Multi-Institutional Breast Cancer Detection Using a Secure On-Boarding Service for Distributed Analytics
Abstract
:1. Introduction
1.1. Objective
1.2. Contribution and Findings
1.3. Overview
2. Related Work
2.1. FAIR Principles
- Findable: To make data usage possible, researchers should be able to find digital assets. Each data object should have a persistent and unique identifier and should include rich metadata. Additionally, the metadata—or the identifier, respectively—should be stored in searchable resources.
- Accessible: Open, free, and universal communication protocols should make data objects accessible by their identifier. Further, metadata should be archivable and available even when the corresponding data is not available.
- Interoperable: Data should be interoperable with other data assets. This can be achieved by using formal, accessible, shared, and broadly applicable languages or vocabularies. Additionally, data should be referenceable from other data.
- Reusable: To enable reusability, data should be equipped with usage licenses and detailed provenance and meet community standards.
2.2. Distributed Analytics
2.3. On-Boarding
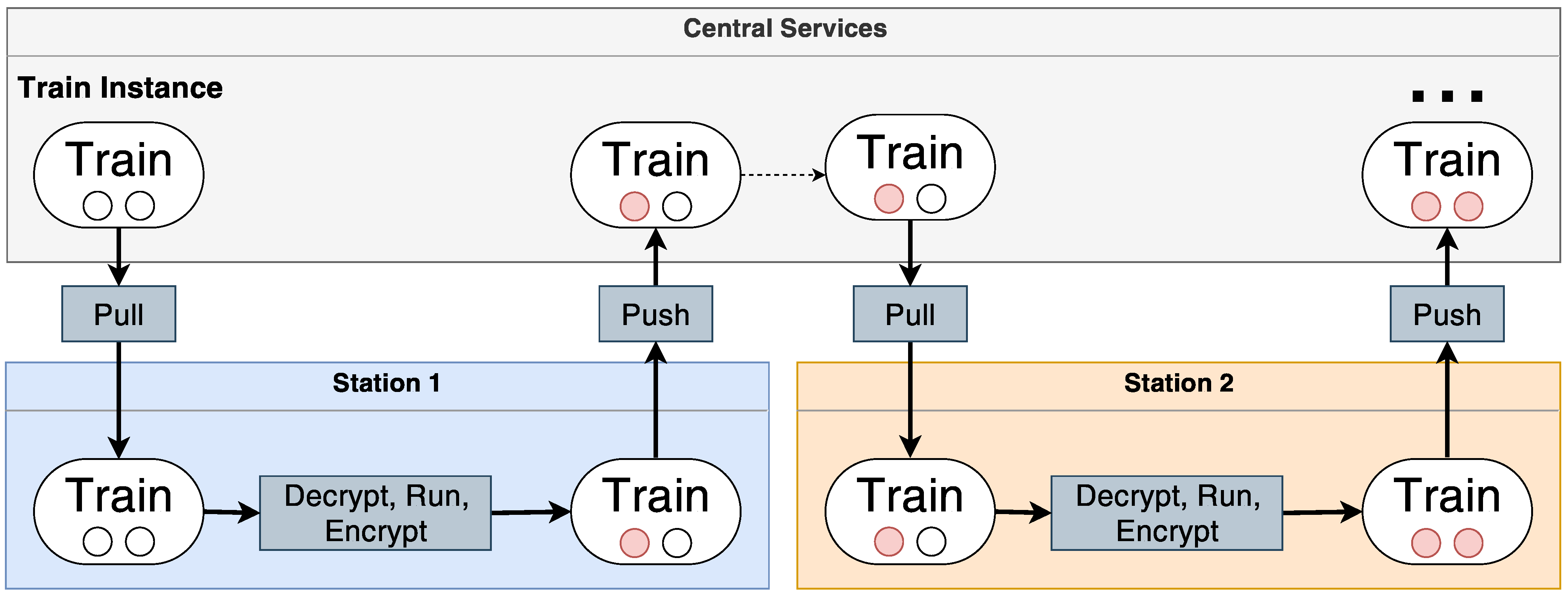
3. On-Boarding Process for Distributed Analysis
- FAIR: The technical solution should be an enabler for FAIR data management. Participating institutions should be findable and equipped with a basic set of metadata.
- Secure: The mandatory key exchange mechanism has to establish a baseline level of secure communication. It should be automatically performed in dedicated channels with no user interaction to reduce the danger of security breaches.
- Configuration time: The configuration time, which might be a bottleneck, should be significantly reduced and automated.
- Usability: The on-boarding service should require less technical knowledge and be based on well-established technologies such that the deployment at each data premise requires less manual effort and technical expertise.
3.1. Central Service
3.2. Station
3.3. On-Boarding Workflow
3.3.1. Station Registry
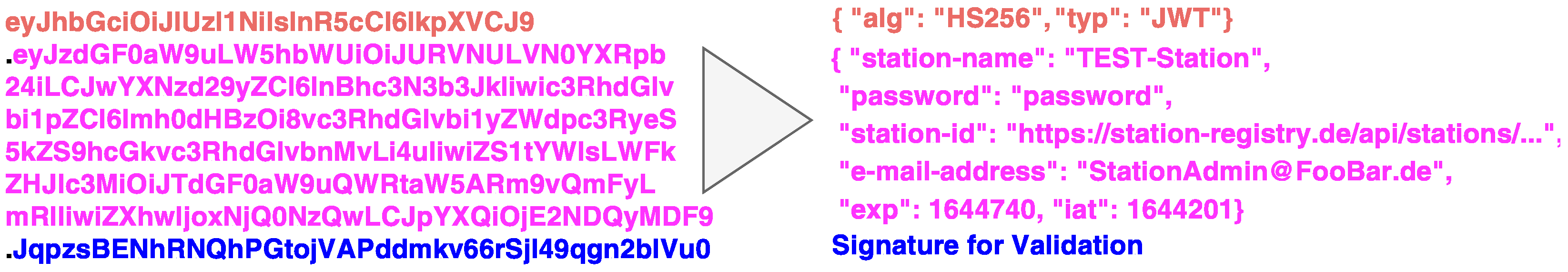
3.4. Secure Station On-Boarding
- 1.
- Authorised person: We assume that each (imminent) participating institution has a dedicated and authorised person—the so-called station admin—who is responsible for the station deployment.
- 2.
- Semi-trusted CS: During the on-boarding process, the CS orchestrates and encrypts the sensitive information (e.g., connection parameters) to the requester after creating them.
- 3.
- Network settings: All network prerequisites have been fulfilled. This especially includes the firewall rules and port configurations.
- 4.
- Base software: As we have mentioned earlier, the station software requires an up and running Docker engine. Since the installation of this prerequisite is negligible for the on-boarding itself, we assume that the installation has been conducted beforehand.
4. Distributed Model Training in a Clinical Study
4.1. Data Set and Characteristics
4.2. Setup
4.2.1. Data Provision
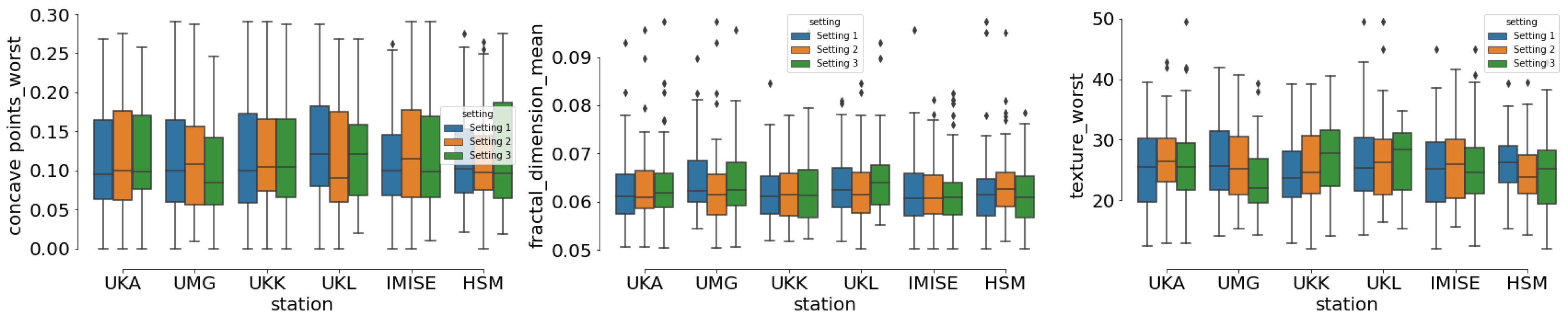
4.2.2. Data Distribution
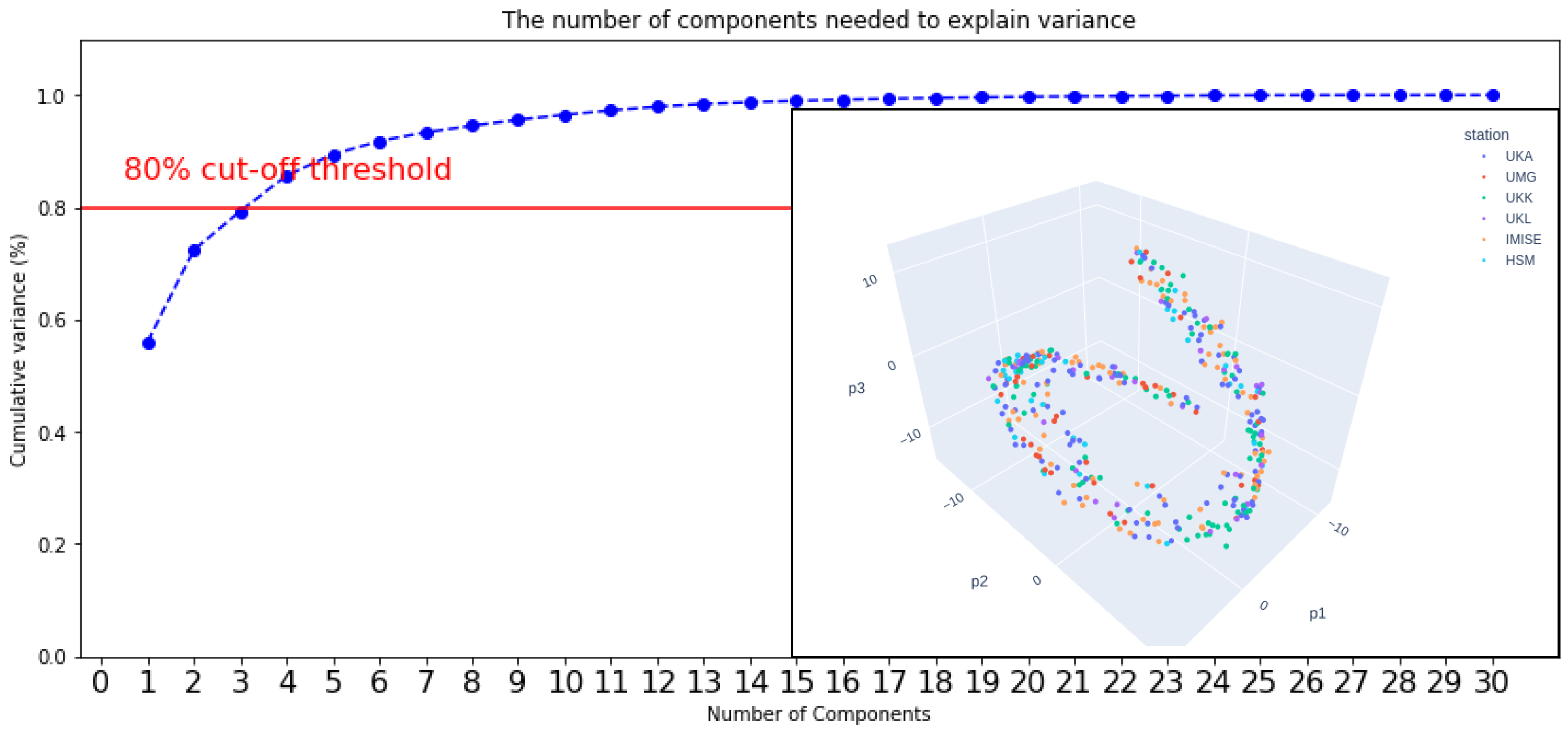
4.3. Methods
4.4. Results
5. Discussion
5.1. FAIRification
5.2. On-Boarding
5.3. Limitations
6. Conclusions and Future Work
Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DA | Distributed Analytics |
| ML | Machine Learning |
| IN | Implementation Networks |
| DS | DataSHIELD |
| SMPC | Secure Multi-Party Computation |
| PHT | Personal Health Train |
| CS | Central Service |
| CLI | Command Line Interface |
| HP | Hewlett Packard |
| IAM | Identity and Access Management |
| DNS | Domain Name Service |
| JWT | JSON Web Token |
| OTP | One-Time-Password |
| FNA | Fine Needle Aspirate |
| FHIR | Fast Healthcare Interoperability Resources |
| GAN | Generative Adversarial Network |
References
- Balicer, R.D.; Cohen-Stavi, C. Advancing Healthcare Through Data-Driven Medicine and Artificial Intelligence. In Healthcare and Artificial Intelligence; Nordlinger, B., Villani, C., Rus, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 9–15. [Google Scholar] [CrossRef]
- Alyass, A.; Turcotte, M.; Meyre, D. From big data analysis to personalized medicine for all: Challenges and opportunities. BMC Med. Genom. 2015, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geifman, N.; Bollyky, J.; Bhattacharya, S.; Butte, A.J. Opening clinical trial data: Are the voluntary data-sharing portals enough? BMC Med. 2015, 13, 280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giger, M.L. Machine Learning in Medical Imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletari, F.; Roth, H.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Rosenblatt, M.; Jain, S.H.; Cahill, M. Sharing of Clinical Trial Data: Benefits, Risks, and Uniform Principles. Ann. Intern. Med. 2015, 162, 306–307. [Google Scholar] [CrossRef]
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes (Workshop); Springer: Cham, Switzerland, 2019; Volume 11383, pp. 92–104. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Welten, S.; Neumann, L.; Yediel, Y.U.; da Silva Santos, L.O.B.; Decker, S.; Beyan, O. DAMS: A Distributed Analytics Metadata Schema. Data Intell. 2021, 3, 528–547. [Google Scholar] [CrossRef]
- Beyan, O.; Choudhury, A.; van Soest, J.; Kohlbacher, O.; Zimmermann, L.; Stenzhorn, H.; Karim, M.R.; Dumontier, M.; Decker, S.; da Silva Santos, L.O.B.; et al. Distributed Analytics on Sensitive Medical Data: The Personal Health Train. Data Intell. 2020, 2, 96–107. [Google Scholar] [CrossRef]
- Chang, K.; Balachandar, N.; Lam, C.; Yi, D.; Brown, J.; Beers, A.; Rosen, B.; Rubin, D.L.; Kalpathy-Cramer, J. Distributed deep learning networks among institutions for medical imaging. J. Am. Med. Inform. Assoc. 2018, 25, 945–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Zhovannik, I.; Traverso, A.; Dankers, F.J.W.M.; Deist, T.M.; Kalendralis, P.; Monshouwer, R.; Bussink, J.; Fijten, R.; Aerts, H.J.W.L.; et al. Distributed radiomics as a signature validation study using the Personal Health Train infrastructure. Sci. Data 2019, 6, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deist, T.M.; Dankers, F.J.W.M.; Ojha, P.; Scott Marshall, M.; Janssen, T.; Faivre-Finn, C.; Masciocchi, C.; Valentini, V.; Wang, J.; Chen, J.; et al. Distributed learning on 20,000+ lung cancer patients—The Personal Health Train. Radiother. Oncol. 2020, 144, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, A.; Kaliyaperumal, R.; da Silva Santos, L.O.B.; Mons, B.; Schultes, E.; Roos, M.; Thompson, M. A Generic Workflow for the Data FAIRification Process. Data Intell. 2020, 2, 56–65. [Google Scholar] [CrossRef]
- Sinaci, A.A.; Núñez-Benjumea, F.J.; Gencturk, M.; Jauer, M.L.; Deserno, T.; Chronaki, C.; Cangioli, G.; Cavero-Barca, C.; Rodríguez-Pérez, J.M.; Pérez-Pérez, M.M.; et al. From Raw Data to FAIR Data: The FAIRification Workflow for Health Research. Methods Inf. Med. 2020, 59, e21–e32. [Google Scholar] [CrossRef]
- Welten, S.; Mou, Y.; Neumann, L.; Jaberansary, M.; Ucer, Y.Y.; Kirsten, T.; Decker, S.; Beyan, O. A Privacy-Preserving Distributed Analytics Platform for Health Care Data. Methods Inf. Med. 2022. [Google Scholar] [CrossRef]
- Gaye, A.; Marcon, Y.; Isaeva, J.; LaFlamme, P.; Turner, A.; Jones, E.M.; Minion, J.; Boyd, A.W.; Newby, C.J.; Nuotio, M.L.; et al. DataSHIELD: Taking the analysis to the data, not the data to the analysis. Int. J. Epidemiol. 2014, 43, 1929–1944. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhao, S.; Zhao, M.; Chen, Z.; Gao, C.Z.; Li, H.; An Tan, Y. Secure Multi-Party Computation: Theory, practice and applications. Inf. Sci. 2019, 476, 357–372. [Google Scholar] [CrossRef]
- Moncada-Torres, A.; Martin, F.; Sieswerda, M.; Van Soest, J.; Geleijnse, G. VANTAGE6: An open source priVAcy preserviNg federaTed leArninG infrastructurE for Secure Insight eXchange. In Proceedings of the AMIA Annual Symposium, Online, 14–18 November 2020; American Medical Informatics Association: Bethesda, MD, USA, 2020; Volume 2020, pp. 870–877. [Google Scholar]
- Wilson, R.C.; Butters, O.W.; Avraam, D.; Baker, J.; Tedds, J.A.; Turner, A.; Murtagh, M.; Burton, P.R. DataSHIELD—New directions and dimensions. Data Sci. J. 2017, 16, 21. [Google Scholar] [CrossRef]
- Mou, Y.; Welten, S.; Jaberansary, M.; Ucer Yediel, Y.; Kirsten, T.; Decker, S.; Beyan, O. Distributed Skin Lesion Analysis Across Decentralised Data Sources. Stud. Health Technol. Inform. 2021, 281, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Warnat-Herresthal, S.; Schultze, H.; Shastry, K.L.; Manamohan, S.; Mukherjee, S.; Garg, V.; Sarveswara, R.; Händler, K.; Pickkers, P.; Aziz, N.A.; et al. Swarm learning for decentralized and confidential clinical machine learning. Nature 2021, 594, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Beutel, D.J.; Topal, T.; Mathur, A.; Qiu, X.; Parcollet, T.; de Gusmão, P.P.; Lane, N.D. Flower: A friendly federated learning research framework. arXiv 2020, arXiv:2007.14390. [Google Scholar]
- van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Bender, D.; Sartipi, K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems, Porto, Portugal, 20–22 June 2013; pp. 326–331. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27, 2672–2680. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]



| Institutions | Setting 1 | Setting 2 | Setting 3 |
|---|---|---|---|
| UKA | 66 (B: 41, M: 25) | 66 (B: 40, M: 26) | 112 (B: 72, M: 40) |
| UMG | 66 (B: 41, M: 25) | 66 (B: 45, M: 21) | 42 (B: 32, M: 10) |
| UKK | 66 (B: 41, M: 25) | 66 (B: 39, M: 27) | 89 (B: 53, M: 36) |
| UKL | 66 (B: 41, M: 25) | 66 (B: 42, M: 24) | 31 (B: 18, M: 13) |
| IMISE | 68 (B: 45, M: 23) | 68 (B: 37, M: 31) | 86 (B: 53, M: 33) |
| HSM | 66 (B: 41, M: 25) | 66 (B: 47, M: 19) | 38 (B: 22, M: 16) |
| Test set | 171 (B: 107, M: 64) | 171 (B: 107, M: 64) | 171 (B: 107, M: 64) |
| Methods | Accuracy | Bal. Accuracy | Precision | Recall | F1-Score | Matt. Cor. Coef. |
|---|---|---|---|---|---|---|
| R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | |
| SST (UKA) | 92.7/90.9/93.1 ±0.008/0.003/0.006 | 90.9/88.4/93.9 ±0.008/0.003/0.004 | 96.4/97.1/86.4 ±0.014/0.011/0.011 | 83.6/78.1/96.9 ±0.009/0.000/0.000 | 89.5/86.6/91.3 ±0.011/0.004/0.006 | 84.5/80.9/86.1 ±0.017/0.008/0.010 |
| SST (UMG) | 92.1/85.4/90.1 ±0.008/0.020/0.008 | 90.1/87.5/89.8 ±0.010/0.021/0.007 | 96.3/73.2/85.4 ±0.001/0.022/0.016 | 82.0/96.1/88.7 ±0.020/0.027/0.008 | 88.6/83.1/87.0 ±0.012/0.023/0.010 | 83.3/72.7/79.0 ±0.016/0.041/0.017 |
| SST (UKK) | 91.4/89.5/92.1 ±0.007/0.000/0.003 | 89.5/87.8/92.0 ±0.009/0.000/0.005 | 94.2/89.7/88.0 ±0.009/0.000/0.001 | 82.0/81.2/91.4 ±0.016/0.000/0.009 | 87.7/85.2/89.7 ±0.011/0.000/0.005 | 81.6/77.3/83.3 ±0.016/0.000/0.008 |
| SST (UKL) | 93.7/94.0/93.9 ±0.003/0.015/0.008 | 91.6/94.3/94.5 ±0.004/0.014/0.006 | 100/ 89.4/ 88.0 ±0.000/0.024/0.016 | 83.2/95.3/96.9 ±0.008/0.013/0.000 | 90.8/92.3/92.2 ±0.005/0.018/0.009 | 87.0/87.5/87.4 ±0.006/0.030/0.014 |
| SST (IMISE) | 93.3/94.7/95.5 ±0.003/0.005/0.003 | 91.0/93.8/95.2 ±0.005/0.005/0.004 | 100/ 95.5/ 93.8 ±0.000/0.008/0.000 | 82.0/90.2/94.1 ±0.009/0.008/0.008 | 90.1/92.8/94.0 ±0.005/0.007/0.004 | 86.1/88.7/90.3 ±0.007/0.010/0.006 |
| SST (HSM) | 92.7/91.1/93.1 ±0.003/0.003/0.006 | 91.2/91.8/92.4 ±0.005/0.004/0.007 | 94.8/83.8/92.0 ±0.001/0.009/0.001 | 85.2/94.5/89.5 ±0.009/0.016/0.015 | 89.7/88.8/90.7 ±0.005/0.004/0.008 | 84.4/81.9/85.3 ±0.007/0.007/0.012 |
| IIL | 92.8/93.6/95.3 ±0.007/0.011/0.005 | 91.2/94.5/94.9 ±0.009/0.010/0.005 | 95.6/86.6/94.1 ±0.010/0.019/0.008 | 84.8/98.0/93.4 ±0.015/0.008/0.008 | 89.9/91.9/93.7 ±0.011/0.013/0.006 | 84.7/87.1/90.0 ±0.016/0.021/0.010 |
| CIIL | 92.4/93.7/93.9 ±0.005/0.009/0.003 | 90.6/94.3/94.5 ±0.005/0.009/0.003 | 95.9/87.6/87.9 ±0.009/0.012/0.007 | 83.2/96.9/96.9 ±0.008/0.013/0.000 | 89.1/92.0/92.2 ±0.007/0.011/0.004 | 83.8/87.2/87.4 ±0.011/0.018/0.006 |
| Methods | Accuracy | Bal. Accuracy | Precision | Recall | F1-Score | Matt. Cor. Coef. |
|---|---|---|---|---|---|---|
| R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | |
| SST (UKA) | 92.4/90.1/90.6 ±0.010/0.000/0.010 | 90.6/88.6/89.8 ±0.010/0.010/0.010 | 95.5/89.8/88.4 ±0.010/0.000/0.010 | 83.6/82.8/86.3 ±0.020/0.010/0.020 | 89.2/86.2/87.3 ±0.010/0.010/0.010 | 83.8/78.6/79.9 ±0.020/0.010/0.020 |
| SST (UMG) | 92.0/90.5/89.5 ±0.010/0.010/0.010 | 89.3/90.9/91.3 ±0.010/0.020/0.010 | 99.5/83.8/78.8 ±0.010/0.020/0.020 | 78.9/92.6/98.4 ±0.010/0.030/0.000 | 88.0/87.9/87.5 ±0.010/0.020/0.010 | 83.3/80.4/80.1 ±0.010/0.030/0.020 |
| SST (UKK) | 93.1/92.1/94.7 ±0.000/0.010/0.000 | 91.8/92.8/94.9 ±0.000/0.010/0.000 | 94.8/85.1/91.1 ±0.000/0.010/0.010 | 86.3/95.7/95.3 ±0.010/0.010/0.000 | 90.4/90.1/93.1 ±0.000/0.010/0.010 | 85.3/84.0/88.9 ±0.010/0.020/0.010 |
| SST (UKL) | 92.1/91.7/94.2 ±0.000/0.000/0.010 | 90.1/90.4/94.3 ±0.000/0.000/0.010 | 96.3/92.0/90.0 ±0.000/0.010/0.020 | 82.0/85.2/94.9 ±0.010/0.010/0.010 | 88.6/88.4/92.4 ±0.010/0.000/0.020 | 83.3/82.1/87.7 ±0.010/0.010/0.030 |
| SST (IMISE) | 95.2/93.0/95.9 ±0.000/0.010/0.000 | 95.4/91.4/95.7 ±0.000/0.010/0.000 | 91.5/95.6/94.2 ±0.010/0.010/0.010 | 96.1/85.2/94.9 ±0.010/0.020/0.010 | 93.7/90.1/94.6 ±0.000/0.010/0.010 | 89.9/85.0/91.3 ±0.010/0.020/0.010 |
| SST (HSM) | 91.2/86.8/83.8 ±0.000/0.010/0.020 | 88.3/89.2/86.7 ±0.010/0.010/0.010 | 100/ 74.6/ 70.2 ±0.000/0.020/0.020 | 76.6/98.4/98.4 ±0.010/0.000/0.000 | 86.7/84.9/82.0 ±0.010/0.010/0.010 | 81.9/75.8/71.2 ±0.010/0.020/0.020 |
| IIL | 92.1/88.9/87.3 ±0.000/0.000/0.000 | 89.5/90.8/89.5 ±0.000/0.000/0.000 | 100/ 77.8/ 75.2 ±0.000/0.010/0.000 | 78.9/98.4/98.4 ±0.010/0.000/0.000 | 88.2/86.9/85.3 ±0.010/0.000/0.000 | 83.7/79.1/76.5 ±0.010/0.010/0.000 |
| CIIL | 92.3/92.3/91.1 ±0.000/0.010/0.010 | 90.3/92.9/92.5 ±0.000/0.010/0.010 | 96.4/85.6/81.8 ±0.010/0.020/0.010 | 82.4/95.3/98.0 ±0.010/0.010/0.010 | 88.8/90.2/89.2 ±0.000/0.020/0.010 | 83.6/84.2/82.7 ±0.010/0.030/0.010 |
| Methods | Accuracy | Bal. Accuracy | Precision | Recall | F1-Score | Matt. Cor. Coef. |
|---|---|---|---|---|---|---|
| R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | R/S/S1000 | |
| SST (UKA) | 92.5/91.4/92.7 ±0.010/0.010/0.010 | 90.3/91.1/92.2 ±0.010/0.010/0.010 | 98.6/87.5/90.2 ±0.020/0.010/0.010 | 81.2/89.8/90.2 ±0.010/0.020/0.010 | 89.1/88.6/90.2 ±0.010/0.010/0.010 | 84.4/81.7/84.4 ±0.020/0.020/0.020 |
| SST (UMG) | 88.6/93.4/91.2 ±0.000/0.010/0.010 | 84.8/92.4/91.0 ±0.000/0.010/0.010 | 100.0/ 93.8/ 86.9 ±0.000/0.020/0.020 | 69.5/88.3/90.2 ±0.010/0.020/0.020 | 82.0/90.9/88.5 ±0.010/0.020/0.020 | 76.7/85.9/81.5 ±0.010/0.020/0.030 |
| SST (UKK) | 92.7/96.8/94.2 ±0.010/0.010/0.010 | 91.1/96.7/93.9 ±0.010/0.010/0.010 | 95.2/95.0/91.5 ±0.010/0.010/0.010 | 84.8/96.5/93.0 ±0.030/0.010/0.020 | 89.7/95.7/92.2 ±0.020/0.010/0.010 | 84.4/93.2/87.6 ±0.020/0.020/0.020 |
| SST (UKL) | 94.7/93.1/94.2 ±0.010/0.010/0.010 | 94.1/92.2/95.0 ±0.010/0.010/0.010 | 94.4/93.1/87.5 ±0.010/0.020/0.020 | 91.4/88.3/98.4 ±0.010/0.020/0.000 | 92.9/90.6/92.7 ±0.010/0.010/0.010 | 88.7/85.3/88.3 ±0.010/0.020/0.020 |
| SST (IMISE) | 94.2/91.4/94.7 ±0.020/0.010/0.000 | 93.0/92.5/94.7 ±0.020/0.010/0.000 | 95.7/83.0/91.7 ±0.020/0.020/0.010 | 88.3/96.9/94.5 ±0.040/0.010/0.020 | 91.8/89.4/93.1 ±0.030/0.020/0.000 | 87.5/82.9/88.9 ±0.040/0.030/0.000 |
| SST (HSM) | 92.1/88.7/94.0 ±0.000/0.010/0.000 | 90.9/90.7/93.5 ±0.000/0.000/0.000 | 92.4/77.5/92.5 ±0.010/0.010/0.010 | 85.9/98.4/91.4 ±0.000/0.000/0.010 | 89.1/86.7/91.9 ±0.000/0.010/0.000 | 83.0/78.9/87.2 ±0.010/0.010/0.010 |
| IIL | 93.3/90.2/95.6 ±0.000/0.010/0.000 | 92.2/91.9/95.6 ±0.000/0.010/0.000 | 93.8/86.0/93.1 ±0.010/0.020/0.010 | 87.9/98.4/95.3 ±0.010/0.000/0.010 | 90.7/88.3/94.2 ±0.000/0.010/0.000 | 85.6/81.3/90.7 ±0.010/0.020/0.010 |
| CIIL | 93.0/93.7/94.6 ±0.010/0.000/0.010 | 91.4/94.6/95.0 ±0.010/0.000/0.000 | 95.6/86.9/89.5 ±0.010/0.010/0.010 | 85.2/98.0/96.9 ±0.020/0.010/0.000 | 90.1/92.1/93.1 ±0.010/0.000/0.010 | 85.0/87.4/88.8 ±0.020/0.010/0.010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welten, S.; Hempel, L.; Abedi, M.; Mou, Y.; Jaberansary, M.; Neumann, L.; Weber, S.; Tahar, K.; Ucer Yediel, Y.; Löbe, M.; et al. Multi-Institutional Breast Cancer Detection Using a Secure On-Boarding Service for Distributed Analytics. Appl. Sci. 2022, 12, 4336. https://doi.org/10.3390/app12094336
Welten S, Hempel L, Abedi M, Mou Y, Jaberansary M, Neumann L, Weber S, Tahar K, Ucer Yediel Y, Löbe M, et al. Multi-Institutional Breast Cancer Detection Using a Secure On-Boarding Service for Distributed Analytics. Applied Sciences. 2022; 12(9):4336. https://doi.org/10.3390/app12094336
Chicago/Turabian StyleWelten, Sascha, Lars Hempel, Masoud Abedi, Yongli Mou, Mehrshad Jaberansary, Laurenz Neumann, Sven Weber, Kais Tahar, Yeliz Ucer Yediel, Matthias Löbe, and et al. 2022. "Multi-Institutional Breast Cancer Detection Using a Secure On-Boarding Service for Distributed Analytics" Applied Sciences 12, no. 9: 4336. https://doi.org/10.3390/app12094336
APA StyleWelten, S., Hempel, L., Abedi, M., Mou, Y., Jaberansary, M., Neumann, L., Weber, S., Tahar, K., Ucer Yediel, Y., Löbe, M., Decker, S., Beyan, O., & Kirsten, T. (2022). Multi-Institutional Breast Cancer Detection Using a Secure On-Boarding Service for Distributed Analytics. Applied Sciences, 12(9), 4336. https://doi.org/10.3390/app12094336








