A Machine Learning Approach for the Non-Destructive Estimation of Leaf Area in Medicinal Orchid Dendrobium nobile L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Process Flow of Selection of Best Leaf Area Model in D. nobile through ML Techniques
2.3. Dataset
2.4. ML Methodologies Used for LA Prediction Modeling
2.4.1. Multiple Linear Regression Analysis (MLR) Models
2.4.2. Support Vector Regression (SVR) Models
2.4.3. Gradient Boosting Regression (GBR) Models
2.4.4. Artificial Neural Networks (ANNs) Models
2.5. Feature Correlation Heatmap and Multicollinearity of the Independent Variables
2.6. Programming Set-Up
2.7. Model Performance Evaluation
2.8. Model Ranking Based on Average Rank (AR) Ranking Methodology
3. Results and Discussion
3.1. Descriptive Statistics of D. nobile Leaves Used for LA Model Building
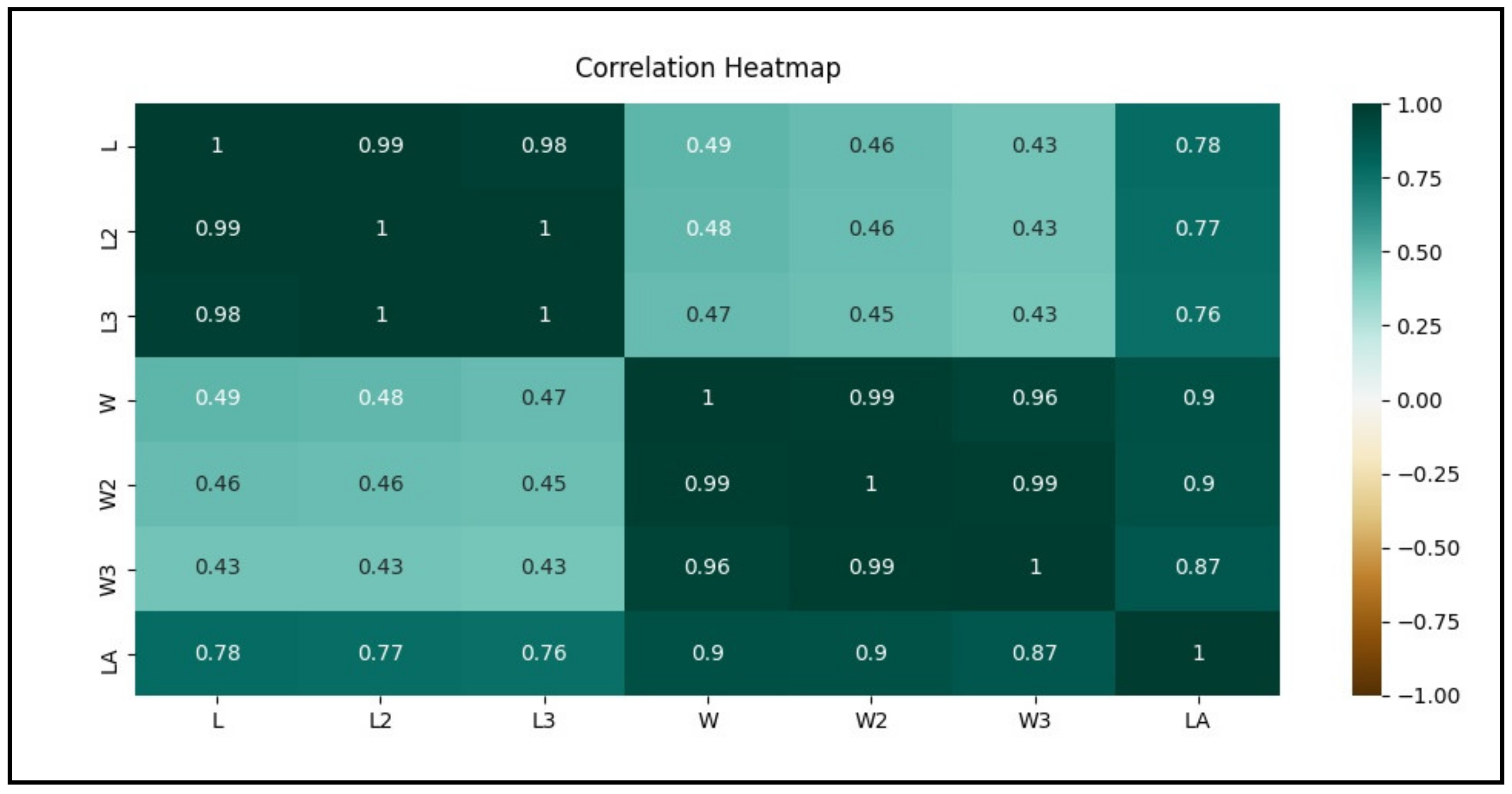
3.2. Feature Correlation Heatmap between the Variables through Correlation Coefficient
- Leaf length (L) and width (W) are linearly least correlated (r = 0.43–0.49) to each other as depicted from the light green color;
- L, L2, L3, and LA are nearly equally correlated (r = 0.76–0.78) to each other as depicted from the medium green color;
- Leaf width (W) shows more correlated to leaf area (LA) with correlation coefficient (r) values ranging between 0.87 and 0.9 than leaf length (L) 0.76–0.78 as depicted from the darker green color in the heatmap;
- Among the variants of leaf width, the best to worst correlation with the leaf area (LA) can be shown in the order of W (r = 0.9) = W2 (r = 0.9) > W3(r = 0.87);
- L, W, and their variants are strongly linearly correlated with leaf area (LA) where r ranges from 0.76 to 0.90 as depicted from dark green color in the heatmap.
3.3. Multicollinearity Status of the Two Independent Variables (L and W)
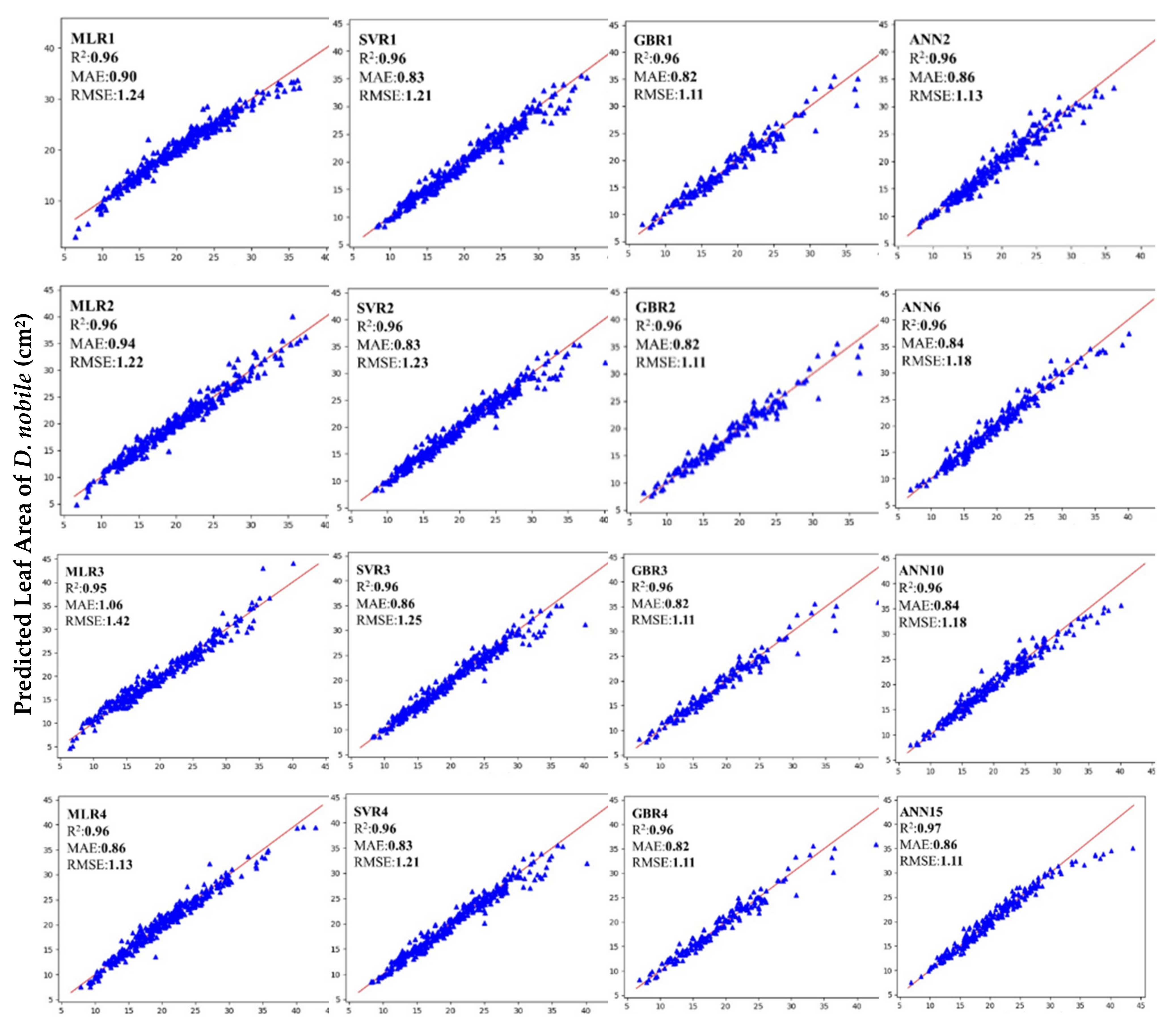
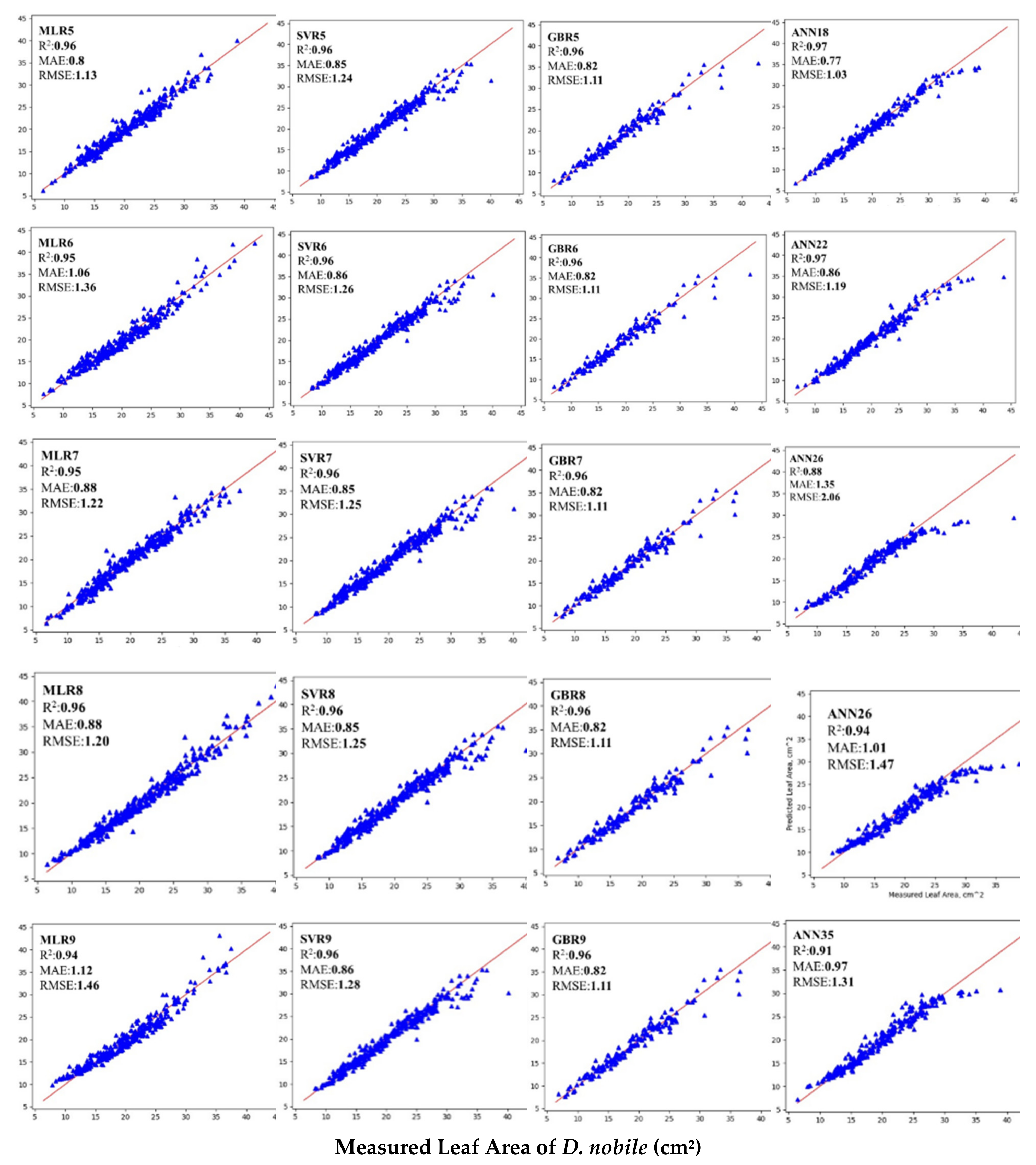
3.4. Selection of Nine Best Performing ANN Models Based on R2, MAE and RMSE
3.5. Comparisons of Different ML Models for Estimating Leaf Area of D. nobile Leaves
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gabel, R. The role of CITES in orchid conservation. Endanger. Species Update 2006, 23, S14. [Google Scholar]
- Li, M.H.; Liu, D.K.; Zhang, G.Q.; Deng, H.; Tu, X.D.; Wang, Y.; Lan, S.R.; Liu, Z.J. A perspective on crassulacean acid metabolism photosynthesis evolution of orchids on different continents: Dendrobium as a case study. J. Exp. Bot. 2019, 70, 6611–6619. [Google Scholar] [CrossRef] [PubMed]
- WCSP. World Checklist of Selected Plant Families. Facilitated by the Royal Botanic Gardens, Kew. Available online: http://apps.kew.org/wcsp/ (accessed on 20 May 2020).
- Zheng, S.; Hu, Y.; Zhao, R.; Zhao, T.; Rao, D.; Chun, Z. Quantitative assessment of secondary metabolites and cancer cell inhibiting activity by high performance liquid chromatography fingerprinting in Dendrobium nobile. J. Chromatogr. B 2020, 1140, 122017. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, Y.; Colla, G. Modeling the transpiration of a greenhouse zucchini crop grown under a Mediterranean climate using the Penman-Monteith equation and its simplified version. Aust. J. Agric. Res. 2004, 55, 931–937. [Google Scholar] [CrossRef]
- Rouphael, Y.; Colla, G. Radiation and water use efficiencies of greenhouse zucchini squash in relation to different climate parameters. Eur. J. Agron. 2005, 23, 183–194. [Google Scholar] [CrossRef]
- De Oliveira Silva, F.M.; Lichtenstein, G.; Alseekh, S.; Rosado-Souza, L.; Conte, M.; Suguiyama, V.F.; Lira, B.S.; Fanourakis, D.; Usadel, B.; Bhering, L.L.; et al. The genetic architecture of photosynthesis and plant growth-related traits in tomato. Plant Cell Environ. 2018, 41, 327–341. [Google Scholar] [CrossRef]
- Qi, Y.; Huang, J.L.; Zhang, S.B. Correlated evolution of leaf and root anatomic traits in Dendrobium (Orchidaceae). AoB Plants 2020, 12, plaa034. [Google Scholar] [CrossRef]
- Basbag, S.; Ekinci, R.; Oktay, G. Relationships between Some Physiomorphological Traits and Cotton (Gossypium hirsutum L.) Yield. In Tenth Regional Meeting; International Cotton Advisory Committee: Washington, DC, USA, 2008. [Google Scholar]
- He, J.; Woon, W.L. Source-to-sink relationship between green leaves and green petals of different ages of the CAM orchid Dendrobium cv. Burana Jade. Photosynthetica 2008, 46, 91–97. [Google Scholar] [CrossRef]
- Kabir, M.I.; Mortuza, M.G.; Islam, M.O. Morphological features growth and development of Dendrobium sp. orchid as influenced by nutrient spray. J. Environ. Sci. Nat. Resour. 2012, 5, 309–318. [Google Scholar] [CrossRef]
- Sun, M.; Feng, C.H.; Liu, Z.Y.; Tian, K. Evolutionary correlation of water-related traits between different structures of Dendrobium plants. Bot. Stud. 2020, 61, 1–14. [Google Scholar] [CrossRef]
- Keramatlou, I.; Sharifani, M.; Sabouri, H.; Alizadeh, M.; Kamkar, B. A simple linear model for leaf area estimation in Persian walnut (Juglans regia L.). Sci. Hortic. 2015, 184, 36–39. [Google Scholar] [CrossRef]
- Demirsoy, H.; Demirsoy, L. A validated leaf area prediction model for some cherry cultivars in Turkey. Pak. J. Bot. 2003, 35, 361–367. [Google Scholar]
- Daughtry, C.S. Direct measurements of canopy structure. Remote Sens. Rev. 1990, 5, 45–60. [Google Scholar] [CrossRef]
- Walia, S.; Kumar, R. Development of the nondestructive leaf area estimation model for valeriana (Valeriana jatamansi Jones). Commun. Soil Sci. Plant Anal. 2017, 48, 83–91. [Google Scholar] [CrossRef]
- Amiri, M.J.; Shabani, A. Application of an adaptive neural-based fuzzy inference system model for predicting leaf area. Commun. Soil Sci. Plant Anal. 2017, 48, 1669–1683. [Google Scholar] [CrossRef]
- Koubouris, G.; Bouranis, D.; Vogiatzis, E.; Nejad, A.R.; Giday, H.; Tsaniklidis, G.; Ligoxigakis, E.K.; Blazakis, K.; Kalaitzis, P.; Fanourakis, D. Leaf area estimation by considering leaf dimensions in olive tree. Sci. Hortic. 2018, 240, 440–445. [Google Scholar] [CrossRef]
- Peksen, E. Non-destructive leaf area estimation model for faba bean (Vicia faba L.). Sci. Hortic. 2007, 113, 322–328. [Google Scholar] [CrossRef]
- Sala, F.; Arsene, G.G.; Iordănescu, O.; Boldea, M. Leaf area constant model in optimizing foliar area measurement in plants: A case study in apple tree. Sci. Hortic. 2015, 193, 218–224. [Google Scholar] [CrossRef]
- Litschmann, T.; Vávra, R.; Falta, V. Non-destructive leaf area assessment of chosen apple cultivars. Vědecké Práce Ovocnářské 2013, 23, 205–212. [Google Scholar]
- Norman, J.M.; Campbell, G.S. Canopy structure. In Plant Physiological Ecology; Springer: Dordrecht, The Netherlands, 1989; pp. 301–325. [Google Scholar]
- Swart, E.D.; Groenwold, R.; Kanne, H.J.; Stam, P.; Marcelis, L.F.; Voorrips, R.E. Non-destructive estimation of leaf area for different plant ages and accessions of Capsicum annuum L. J. Hortic. Sci. Biotechnol. 2004, 79, 764–770. [Google Scholar] [CrossRef]
- Zizka, A.; Silvestro, D.; Vitt, P.; Knight, T.M. Automated conservation assessment of the orchid family with deep learning. Conserv. Biol. 2021, 35, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V.; Guyon, I.; Hastie, T. Support vector machines. Mach. Learn. 1995, 20, 273–297. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Johnson, R.; Zhang, T. Learning nonlinear functions using regularized greedy forest. arXiv 2011, arXiv:1109.0887. [Google Scholar] [CrossRef]
- Hutchinson, R.; Liu, L.P.; Dietterich, T. Incorporating boosted regression trees into ecological latent variable models. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 7–11 August 2011; pp. 1343–1348. [Google Scholar]
- Pittman, S.J.; Brown, K.A. Multi-scale approach for predicting fish species distributions across coral reef seascapes. PLoS ONE 2011, 6, e20583. [Google Scholar] [CrossRef]
- Dyer, A.S.; Zaengle, D.; Nelson, J.R.; Duran, R.; Wenzlick, M.; Wingo, P.C.; Bauer, J.R.; Rose, K.; Romeo, L. Applied machine learning model comparison: Predicting offshore platform integrity with gradient boosting algorithms and neural networks. Mar. Struct. 2022, 83, 103152. [Google Scholar] [CrossRef]
- Cai, J.; Xu, K.; Zhu, Y.; Hu, F.; Li, L. Prediction and analysis of net ecosystem carbon exchange based on gradient boosting regression and random forest. Appl. Energy 2020, 262, 114566. [Google Scholar] [CrossRef]
- Yang, F.; Wang, D.; Xu, F.; Huang, Z.; Tsui, K.L. Lifespan prediction of lithium-ion batteries based on various extracted features and gradient boosting regression tree model. J. Power Sources 2020, 476, 228654. [Google Scholar] [CrossRef]
- Cemek, B.; Ünlükara, A.; Kurunç, A.; Küçüktopcu, E. Leaf area modeling of bell pepper (Capsicum annuum L.) grown under different stress conditions by soft computing approaches. Comput. Electron. Agric. 2020, 174, 105514. [Google Scholar] [CrossRef]
- Odabas, M.S.; Ergun, E.; Oner, F. Artificial neural network approach for the predicition of the corn (Zea mays L.) leaf area. Bulg. J. Agric. Sci. 2013, 19, 766–769. [Google Scholar]
- Wright, S. Correlation and causation. J. Agric. Res. 1921, XX, 557–585. [Google Scholar]
- Sammut, C.; Webb, G.I. Mean absolute error. In Encyclopedia of Machine Learning; Springer Science & Business Media: Berlin, Germany, 2010; p. 652. [Google Scholar]
- Chicco, D.; Warrens, M.J.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef] [PubMed]
- Neave, H.R.; Worthington, P.L. Distribution-Free Tests; Routledge: London, UK, 1998. [Google Scholar]
- Kishore, D.K.; Pramanick, K.K.; Verma, J.K.; Singh, R. Non-destructive estimation of apple (Malus domestica Borkh.) leaf area. J. Hortic. Sci. Biotechnol. 2012, 87, 388–390. [Google Scholar] [CrossRef]
- Cirillo, C.; Pannico, A.; Basile, B.; Rivera, C.M.; Giaccone, M.; Colla, G.; De Pascale, S. Rouphael, Y. A simple and accurate allometric model to predict single leaf area of twenty-one European apricot cultivars. Eur. J. Hortic. Sci. 2017, 82, 65–71. [Google Scholar] [CrossRef]
- Córcoles, J.I.; Ortega, J.F.; Hernández, D.; Moreno, M.A. Estimation of leaf area index in onion (Allium cepa L.) using an unmanned aerial vehicle. Biosyst. Eng. 2013, 115, 31–42. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Antunes, W.C.; Ferreira, D.T.R.G.; Cavalcante, P.G.S.; Wanderley-Filho, H.C.L.; Endres, L. Allometric models for non-destructive leaf area estimation of Jatropha curcas. Biomass Bioenergy 2012, 36, 77–85. [Google Scholar] [CrossRef]
- Salazar, J.C.S.; Melgarejo, L.M.; Bautista, E.H.D.; Di Rienzo, J.A.; Casanoves, F. Non-destructive estimation of the leaf weight and leaf area in cacao (Theobroma cacao L.). Sci. Hortic. 2018, 229, 19–24. [Google Scholar] [CrossRef]
- Serdar, Ü.; Demirsoy, H. Non-destructive leaf area estimation in chestnut. Sci. Hortic. 2006, 108, 227–230. [Google Scholar] [CrossRef]
- Montero, F.J.; De Juan, J.A.; Cuesta, A.; Brasa, A. Nondestructive methods to estimate leaf area in Vitis vinifera L. HortScience 2000, 35, 696–698. [Google Scholar] [CrossRef]
- Tsialtas, J.T.; Koundouras, S.; Zioziou, E. Leaf area estimation by simple measurements and evaluation of leaf area prediction models in Cabernet-Sauvignon grapevine leaves. Photosynthetica 2008, 46, 452–456. [Google Scholar] [CrossRef]
- Ahmadian-Moghadam, H. Prediction of pepper (Capsicum annuum L.) leaf area using group method of data handling-type neural networks. Int. J. AgriSci. 2012, 2, 993–999. [Google Scholar]
- Cemek, B.; Unlukara, A.; Kurunç, A. Nondestructive leaf-area estimation and validation for green pepper (Capsicum annuum L.) grown under different stress conditions. Photosynthetica 2011, 49, 98. [Google Scholar] [CrossRef]
- Kandiannan, K.; Parthasarathy, U.; Krishnamurthy, K.S.; Thankamani, C.K.; Srinivasan, V. Modeling individual leaf area of ginger (Zingiber officinale Roscoe) using leaf length and width. Sci. Hortic. 2009, 120, 532–537. [Google Scholar] [CrossRef]
- Teobaldelli, M.; Basile, B.; Giuffrida, F.; Romano, D.; Toscano, S.; Leonardi, C.; Rivera, C.M.; Colla, G.; Rouphael, Y. Analysis of Cultivar-Specific Variability in Size-Related Leaf Traits and Modeling of Single Leaf Area in Three Medicinal and Aromatic Plants: Ocimum basilicum L., Mentha Spp., and Salvia Spp. Plants 2020, 9, 13. [Google Scholar] [CrossRef]
- Williams III, L.; Martinson, T.E. Nondestructive leaf area estimation of ‘Niagara’and ‘DeChaunac’grapevines. Sci. Hortic. 2003, 98, 493–498. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Tikader, A.; Das, N.K. Nondestructive, simple, and accurate model for estimation of the individual leaf area of som (Persea bombycina). Photosynthetica 2011, 49, 627–632. [Google Scholar] [CrossRef]
- Ghoreishi, M.; Hossini, Y.; Maftoon, M. Simple models for predicting leaf area of mango (L.). Mangifera Indicaj. 2012, 2, 45–53. [Google Scholar]
- Vazquez-Cruz, M.A.; Luna-Rubio, R.; Contreras-Medina, L.M.; Torres-Pacheco, I.; Guevara-Gonzalez, R.G. Estimating the response of tomato (Solanum lycopersicum) leaf area to changes in climate and salicylic acid applications by means of artificial neural networks. Biosyst. Eng. 2012, 112, 319–327. [Google Scholar] [CrossRef]
- Aboukarima, A.M.; Elsoury, H.A.; Menyawi, M. Artificial neural network model for the prediction of the cotton crop leaf area. Int. J. Plant Soil Sci. 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Aboukarima, A.M.; Zayed, M.F.; Minyawi, M.; Elsoury, H.A.; Tarabye, H.H.H. Image analysis-based system for estimating cotton leaf area. Asian Res. J. Agric. 2017, 5, 1–8. [Google Scholar] [CrossRef][Green Version]
- Aboukarima, A.; Elsoury, H.; Minyawi, M. Simple mathematical models for predicting leaf area of cotton plant. J. Soil Sci. Agric. Eng. 2015, 6, 275–294. [Google Scholar] [CrossRef]
- Shabani, A.; Sepaskhah, A.R. Leaf area estimation by a simple and non-destructive method. Iran Agric. Res. 2017, 36, 101–105. [Google Scholar]
- Mendoza-de Gyves, E.; Rouphael, Y.; Cristofori, V.; Mira, F.R. A non-destructive, simple and accurate model for estimating the individual leaf area of kiwi (Actinidia deliciosa). Fruits 2007, 62, 171–176. [Google Scholar] [CrossRef]
- Sankar, V.; Sakthivel, T.; Karunakaran, G.; Tripathi, P.C. Non-destructive estimation of leaf area of durian (Durio zibethinus)—An artificial neural network approach. Sci. Hortic. 2017, 219, 319–325. [Google Scholar]
- Torri, S.I.; Descalzi, C.; Frusso, E. Estimation of leaf area in pecan cultivars (Carya illinoinensis). Cienc. Investig. Agrar. 2009, 36, 53–58. [Google Scholar] [CrossRef]
- Ambebe, T.F.; Zee, F.G.; Shu, M.A.; Ambebe, T.F. Modeling of leaf area of three Afromontane forest tree species through linear measurements. J. Res. Ecol. 2018, 6, 2334–2341. [Google Scholar]
- Cristofori, V.; Fallovo, C.; Mendoza-de Gyves, E.; Rivera, C.M.; Bignami, C.; Rouphael, Y. Non-destructive, analogue model for leaf area estimation in persimmon (Diospyros kaki L. f.) based on leaf length and width measurement. Eur. J. Hortic. Sci. 2008, 73, 216. [Google Scholar]
- Pinto, A.C.R.; Rodrigues, T.D.J.D.; Barbosa, J.C.; Leite, I.C. Leaf area prediction models for Zinnia elegans Jacq., Zinnia haageana Regel and ‘Profusion Cherry’. Sci. Agric. 2004, 61, 47–52. [Google Scholar] [CrossRef][Green Version]
- Rouphael, Y.; Colla, G.; Fanasca, S.; Karam, F. Leaf area estimation of sunflower leaves from simple linear measurements. Photosynthetica 2007, 45, 306–308. [Google Scholar] [CrossRef]
- Rouphael, Y.; Mouneimne, A.H.; Ismail, A.; Mendoza-De Gyves, E.; Rivera, C.M.; Colla, G. Modeling individual leaf area of rose (Rosa hybrida L.) based on leaf length and width measurement. Photosynthetica 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Fascella, G.; Rouphael, Y.; Cirillo, C.; Mammano, M.M.; Pannico, A.; De Pascale, S. Allometric model for leaf area estimation in Bougainvillea genotypes. In Proceedings of the International Symposium on Greener Cities for More Efficient Ecosystem Services in a Climate Changing World, Bologna, Italy, 12–15 September 2017; pp. 449–452. [Google Scholar]
- Giuffrida, F.; Rouphael, Y.; Toscano, S.; Scuderi, D.; Romano, D.; Rivera, C.M.; Colla, G.; Leonardi, C. A simple model for nondestructive leaf area estimation in bedding plants. Photosynthetica 2011, 49, 380. [Google Scholar] [CrossRef]
- Fascella, G.; Maggiore, P.; Rouphael, Y.; Colla, G.; Zizzo, G.V. A simple and low-cost method for leaf area measurement in Euphorbia × lomi Thai hybrids. In Advances in Horticultural Science; Firenze University Press: Florence, Italy, 2009; pp. 1000–1004. [Google Scholar]
- Chen, C. Nondestructive estimation of dry weight and leaf area of Phalaenopsis leaves. Appl. Eng. Agric. 2004, 20, 467. [Google Scholar] [CrossRef]
- Fay, M.F. Orchid conservation: How can we meet the challenges in the twenty-first century? Bot. Stud. 2018, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, Y.P.; Hoffmann, S.; Kunwar, R.M.; Bobrowski, M.; Jentsch, A.; Beierkuhnlein, C. Vascular epiphyte diversity and host tree architecture in two forest management types in the Himalaya. Glob. Ecol. Conserv. 2021, 27, e01544. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- McLachlan, G.J.; Do, K.A.; Ambroise, C. Analyzing Microarray Gene Expression Data; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2005; Volume 422. [Google Scholar]
- Ferreira, T.; Rasband, W. ImageJ user guide. ImageJ/Fiji 2012, 1, 155–161. [Google Scholar]
- Ashtiani, S.H.M.; Rohani, A.; Aghkhani, M.H. Soft computing-based method for estimation of almond kernel mass from its shell features. Sci. Hortic. 2020, 262, 109071. [Google Scholar] [CrossRef]
- Niu, W.J.; Feng, Z.K.; Feng, B.F.; Min, Y.W.; Cheng, C.T.; Zhou, J.Z. Comparison of multiple linear regression, artificial neural network, extreme learning machine, and support vector machine in deriving operation rule of hydropower reservoir. Water 2019, 11, 88. [Google Scholar] [CrossRef]
- Kayabasi, A.; Toktas, A.; Sabanci, K.; Yigit, E. Automatic classification of agricultural grains: Comparison of neural networks. Neural Netw. World 2018, 28, 213–224. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support vector regression. In Efficient Learning Machines; Apress: Berkeley, CA, USA, 2015; pp. 67–80. [Google Scholar]
- Géron, A. Hands-on Machine Learning with Scikit-Learn and Tensorflow: Concepts. Tools, and Techniques to Build Intelligent Systems; O’reilly Media: Newton, MA, USA, 2017. [Google Scholar]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobotics 2013, 7, 21. [Google Scholar] [CrossRef]
- Maity, K.; Mishra, H. ANN modeling and Elitist teaching learning approach for multi-objective optimization of $$\upmu $$ μ-EDM. J. Intell. Manuf. 2018, 29, 1599–1616. [Google Scholar] [CrossRef]
- Hashim, N.; Adebayo, S.E.; Abdan, K.; Hanafi, M. Comparative study of transform-based image texture analysis for the evaluation of banana quality using an optical backscattering system. Postharvest Biol. Technol. 2018, 135, 38–50. [Google Scholar] [CrossRef]
- Zareei, J.; Rohani, A.; Mahmood, W.M.F.W. Simulation of a hydrogen/natural gas engine and modeling of engine operating parameters. Int. J. Hydrogen Energy 2018, 43, 11639–11651. [Google Scholar] [CrossRef]
- Grimm, L.G.; Nesselroade, K.P. Statistical Applications for the Behavioral and Social Sciences; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2019. [Google Scholar]
- Fallovo, C.; Cristofori, V.; De-Gyves, E.M.; Rivera, C.M.; Rea, R.; Fanasca, S.; Bignami, C.; Sassine, Y.; Rouphael, Y. Leaf area estimation model for small fruits from linear measurements. HortScience 2008, 43, 2263–2267. [Google Scholar] [CrossRef]
- Marquaridt, D.W. Generalized inverses, ridge regression, biased linear estimation, and nonlinear estimation. Technometrics 1970, 12, 591–612. [Google Scholar] [CrossRef]
- Gill, J.L. Outliers, residuals, and influence in multiple regression. Z. Tierzuechtung Zuechtungsbiologie 1986, 103, 161–175. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Suárez, J.C.; Casanoves, F.; Di Rienzo, J. Non-Destructive Estimation of the Leaf Weight and Leaf Area in Common Bean. Agronomy 2022, 12, 711. [Google Scholar] [CrossRef]
- Cheng, J.; Dang, P.P.; Zhao, Z.; Yuan, L.C.; Zhou, Z.H.; Wolf, D.; Luo, Y.B. An assessment of the Chinese medicinal Dendrobium industry: Supply, demand and sustainability. J. Ethnopharmacol. 2019, 229, 81–88. [Google Scholar] [CrossRef]
- Tang, X.; Yuan, Y.; Zhang, J. How climate change will alter the distribution of suitable Dendrobium habitats. Front. Ecol. Evol. 2020, 8, 320. [Google Scholar] [CrossRef]
- Hassan, M.A.; Khalil, A.; Kaseb, S.; Kassem, M.A. Exploring the potential of tree-based ensemble methods in solar radiation modeling. Appl. Energy 2017, 203, 897–916. [Google Scholar] [CrossRef]
- Díaz, G.; Coto, J.; Gómez-Aleixandre, J. Prediction and explanation of the formation of the Spanish day-ahead electricity price through machine learning regression. Appl. Energy 2019, 239, 610–625. [Google Scholar] [CrossRef]
- Gong, M.; Bai, Y.; Qin, J.; Wang, J.; Yang, P.; Wang, S. Gradient boosting machine for predicting return temperature of district heating system: A case study for residential buildings in Tianjin. J. Build. Eng. 2020, 27, 100950. [Google Scholar] [CrossRef]
- Yang, X.; Bian, J.; Fang, R.; Bjarnadottir, R.I.; Hogan, W.R.; Wu, Y. Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. J. Am. Med. Inform. Assoc. 2020, 27, 65–72. [Google Scholar] [CrossRef]
- Devine, S.M.; O’Geen, A.T.; Liu, H.; Jin, Y.; Dahlke, H.E.; Larsen, R.E.; Dahlgren, R.A. Terrain attributes and forage productivity predict catchment-scale soil organic carbon stocks. Geoderma 2020, 368, 114286. [Google Scholar] [CrossRef]
- Bonfatti, B.R.; Hartemink, A.E.; Giasson, E.; Tornquist, C.G.; Adhikari, K. Digital mapping of soil carbon in a viticultural region of Southern Brazil. Geoderma 2016, 261, 204–221. [Google Scholar] [CrossRef]
- Abrougui, K.; Gabsi, K.; Mercatoris, B.; Khemis, C.; Amami, R.; Chehaibi, S. Prediction of organic potato yield using tillage systems and soil properties by artificial neural network (ANN) and multiple linear regressions (MLR). Soil Tillage Res. 2019, 190, 202–208. [Google Scholar] [CrossRef]
- Liu, M.; Liu, X.; Li, M.; Fang, M.; Chi, W. Neural-network model for estimating leaf chlorophyll concentration in rice under stress from heavy metals using four spectral indices. Biosyst. Eng. 2010, 106, 223–233. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Dou, X.; Yang, Y. Estimating forest carbon fluxes using four different data-driven techniques based on long-term eddy covariance measurements: Model comparison and evaluation. Sci. Total Environ. 2018, 627, 78–94. [Google Scholar] [CrossRef]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Brachman, R.J.; Anand, T. The process of knowledge discovery in databases In Advances in Knowledge Discovery and Data Mining; The MIT Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Sharkey, T.D. Advances in Photosynthesis and Respiration; Springer: Berlin, Germany, 2012; pp. 327–329. [Google Scholar]
- Buttaro, D.; Rouphael, Y.; Rivera, C.M.; Colla, G.; Gonnella, M. Simple and accurate allometric model for leaf area estimation in Vitis vinifera L. genotypes. Photosynthetica 2015, 53, 342–348. [Google Scholar] [CrossRef]
- Waller, D.L. Operations Management. A Supply Chain Approach; Cengage Learning Business Press: Boston, MA, USA, 2003. [Google Scholar]
- Brazdil, P.B.; Soares, C. A comparison of ranking methods for classification algorithm selection. In European Conference on Machine Learning; Springer: Berlin/Heidelberg, Germany, 2000; pp. 63–75. [Google Scholar]
- Unigarro-Muñoz, C.A.; Hernández-Arredondo, J.D.; Montoya-Restrepo, E.C.; Medina-Rivera, R.D.; Ibarra-Ruales, L.N.; Carmona-González, C.Y.; Flórez-Ramos, C.P. Estimation of leaf area in coffee leaves (Coffea arabica L.) of the Castillo® variety. Bragantia 2015, 74, 412–416. [Google Scholar] [CrossRef]




| Inputs | W | W2 | W3 |
|---|---|---|---|
| L | L, W (MLR1, SVR1, GBR1) | L, W2 (MLR2, SVR2, GBR2) | L, W3 (MLR3, SVR3, GBR3) |
| L2 | L2, W (MLR4, SVR4, GBR4) | L2, W2 (MLR5, SVR5, GBR5) | L2, W3 (MLR6, SVR6, GBR6) |
| L3 | L3, W (MLR7, SVR7, GBR7) | L3, W2 (MLR8, SVR8, GBR8) | L3, W3 (MLR9, SVR9, GBR9) |
| Models | Input Variables | Output Variable | Layers (Input-Hidden-Output) |
|---|---|---|---|
| ANN1 | L, W | LA | 2-3-1 |
| ANN2 | L, W | LA | 2-5-1 |
| ANN3 | L, W | LA | 2-10-1 |
| ANN4 | L, W | LA | 2-3-3-1 |
| ANN5 | L, W2 | LA | 2-3-1 |
| ANN6 | L, W2 | LA | 2-5-1 |
| ANN7 | L, W2 | LA | 2-10-1 |
| ANN8 | L, W2 | LA | 2-3-3-1 |
| ANN9 | L, W3 | LA | 2-3-1 |
| ANN10 | L, W3 | LA | 2-5-1 |
| ANN11 | L, W3 | LA | 2-10-1 |
| ANN12 | L, W3 | LA | 2-3-3-1 |
| ANN13 | L2,W | LA | 2-3-1 |
| ANN14 | L2,W | LA | 2-5-1 |
| ANN15 | L2,W | LA | 2-10-1 |
| ANN16 | L2,W | LA | 2-3-3-1 |
| ANN17 | L2, W2 | LA | 2-3-1 |
| ANN18 | L2, W2 | LA | 2-5-1 |
| ANN19 | L2, W2 | LA | 2-10-1 |
| ANN20 | L2, W2 | LA | 2-3-3-1 |
| ANN21 | L2, W3 | LA | 2-3-1 |
| ANN22 | L2, W3 | LA | 2-5-1 |
| ANN23 | L2, W3 | LA | 2-10-1 |
| ANN24 | L2, W3 | LA | 2-3-3-1 |
| ANN25 | L3, W | LA | 2-3-1 |
| ANN26 | L3, W | LA | 2-5-1 |
| ANN27 | L3, W | LA | 2-10-1 |
| ANN28 | L3, W | LA | 2-3-3-1 |
| ANN29 | L3, W2 | LA | 2-3-1 |
| ANN30 | L3, W2 | LA | 2-5-1 |
| ANN31 | L3, W2 | LA | 2-10-1 |
| ANN32 | L3, W2 | LA | 2-3-3-1 |
| ANN33 | L3, W3 | LA | 2-3-1 |
| ANN34 | L3, W3 | LA | 2-5-1 |
| ANN35 | L3, W3 | LA | 2-10-1 |
| ANN36 | L3, W3 | LA | 2-3-3-1 |
| GBR | ANN | ||||
|---|---|---|---|---|---|
| Hyper Parameters | Values | Descriptions | Hyper Parameters | Values | Descriptions |
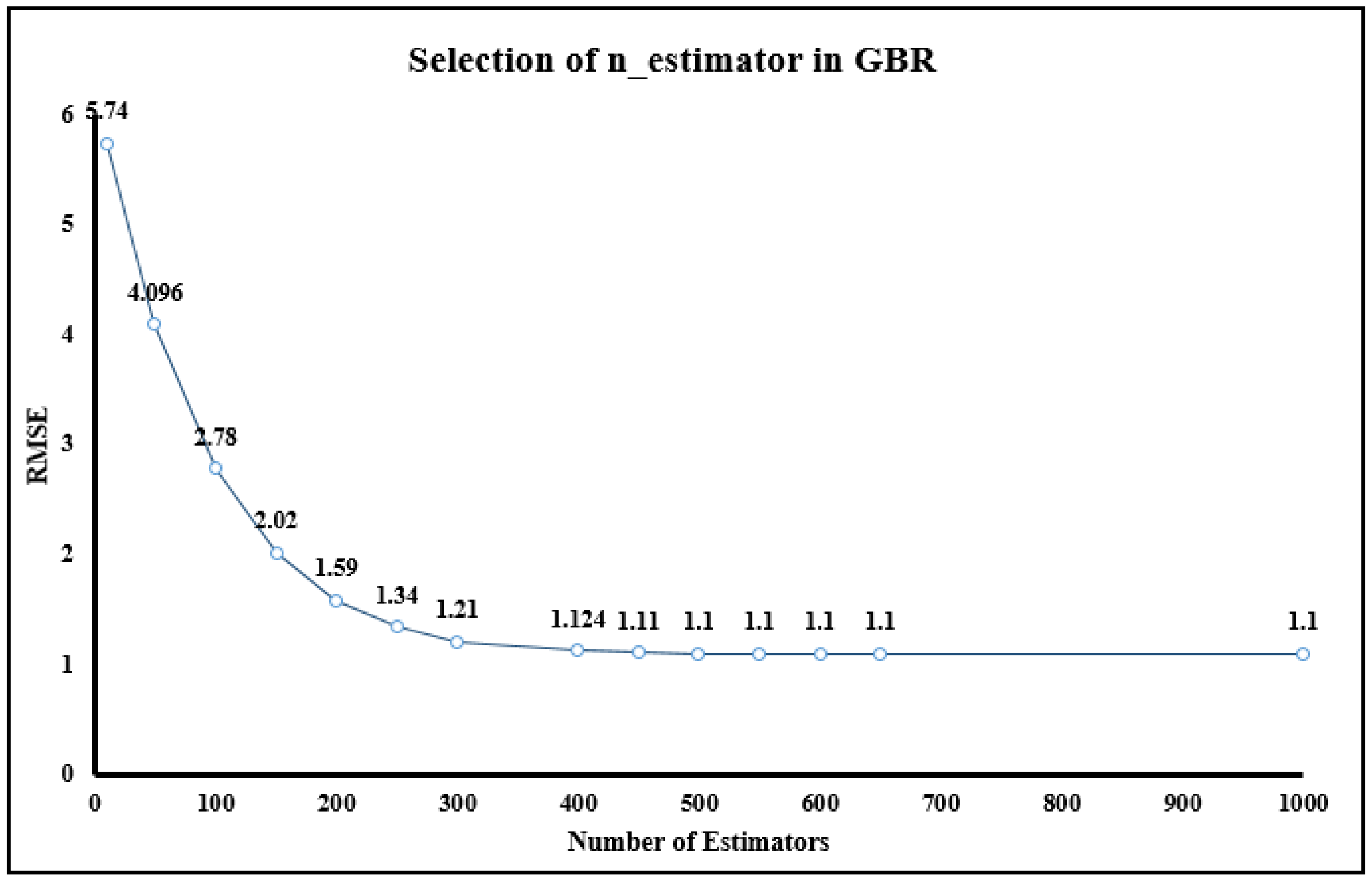
| n_estimators | 500 | No of decision tree in the ensemble | alpha | 0.001 | Regularization parameter |
| max_depth | 4 | Maximum depth of the decision tree | hidden_layer_sizes | (3,); (5,); (10,); (3,3) | Four architecture 2-3-1, 2-5-1, 2-10-1, 2-3-3-1 |
| min_samples_split | 5 | Minimum number if sample required to split an internal node | max_iter | 1000 | No. of iteration |
| learning_rate | 0.01 | Determine the impact of each tree on final outcome. | activation | ‘logistic’ | Logistic sigmoid function |
| loss | ‘ls’ | Least square loss function | learning_rate | ‘adaptive’ | Keep learning rate constant as the initial learning rate |
| Parameters | Maximum | Minimum | Mean and Standard Deviation |
|---|---|---|---|
| Leaf Length (L) cm | 16.30 | 5.24 | 10.68 and 1.57 |
| Leaf Width (W) cm | 4.34 | 1.30 | 2.39 and 0.51 |
| Leaf Area (LA) cm2 | 43.90 | 6.39 | 20.04 and 6.26 |
| Methods | Input Combinations | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| L, W | L, W2 | L, W3 | L2, W | L2, W2 | L2, W3 | L3, W | L3, W2 | L3, W3 | |
| Variance Inflation Factor (VIF) | 1.31 | 1.27 | 1.23 | 1.30 | 1.27 | 1.23 | 1.28 | 1.25 | 1.22 |
| Tolerance (T) | 0.76 | 0.78 | 0.81 | 0.77 | 0.79 | 0.81 | 0.78 | 0.80 | 0.82 |
| Inputs | Model | Training | Testing | ||||
|---|---|---|---|---|---|---|---|
| R2 | MAE | RMSE (cm2) | R2 | MAE | RMSE (cm2) | ||
| LW | ANN1 | 0.96 | 0.93 | 1.29 | 0.96 | 0.89 | 1.21 |
| LW | ANN2 | 0.96 | 0.87 | 1.27 | 0.96 | 0.86 | 1.13 |
| LW | ANN3 | 0.96 | 0.86 | 1.26 | 0.96 | 0.93 | 1.32 |
| LW | ANN4 | 0.85 | 1.03 | 2.51 | 0.96 | 0.98 | 1.32 |
| LW2 | ANN5 | 0.96 | 0.96 | 1.30 | 0.96 | 0.88 | 1.23 |
| LW2 | ANN6 | 0.96 | 0.90 | 1.25 | 0.96 | 0.84 | 1.18 |
| LW2 | ANN7 | 0.96 | 0.91 | 1.28 | 0.96 | 0.84 | 1.21 |
| LW2 | ANN8 | 0.93 | 1.13 | 1.50 | 0.94 | 1.18 | 1.62 |
| LW3 | ANN9 | 0.96 | 0.92 | 1.28 | 0.95 | 0.91 | 1.35 |
| LW3 | ANN10 | 0.96 | 0.90 | 1.27 | 0.97 | 0.87 | 1.18 |
| LW3 | ANN11 | 0.96 | 0.90 | 1.23 | 0.96 | 0.91 | 1.29 |
| LW3 | ANN12 | 0.95 | 1.12 | 1.63 | 0.96 | 0.88 | 1.20 |
| L2W | ANN13 | 0.94 | 1.00 | 1.55 | 0.94 | 1.12 | 1.60 |
| L2W | ANN14 | 0.95 | 0.96 | 1.48 | 0.96 | 0.92 | 1.40 |
| L2W | ANN15 | 0.95 | 0.94 | 1.41 | 0.97 | 0.86 | 1.11 |
| L2W | ANN16 | 0.92 | 2.28 | 5.17 | 0.95 | 1.07 | 1.45 |
| L2W2 | ANN17 | 0.95 | 1.00 | 1.44 | 0.96 | 0.91 | 1.34 |
| L2W2 | ANN18 | 0.94 | 0.93 | 1.36 | 0.97 | 0.77 | 1.11 |
| L2W2 | ANN19 | 0.95 | 0.94 | 1.39 | 0.96 | 0.89 | 1.24 |
| L2W2 | ANN20 | 0.74 | 1.60 | 4.05 | 0.00 | 4.62 | 5.84 |
| L2W3 | ANN21 | 0.94 | 1.03 | 1.47 | 0.95 | 0.88 | 1.32 |
| L2W3 | ANN22 | 0.95 | 0.97 | 1.39 | 0.97 | 0.86 | 1.19 |
| L2W3 | ANN23 | 0.95 | 0.90 | 1.32 | 0.96 | 0.92 | 1.34 |
| L2W3 | ANN24 | 0.83 | 1.67 | 3.19 | 0.95 | 0.93 | 1.26 |
| L3W | ANN25 | 0.47 | 2.73 | 3.74 | 0.88 | 1.42 | 2.06 |
| L3W | ANN26 | 0.73 | 2.77 | 3.78 | 0.88 | 1.35 | 2.06 |
| L3W | ANN27 | 0.70 | 2.46 | 4.32 | 0.88 | 1.38 | 2.17 |
| L3W | ANN28 | 0.39 | 3.19 | 4.41 | 0.83 | 1.61 | 2.68 |
| L3W2 | ANN29 | 0.86 | 1.63 | 3.46 | 0.93 | 1.07 | 1.61 |
| L3W2 | ANN30 | 0.81 | 1.36 | 2.15 | 0.89 | 1.13 | 1.97 |
| L3W2 | ANN31 | 0.88 | 1.41 | 2.22 | 0.94 | 1.01 | 1.47 |
| L3W2 | ANN32 | 0.75 | 1.90 | 3.02 | 0.88 | 1.54 | 2.06 |
| L3W3 | ANN33 | 0.87 | 2.04 | 2.32 | 0.85 | 1.58 | 2.45 |
| L3W3 | ANN34 | 0.77 | 1.57 | 2.54 | 0.86 | 1.41 | 2.38 |
| L3W3 | ANN35 | 0.88 | 1.29 | 2.34 | 0.96 | 0.97 | 1.31 |
| L3W3 | ANN36 | 0.85 | 1.92 | 3.58 | 0.89 | 1.38 | 1.99 |
| Model Input Combinations | Models |
|---|---|
| MLR1 | LA = −18.72 + L × 1.74 + W × 8.41 |
| MLR2 | LA = −9.01 + L × 1.79 + W2 × 1.65 |
| MLR3 | LA = −6.76 + L × 1.92 + W3 × 0.40 |
| MLR4 | LA = −9.56 + L2 × 0.08 + W2 × 8.34 |
| MLR5 | LA = 0.28 + L2 × 0.75 + W2 × 0.98 |
| MLR6 | LA = 3.37 + L2 × 0.089 + W3 × 0.40 |
| MLR7 | LA = −6.6 + L3 × 0.00049 + W × 8.45 |
| MLR8 | LA = 3.5 + L × 1.79 + W2 × 1.65 |
| MLR9 | LA = 6.8 + L3 × 0.0054 + W3 × 0.40 |
| Inputs | Model | Training | Testing | ||||
|---|---|---|---|---|---|---|---|
| R2 | MAE | RMSE (cm2) | R2 | MAE | RMSE (cm2) | ||
| LW | ANN2 | 0.96 | 0.87 | 1.27 | 0.96 | 0.86 | 1.13 |
| LW | GBR1 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.11 |
| LW | SVR1 | 0.94 | 0.87 | 1.47 | 0.96 | 0.83 | 1.21 |
| LW | MLR1 | 0.96 | 0.90 | 1.2 | 0.96 | 0.9 | 1.24 |
| LW2 | ANN6 | 0.96 | 0.90 | 1.25 | 0.96 | 0.84 | 1.18 |
| LW2 | GBR2 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.11 |
| LW2 | SVR2 | 0.94 | 0.89 | 1.52 | 0.96 | 0.84 | 1.23 |
| LW2 | MLR2 | 0.97 | 0.87 | 1.15 | 0.96 | 0.94 | 1.22 |
| LW3 | ANN10 | 0.96 | 0.90 | 1.27 | 0.97 | 0.87 | 1.18 |
| LW3 | GBR3 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.11 |
| LW3 | SVR3 | 0.94 | 0.93 | 1.58 | 0.96 | 0.86 | 1.25 |
| LW3 | MLR3 | 0.95 | 1.05 | 1.34 | 0.95 | 1.06 | 1.42 |
| L2W | ANN15 | 0.95 | 0.94 | 1.41 | 0.97 | 0.86 | 1.11 |
| L2W | GBR4 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.10 |
| L2W | SVR4 | 0.94 | 0.88 | 1.48 | 0.96 | 0.83 | 1.21 |
| L2W | MLR4 | 0.96 | 0.86 | 1.15 | 0.96 | 0.86 | 1.13 |
| L2W2 | ANN18 | 0.95 | 0.93 | 1.36 | 0.97 | 0.77 | 1.03 |
| L2W2 | GBR5 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.10 |
| L2W2 | SVR5 | 0.94 | 0.89 | 1.53 | 0.96 | 0.85 | 1.24 |
| L2W2 | MLR5 | 0.97 | 0.84 | 1.16 | 0.96 | 0.85 | 1.13 |
| L2W3 | ANN22 | 0.95 | 0.97 | 1.39 | 0.97 | 0.86 | 1.19 |
| L2W3 | GBR6 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.10 |
| L2W3 | SVR6 | 0.94 | 0.92 | 1.57 | 0.96 | 0.86 | 1.26 |
| L2W3 | MLR6 | 0.95 | 1.04 | 1.36 | 0.95 | 1.06 | 1.36 |
| L3W | ANN26 | 0.73 | 2.77 | 3.78 | 0.88 | 1.35 | 2.06 |
| L3W | GBR7 | 0.96 | 0.86 | 1.18 | 0.96 | 0.82 | 1.10 |
| L3W | SVR7 | 0.94 | 0.88 | 1.49 | 0.96 | 0.85 | 1.25 |
| L3W | MLR7 | 0.96 | 0.89 | 1.17 | 0.95 | 0.88 | 1.22 |
| L3W2 | ANN31 | 0.88 | 1.41 | 2.22 | 0.94 | 1.01 | 1.47 |
| L3W2 | GBR8 | 0.96 | 0.86 | 1.18 | 0.96 | 0.91 | 1.10 |
| L3W2 | SVR8 | 0.94 | 0.89 | 1.53 | 0.96 | 0.85 | 1.26 |
| L3W2 | MLR8 | 0.96 | 0.88 | 1.17 | 0.96 | 0.88 | 1.2 |
| L3W3 | ANN35 | 0.91 | 1.29 | 2.34 | 0.91 | 0.97 | 1.31 |
| L3W3 | GBR9 | 0.96 | 0.86 | 1.18 | 0.96 | 0.91 | 1.10 |
| L3W3 | SVR9 | 0.94 | 0.92 | 1.56 | 0.96 | 0.86 | 1.28 |
| L3W3 | MLR9 | 0.95 | 1.11 | 1.42 | 0.94 | 1.12 | 1.46 |
| Models | Ranks Based on Training Results | Ranks Based on Testing Results | Average Rank (AR) | Final Rank | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | MAE | RMSE (cm2) | R2 | MAE | RMSE (cm2) | |||
| GBR7 | 7 | 4 | 8 | 11 | 2 | 2 | 5.67 | 1 |
| MLR5 | 1 | 1 | 3 | 7 | 13 | 12 | 6.17 | 2 |
| GBR6 | 8 | 5 | 9 | 12 | 3 | 3 | 6.67 | 3 |
| GBR5 | 9 | 6 | 10 | 13 | 4 | 4 | 7.67 | 4 |
| GBR4 | 11 | 8 | 11 | 14 | 5 | 5 | 9.00 | 5 |
| GBR9 | 3 | 2 | 6 | 9 | 28 | 6 | 9.00 | 6 |
| MLR4 | 10 | 7 | 1 | 5 | 19 | 13 | 9.17 | 7 |
| GBR8 | 5 | 3 | 7 | 10 | 29 | 7 | 10.17 | 8 |
| GBR3 | 12 | 9 | 12 | 15 | 6 | 8 | 10.33 | 9 |
| GBR2 | 14 | 10 | 13 | 16 | 7 | 9 | 11.50 | 10 |
| ANN18 | 22 | 27 | 20 | 2 | 1 | 1 | 12.17 | 11 |
| MLR2 | 2 | 12 | 2 | 6 | 30 | 22 | 12.33 | 12 |
| MLR8 | 4 | 15 | 4 | 8 | 25 | 18 | 12.33 | 13 |
| GBR1 | 17 | 11 | 14 | 17 | 8 | 10 | 12.83 | 14 |
| ANN10 | 13 | 22 | 18 | 1 | 24 | 16 | 15.67 | 15 |
| ANN6 | 15 | 23 | 16 | 19 | 11 | 15 | 16.50 | 16 |
| ANN2 | 18 | 13 | 17 | 20 | 20 | 14 | 17.00 | 17 |
| MLR7 | 6 | 18 | 5 | 30 | 26 | 21 | 17.67 | 18 |
| ANN15 | 23 | 29 | 23 | 4 | 18 | 11 | 18.00 | 19 |
| ANN22 | 21 | 30 | 22 | 3 | 17 | 17 | 18.33 | 20 |
| SVR1 | 33 | 14 | 25 | 21 | 9 | 19 | 20.17 | 21 |
| MLR1 | 16 | 24 | 15 | 18 | 27 | 25 | 20.83 | 22 |
| SVR4 | 30 | 17 | 26 | 22 | 10 | 20 | 20.83 | 23 |
| SVR7 | 27 | 16 | 27 | 23 | 14 | 26 | 22.17 | 24 |
| SVR2 | 32 | 21 | 28 | 24 | 12 | 23 | 23.33 | 25 |
| SVR8 | 26 | 19 | 29 | 25 | 15 | 28 | 23.67 | 26 |
| SVR5 | 29 | 20 | 30 | 26 | 16 | 24 | 24.17 | 27 |
| SVR9 | 25 | 25 | 31 | 27 | 21 | 30 | 26.50 | 28 |
| SVR6 | 28 | 26 | 32 | 28 | 22 | 29 | 27.50 | 29 |
| MLR6 | 20 | 31 | 21 | 32 | 34 | 32 | 28.33 | 30 |
| SVR3 | 31 | 28 | 33 | 29 | 23 | 27 | 28.50 | 31 |
| MLR3 | 24 | 32 | 19 | 31 | 33 | 33 | 28.67 | 32 |
| MLR9 | 19 | 33 | 24 | 33 | 35 | 34 | 29.67 | 33 |
| ANN35 | 34 | 34 | 35 | 35 | 31 | 31 | 33.33 | 34 |
| ANN31 | 35 | 35 | 34 | 34 | 32 | 35 | 34.17 | 35 |
| ANN26 | 36 | 36 | 36 | 36 | 36 | 36 | 36.00 | 36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, M.; Deb, C.K.; Pal, R.; Marwaha, S. A Machine Learning Approach for the Non-Destructive Estimation of Leaf Area in Medicinal Orchid Dendrobium nobile L. Appl. Sci. 2022, 12, 4770. https://doi.org/10.3390/app12094770
Das M, Deb CK, Pal R, Marwaha S. A Machine Learning Approach for the Non-Destructive Estimation of Leaf Area in Medicinal Orchid Dendrobium nobile L. Applied Sciences. 2022; 12(9):4770. https://doi.org/10.3390/app12094770
Chicago/Turabian StyleDas, Madhurima, Chandan Kumar Deb, Ram Pal, and Sudeep Marwaha. 2022. "A Machine Learning Approach for the Non-Destructive Estimation of Leaf Area in Medicinal Orchid Dendrobium nobile L." Applied Sciences 12, no. 9: 4770. https://doi.org/10.3390/app12094770
APA StyleDas, M., Deb, C. K., Pal, R., & Marwaha, S. (2022). A Machine Learning Approach for the Non-Destructive Estimation of Leaf Area in Medicinal Orchid Dendrobium nobile L. Applied Sciences, 12(9), 4770. https://doi.org/10.3390/app12094770






