosa, N.; Tripon, S.; Ciorîță, A.; Mihaila, R.M.; Mociran, B.; Costache, C.; Popa, C.O. Study of the Influence of the Dielectrophoretic Force on the Preferential Growth of Bacterial Biofilms in 3D Printed Microfluidic Devices. Appl. Sci. 2023, 13, 60. https://doi.org/10.3390/app13010060
Csapai A, Toc DA, Pascalau V, Tosa N, Tripon S, Ciorîță A, Mihaila RM, Mociran B, Costache C, Popa CO. Study of the Influence of the Dielectrophoretic Force on the Preferential Growth of Bacterial Biofilms in 3D Printed Microfluidic Devices. Applied Sciences. 2023; 13(1):60. https://doi.org/10.3390/app13010060
Chicago/Turabian StyleCsapai, Alexandra, Dan Alexandru Toc, Violeta Pascalau, Nicoleta Tosa, Septimiu Tripon, Alexandra Ciorîță, Razvan Marian Mihaila, Bogdan Mociran, Carmen Costache, and Catalin O. Popa. 2023. "Study of the Influence of the Dielectrophoretic Force on the Preferential Growth of Bacterial Biofilms in 3D Printed Microfluidic Devices" Applied Sciences 13, no. 1: 60. https://doi.org/10.3390/app13010060
APA StyleCsapai, A., Toc, D. A., Pascalau, V., Tosa, N., Tripon, S., Ciorîță, A., Mihaila, R. M., Mociran, B., Costache, C., & Popa, C. O. (2023). Study of the Influence of the Dielectrophoretic Force on the Preferential Growth of Bacterial Biofilms in 3D Printed Microfluidic Devices. Applied Sciences, 13(1), 60. https://doi.org/10.3390/app13010060








